In Vitro Production and Exudation of 20-Hydroxymaytenin from Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. Cell Culture
Abstract
1. Introduction
2. Results
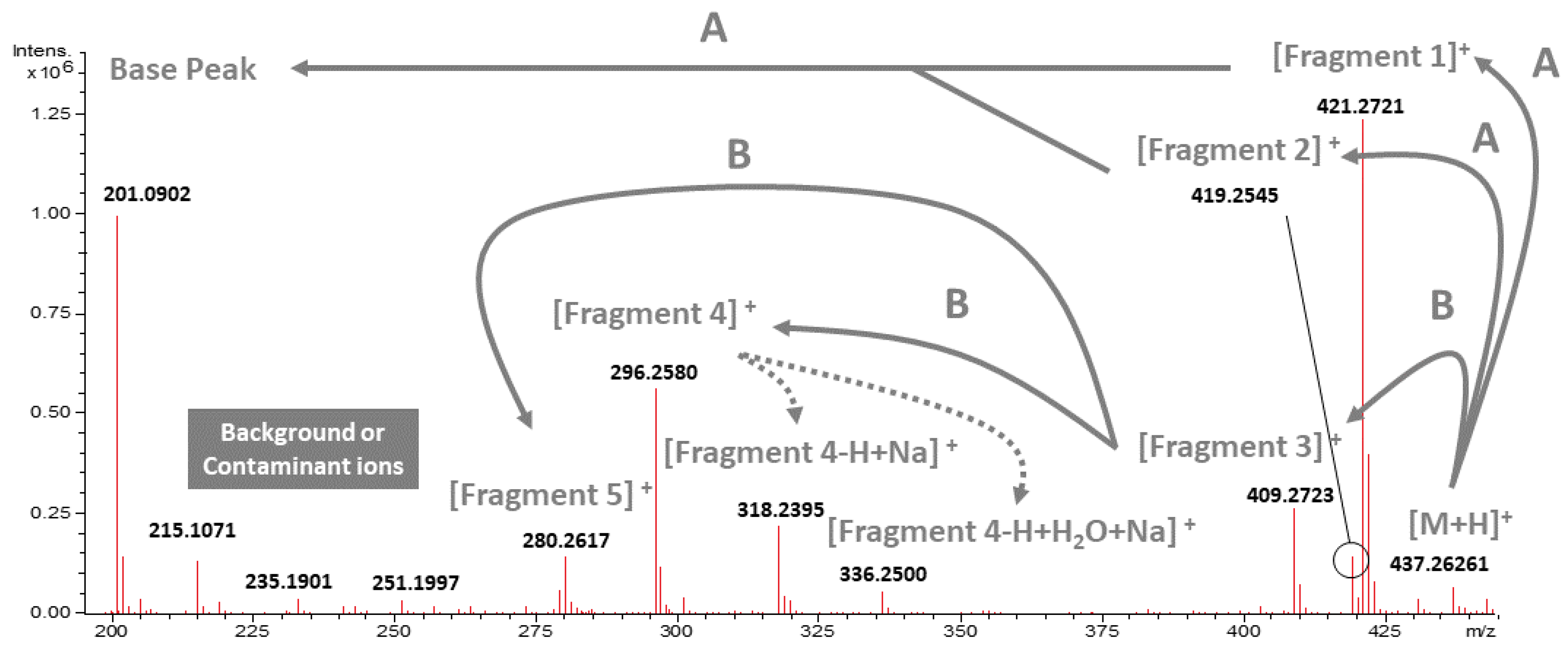
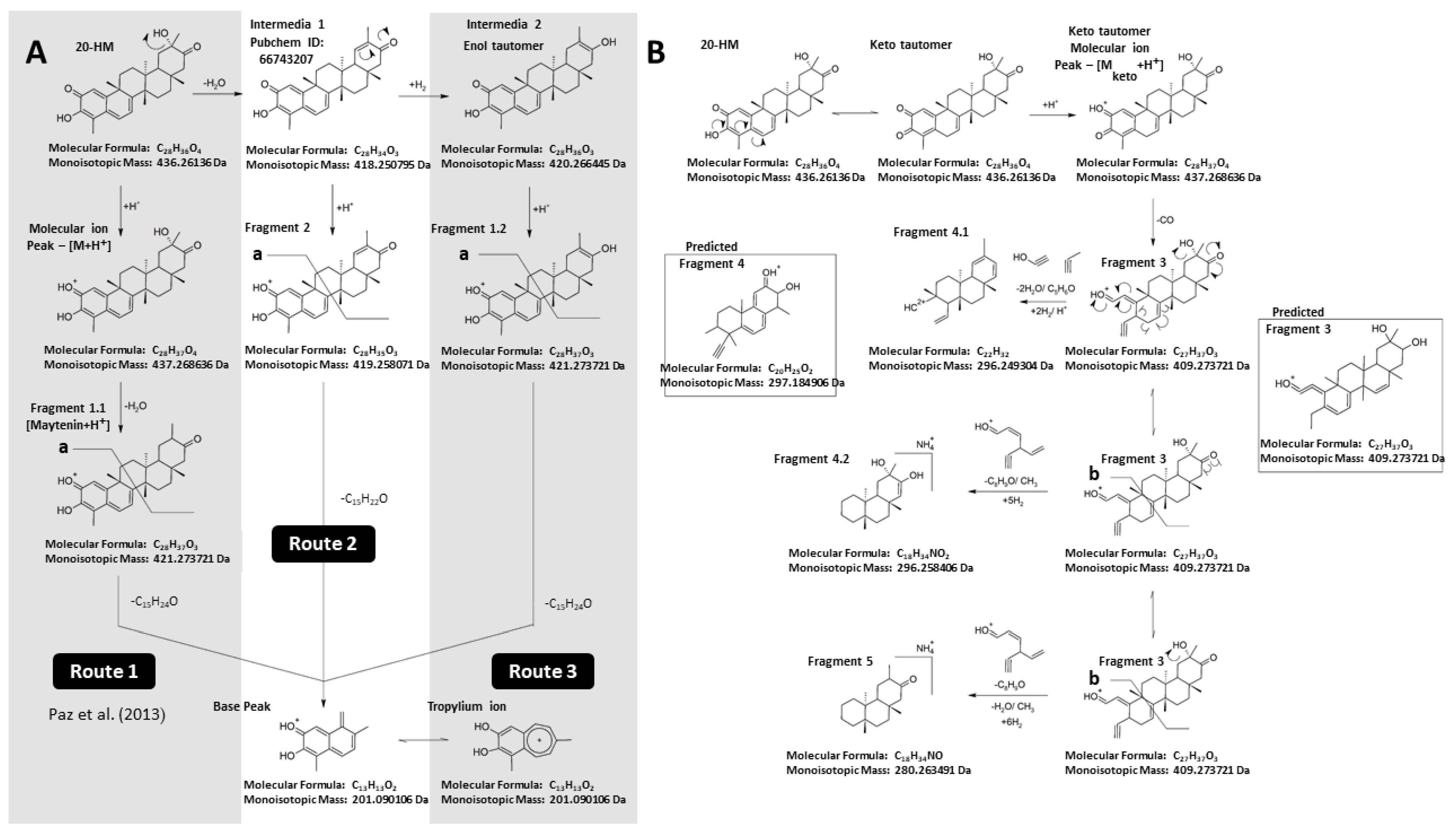
2.1. Isolation, Identification, and Proposed Mass Fragmentation of 20-HM from G. heterophylla
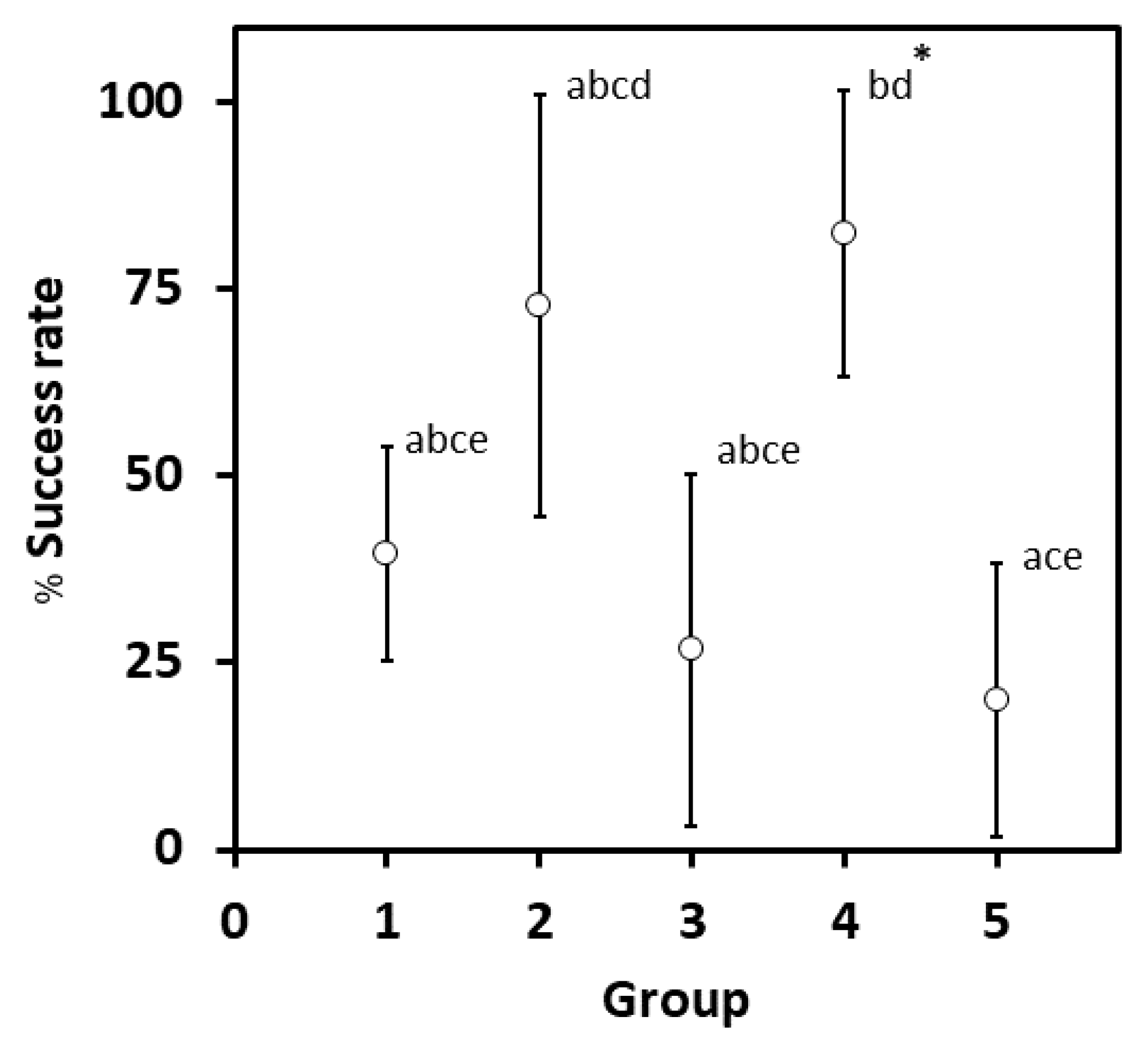
2.2. Callus Induction Medium Optimization for Leaves of G. heterophylla
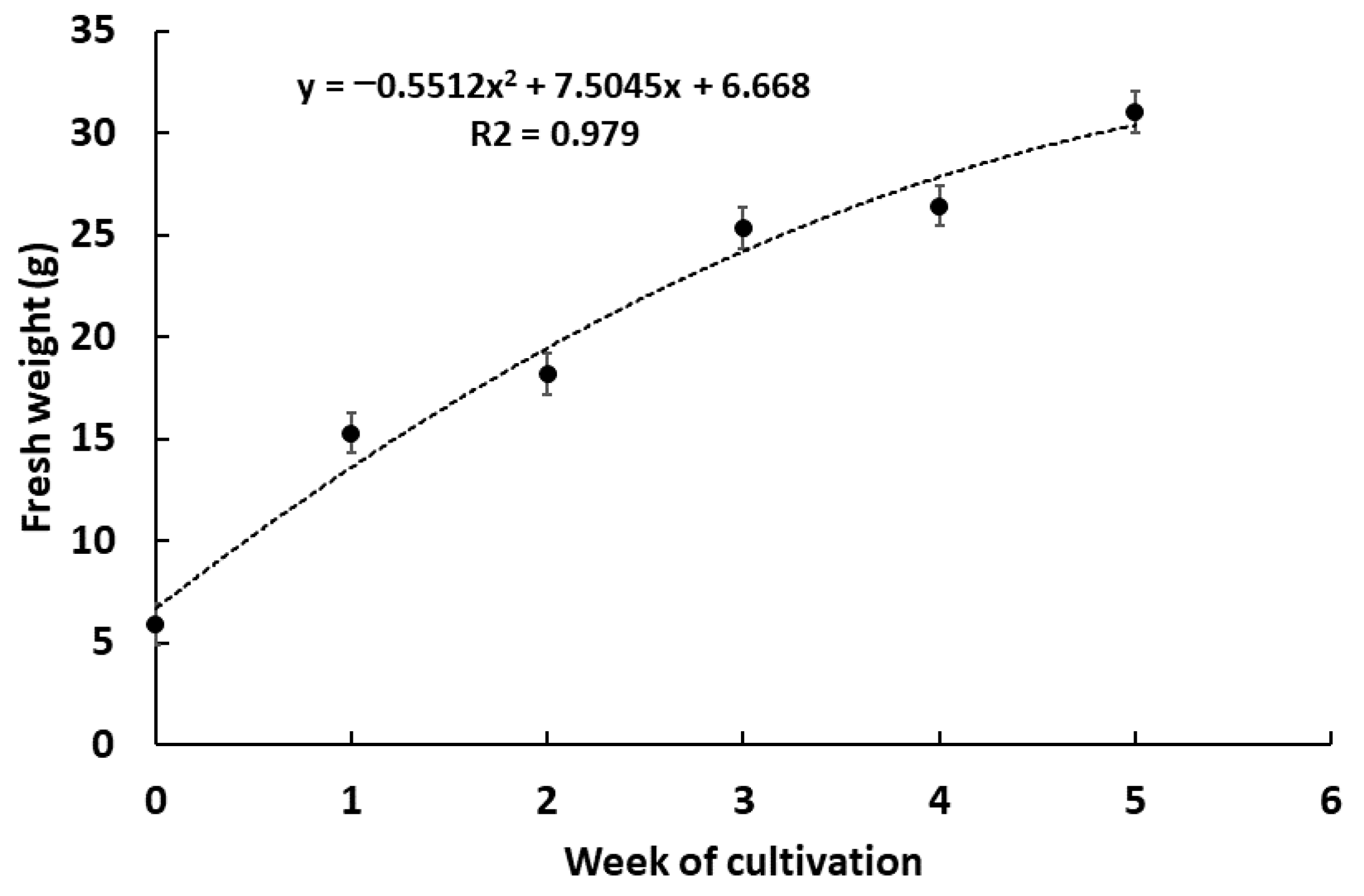
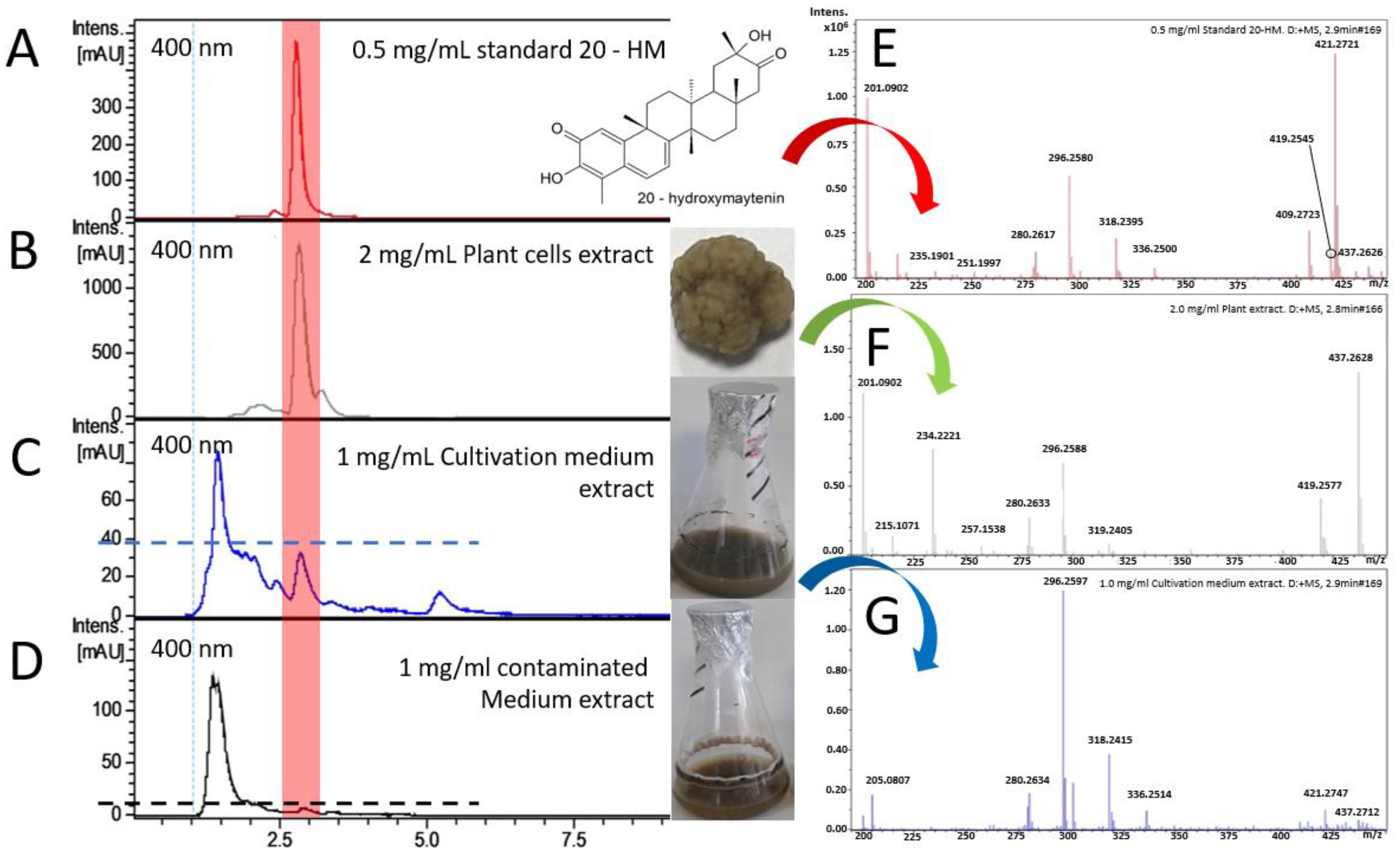
2.3. Cell Suspension Culture of G. heterophylla and Evaluation of the Presence of 20-HM in Both Plant Cells and Cultivation Medium by HPLC-ESI-MS Analysis
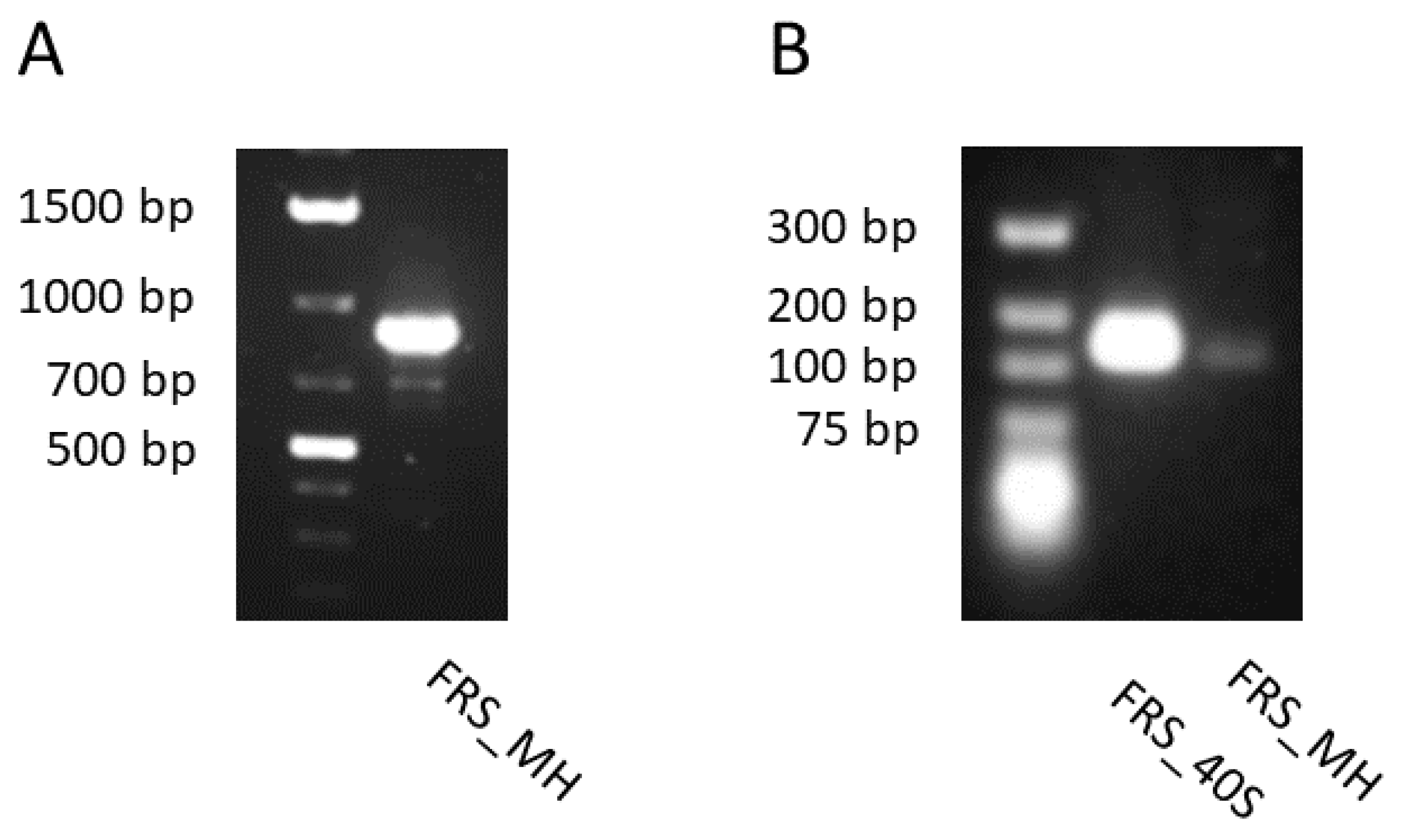
2.4. Partial Sequences and Gene Expression of the Hypothetical FRS Gene from G. heterophylla
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Callus Induction and Cell Suspension Culture of G. heterophylla
4.3. HLPC-ESI-MS Analysis
4.4. Isolation, Purification, and Structural Determination of 20—HM
4.5. Isolation and Identification of the Parasitic Fungus
4.6. Primer Design and PCR Amplification of Partial Sequences of the Hypothetical FRS Gene from G. heterophylla
4.7. DNA Barcoding Identification of G. heterophylla
4.8. Data Availability
4.9. Bioinformatic and Statistic Softwares
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, E.A.; Santos, L.R.; Pontes, I.S.; Matos, L.G.; Silva, G.A.; Lião, L.M. Analgesic and Anti-Inflammatory Effects of Cheiloclinium cognatum Root Barks. Braz. J. Pharm. 2007, 17, 508–513. [Google Scholar] [CrossRef]
- Aparecida de Freitas Formenton Macedo dos Santos, V.; Pereira dos Santos, D.; Castro-Gamboa, I.; Zanoni, M.V.B.; Furlan, M. Evaluation of Antioxidant Capacity and Synergistic Associations of Quinonemethide Triterpenes and Phenolic Substances from Maytenus ilicifolia (Celastraceae). Molecules 2010, 15, 6956. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.R.; Seo, W.D.; Pyun, B.-J.; Lee, B.W.; Jin, Y.B.; Park, K.H.; Seo, E.-K.; Lee, Y.-J.; Lee, Y.-S. Radiosensitization by Celastrol Is Mediated by Modification of Antioxidant Thiol Molecules. Chem. Biol. Interact. 2011, 193, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, G.M.; Kim, J.-K. Anti-Inflammatory Effect of Pristimerin on Lipopolysaccharide-Induced Inflammatory Responses in Murine Macrophages. Arch. Pharm. Res. 2013, 36, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Deeb, D.; Gao, X.; Liu, Y.B.; Pindolia, K.; Gautam, S.C. Pristimerin, a Quinonemethide Triterpenoid, Induces Apoptosis in Pancreatic Cancer Cells through the Inhibition of pro-Survival Akt/NF-ΚB/MTOR Signaling Proteins and Anti-Apoptotic Bcl-2. Int. J. Oncol. 2014, 44, 1707–1715. [Google Scholar] [CrossRef]
- López, M.R.; de León, L.; Moujir, L. Antibacterial Properties of Phenolic Triterpenoids against Staphylococcus epidermidis. Planta Med. 2011, 77, 726–729. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, V.G.; Duarte, L.P.; Silva, G.D.F.; Silva, F.C.; Góes, J.V.; Takahashi, J.A.; Pimenta, L.P.S.; Vieira Filho, S.A. Evaluation of Antimicrobial Activity and Toxic Potential of Extracts and Triterpenes Isolated from Maytenus imbricata. Quím. Nova. 2012, 35, 1375–1380. [Google Scholar] [CrossRef]
- Vinayagam, R.; Xiao, J.; Xu, B. An Insight into Anti-Diabetic Properties of Dietary Phytochemicals. Phytochem. Rev. 2017, 16, 535–553. [Google Scholar] [CrossRef]
- El-Tanbouly, G.S.; El-Awady, M.S.; Megahed, N.A.; Salem, H.A.; El-Kashef, H.A. The NF-ΚB Inhibitor Celastrol Attenuates Acute Hepatic Dysfunction Induced by Cecal Ligation and Puncture in Rats. Environ. Toxicol. Pharm. 2017, 50, 175–182. [Google Scholar] [CrossRef]
- Aceros, H.; Der Sarkissian, S.; Borie, M.; Stevens, L.-M.; Mansour, S.; Noiseux, N. Celastrol-Type HSP90 Modulators Allow for Potent Cardioprotective Effects. Life Sci. 2019, 227, 8–19. [Google Scholar] [CrossRef]
- Salvador, J.A.R.; Leal, A.S.; Valdeira, A.S.; Gonçalves, B.M.F.; Alho, D.P.S.; Figueiredo, S.A.C.; Silvestre, S.M.; Mendes, V.I.S. Oleanane-, Ursane-, and Quinone Methide Friedelane-Type Triterpenoid Derivatives: Recent Advances in Cancer Treatment. Eur. J. Med. Chem. 2017, 142, 95–130. [Google Scholar] [CrossRef]
- Pang, C.; Luo, J.; Liu, C.; Wu, X.; Wang, D. Synthesis and Biological Evaluation of a Series of Novel Celastrol Derivatives with Amino Acid Chain. Chem. Biodivers. 2018, 15, e1800059. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Han, L.Z.; Liang, H.; Mi, C.; Shi, H.; Lee, J.J.; Jin, X. Celastrol Inhibits the HIF-1α Pathway by Inhibition of MTOR/P70S6K/EIF4E and ERK1/2 Phosphorylation in Human Hepatoma Cells. Oncol. Rep. 2014, 32, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hernandes, C.; Miguita, L.; de Sales, R.O.; Silva, E.D.; Mendonça, P.O.; Lorencini da Silva, B.; Klingbeil, M.D.; Mathor, M.B.; Rangel, E.B.; Marti, L.C.; et al. Anticancer Activities of the Quinone-Methide Triterpenes Maytenin and 22-β-Hydroxymaytenin Obtained from Cultivated Maytenus ilicifolia Roots Associated with Down-Regulation of MiRNA-27a and MiR-20a/MiR-17-5p. Molecules 2020, 25, 760. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cheng, J.; Sun, G.; Wu, S.; Li, M.; Gao, Z.; Zhai, S.; Li, P.; Su, D.; Wang, X. P70S6K Promotes IL-6-Induced Epithelial-Mesenchymal Transition and Metastasis of Head and Neck Squamous Cell Carcinoma. Oncotarget 2016, 7, 36539. [Google Scholar] [CrossRef] [PubMed]
- Sannigrahi, M.; Sharma, R.; Panda, N.; Khullar, M. Role of Non-Coding RNAs in Head and Neck Squamous Cell Carcinoma: A Narrative Review. Oral Dis. 2018, 24, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Paz, T.A.; dos Santos, V.A.F.F.M.; Inácio, M.C.; Pina, E.S.; Pereira, A.M.S.; Furlan, M. Production of the Quinone-Methide Triterpene Maytenin by In Vitro Adventitious Roots of Peritassa campestris (Cambess.) A.C.Sm. (Celastraceae) and Rapid Detection and Identification by APCI-IT-MS/MS. Biomed. Res. Int. 2013, 2013, 485837. [Google Scholar] [CrossRef]
- Inácio, M.C.; Paz, T.A.; Pereira, A.M.S.; Furlan, M. Endophytic Bacillus megaterium and Exogenous Stimuli Affect the Quinonemethide Triterpenes Production in Adventitious Roots of Peritassa campestris (Celastraceae). Plant Cell Tissue Organ Cult. 2017, 131, 15–26. [Google Scholar] [CrossRef]
- Taddeo, V.A.; Castillo, U.G.; Martínez, M.L.; Menjivar, J.; Jiménez, I.A.; Núñez, M.J.; Bazzocchi, I.L. Development and Validation of an HPLC-PDA Method for Biologically Active Quinonemethide Triterpenoids Isolated from Maytenus chiapensis. Medicines 2019, 6, 36. [Google Scholar] [CrossRef]
- Yelani, T.; Hussein, A.A.; Meyer, J.J.M. Isolation and Identification of Poisonous Triterpenoids from Elaeodendron croceum. Nat. Prod. Res. 2010, 24, 1418–1425. [Google Scholar] [CrossRef]
- Coppede, J.S.; Pina, E.S.; Paz, T.A.; Fachin, A.L.; Marins, M.A.; Bertoni, B.W.; França, S.C.; Pereira, A.M.S. Cell Cultures of Maytenus ilicifolia Mart. Are Richer Sources of Quinone-Methide Triterpenoids than Plant Roots in Natura. Plant Cell Tissue Organ Cult. 2014, 118, 33–43. [Google Scholar] [CrossRef]
- Paz, T.A.; dos Santos, V.A.F.F.M.; Inácio, M.C.; Dias, N.B.; Palma, M.S.; Pereira, A.M.S.; Furlan, M. Proteome Profiling Reveals Insights into Secondary Metabolism in Maytenus ilicifolia (Celastraceae) Cell Cultures Producing Quinonemethide Triterpenes. Plant Cell Tissue Organ Cult. 2017, 130, 405–416. [Google Scholar] [CrossRef]
- Inácio, M.C.; Paz, T.A.; Pereira, A.M.S.; Furlan, M. Maytenin Plays a Special Role in the Regulation of the Endophytic Bacillus megaterium in Peritassa campestris Adventitious Roots. J. Chem. Ecol. 2019, 45, 789–797. [Google Scholar] [CrossRef]
- Pina, E.S.; Coppede, J.S.; Contini, S.H.T.; Crevelin, E.J.; Lião, L.M.; Bertoni, B.W.; França, S.C.; Pereira, A.M.S. Improved Production of Quinone-Methide Triterpenoids by Cheiloclinium cognatum Root Cultures: Possibilities for a Non-Destructive Biotechnological Process. Plant Cell Tissue Organ Cult. 2017, 128, 705–714. [Google Scholar] [CrossRef]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What These Terms Mean in the Era of Molecular Plant Biology? Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Gunatilaka, A.A.L.; Fernando, H.C.; Kikuchi, T.; Tezuka, Y. 1H and 13C NMR Analysis of Three Quinone-Methide Triterpenoids. Magn. Reson. Chem. 1989, 27, 803–807. [Google Scholar] [CrossRef]
- Fisher Chemical Common Background Contamination Ions in Mass Spectrometry. Available online: https://www.fishersci.de/de/de/brands/IFIBTC31/resources.html (accessed on 5 June 2021).
- Tsvetkov, D.E.; Dmitrenok, A.S.; Tsvetkov, Y.E.; Chizhov, A.O.; Nifantiev, N.E. Polyphenol Components of the Knotwood Extracts of Salix capreal. Russ. Chem. Bull. 2020, 69, 2390–2395. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Pon, A.; Karu, N.; Zheng, J.; Li, C.; Arndt, D.; Gautam, M.; Allen, F.; Wishart, D.S. CFM-ID 3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Watt, A.P.; Pike, A.; Morrison, D. Determination of the Collisionally Activated Dissociation of a Substituted Indole by Orthogonal Acceleration Quadrupole Time-of-Flight Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 1145–1152. [Google Scholar] [CrossRef][Green Version]
- Zhang, D.; Gao, Y.; Jiang, S.; Chen, Y.; Zhang, Y.; Pan, Z. The Similarity and Variability of the Iridoid Glycoside Profile and Antioxidant Capacity of Aerial and Underground Parts of Lamiophlomis rotata According to UPLC-TOF-MS and Multivariate Analyses. RSC Adv. 2018, 8, 2459–2468. [Google Scholar] [CrossRef]
- Aygun, A.; Dumanoglu, H. In Vitro Shoot Proliferation and in Vitro and Ex Vitro Root Formation of Pyrus elaeagrifolia Pallas. Front. Plant Sci. 2015, 6, 225. [Google Scholar] [CrossRef]
- Li, S.; Huang, P.; Ding, G.; Zhou, L.; Tang, P.; Sun, M.; Zheng, Y.; Lin, S. Optimization of Hormone Combinations for Root Growth and Bud Germination in Chinese Fir (Cunninghamia lanceolata) Clone Leaf Cuttings. Sci. Rep. 2017, 7, 5046. [Google Scholar] [CrossRef]
- Elmongy, M.S.; Cao, Y.; Zhou, H.; Xia, Y. Root Development Enhanced by Using Indole-3-Butyric Acid and Naphthalene Acetic Acid and Associated Biochemical Changes of In Vitro Azalea Microshoots. J. Plant Growth Regul. 2018, 37, 813–825. [Google Scholar] [CrossRef]
- Dakah, A.; Zaid, S.; Suleiman, M.; Abbas, S.; Wink, M. In Vitro Propagation of the Medicinal Plant Ziziphora tenuior L. and Evaluation of Its Antioxidant Activity. Saudi J. Biol. Sci. 2014, 21, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiar, Z.; Mirjalili, M.H.; Sonboli, A. In Vitro Callus Induction and Micropropagation of Thymus persicus (Lamiaceae), an Endangered Medicinal Plant Crop. Breed. Appl. Biotechnol. 2016, 16, 48–54. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Cheng, Q.; Niu, M.; Liang, H.; Yan, H.; Zhang, X.; Teixeira da Silva, J.A.; Ma, G. Plant Regeneration via Direct and Callus-Mediated Organogenesis from Leaf Explants of Chirita swinglei (Merr.) W. T. Wang. In Vitro Cell. Dev. Biol. Plant 2016, 52, 521–529. [Google Scholar] [CrossRef]
- Das, P.; Tanti, B.; Borthakur, S.K. In Vitro Callus Induction and Indirect Organogenesis of Brucea mollis Wall. Ex Kurz—A Potential Medicinal Plant of Northeast India. S. Afr. J. Bot. 2018, 119, 203–211. [Google Scholar] [CrossRef]
- Trimpin, S.; Inutan, E.D.; Karki, S.; Elia, E.A.; Zhang, W.-J.; Weidner, S.M.; Marshall, D.D.; Hoang, K.; Lee, C.; Davis, E.T.J.; et al. Fundamental Studies of New Ionization Technologies and Insights from IMS-MS. J. Am. Soc. Mass Spectrom. 2019, 30, 1133–1147. [Google Scholar] [CrossRef] [PubMed]
- Latz, M.A.C.; Jensen, B.; Collinge, D.B.; Lyngs Jørgensen, H.J. Identification of Two Endophytic Fungi That Control Septoria tritici Blotch in the Field, Using a Structured Screening Approach. Biol. Control 2020, 141, 104128. [Google Scholar] [CrossRef]
- Punja, Z.K.; Collyer, D.; Scott, C.; Lung, S.; Holmes, J.; Sutton, D. Pathogens and Molds Affecting Production and Quality of Cannabis sativa L. Front. Plant Sci. 2019, 10, 1120. [Google Scholar] [CrossRef]
- Kogel, K.-H.; Franken, P.; Hückelhoven, R. Endophyte or Parasite—What Decides? Curr. Opin. Plant Biol. 2006, 9, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Souza-Moreira, T.M.; Alves, T.B.; Pinheiro, K.A.; Felippe, L.G.; De Lima, G.M.A.; Watanabe, T.F.; Barbosa, C.C.; Santos, V.A.F.F.M.; Lopes, N.P.; Valentini, S.R.; et al. Friedelin Synthase from Maytenus ilicifolia: Leucine 482 Plays an Essential Role in the Production of the Most Rearranged Pentacyclic Triterpene. Sci. Rep. 2016, 6, 36858. [Google Scholar] [CrossRef] [PubMed]
- Alves, T.B.; Souza-Moreira, T.M.; Valentini, S.R.; Zanelli, C.F.; Furlan, M. Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms. Molecules 2018, 23, 700. [Google Scholar] [CrossRef]
- Busta, L.; Serra, O.; Kim, O.T.; Molinas, M.; Peré-Fossoul, I.; Figueras, M.; Jetter, R. Oxidosqualene Cyclases Involved in the Biosynthesis of Triterpenoids in Quercus suber Cork. Sci Rep. 2020, 10, 8011. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.-Q.; Wang, H.; Tian, X.; Shao, H.-J.; Liu, J.-K. Antifungal Properties of Pristimerin and Celastrol Isolated from Celastrus hypoleucus. Pest Manag. Sci. 2005, 61, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Mahlo, S.M.; Eloff, J.N. Acetone Leaf Extracts of Breonadia salicina (Rubiaceae) and Ursolic Acid Protect Oranges against Infection by Penicillium species. S. Afr. J. Bot. 2014, 93, 48–53. [Google Scholar] [CrossRef]
- Nikolova, M.; Dzhurmanski, A.; Berkov, S. Exudate Compounds of Origanum Species. In Proceedings of the 1st International Electronic Conference on Biological Diversity, Ecology and Evolution (BDEE 2021); MDPI: Basel, Switzerland, 2021. [Google Scholar] [CrossRef]
- Tyndall, J.D.A.; Sabherwal, M.; Sagatova, A.A.; Keniya, M.V.; Negroni, J.; Wilson, R.K.; Woods, M.A.; Tietjen, K.; Monk, B.C. Structural and Functional Elucidation of Yeast Lanosterol 14α-Demethylase in Complex with Agrochemical Antifungals. PLoS ONE 2016, 11, e0167485. [Google Scholar] [CrossRef]
- Parker, J.E.; Warrilow, A.G.S.; Price, C.L.; Mullins, J.G.L.; Kelly, D.E.; Kelly, S.L. Resistance to Antifungals That Target CYP51. J. Chem. Biol. 2014, 7, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Dabiri, M.; Majdi, M.; Bahramnejad, B. Partial Sequence Isolation of DXS and AOS Genes and Gene Expression Analysis of Terpenoids and Pyrethrin Biosynthetic Pathway of Chrysanthemum cinerariaefolium under Abiotic Elicitation. Acta Physiol. Plant 2020, 42, 30. [Google Scholar] [CrossRef]
- Creyaufmüller, F.C.; Chassignet, I.; Delb, H.; Dounavi, A.; Gailing, O.; Leinemann, L.; Kreuzwieser, J.; Teply-Szymanski, J.; Vornam, B. Terpene Synthase Genes in Quercus robur—Gene Characterization, Expression and Resulting Terpenes Due to Cockchafer Feeding. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Kusari, P.; Kusari, S.; Eckelmann, D.; Zühlke, S.; Kayser, O.; Spiteller, M. Cross-Species Biosynthesis of Maytansine in Maytenus serrata. RSC Adv. 2016, 6, 10011–10016. [Google Scholar] [CrossRef]
- Sester, A.; Stüer-Patowsky, K.; Hiller, W.; Kloss, F.; Lütz, S.; Nett, M. Biosynthetic Plasticity Enables Production of Fluorinated Aurachins. ChemBioChem 2020, 21, 2268–2273. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Hubmann, G.; Rosenthal, K.; Lütz, S. Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria. Biomolecules 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Pitakbut, T.; Kusari, S.; Kayser, O.; Spiteller, M. Isolation, Purification and Identification of 20—Hydroxymaytenin as a Bioactive Metabolite from Maytenus heterophylla Liquid Cell Culture. Planta Med. 2019, 85, 1490. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Badotti, F.; de Oliveira, F.S.; Garcia, C.F.; Vaz, A.B.M.; Fonseca, P.L.C.; Nahum, L.A.; Oliveira, G.; Góes-Neto, A. Effectiveness of ITS and Sub-Regions as DNA Barcode Markers for the Identification of Basidiomycota (Fungi). BMC Microbiol. 2017, 17, 42. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Hosein, F.N.; Austin, N.; Maharaj, S.; Johnson, W.; Rostant, L.; Ramdass, A.C.; Rampersad, S.N. Utility of DNA Barcoding to Identify Rare Endemic Vascular Plant Species in Trinidad. Ecol. Evol. 2017, 7, 7311–7333. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Evolution 2008, 62, 1103–1118. [Google Scholar]
- Project, J. Jamovi (Version 1.2.27) [Computer Software]. Available online: https://www.jamovi.org (accessed on 21 June 2021).

| Position | 13C—NMR | Position | 1H—NMR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Reference * | Isolated | Reference * | Isolated | ||||||
| δ (ppm) | δ (ppm) | δ (ppm) | Multiplicity | J value (Hz) | δ | Multiplicity | J Value (Hz) | ||
| 1 | 119.8 | 119.8 | 1 | 6.53 | d | 1.0 | 6.55 | d | 1.0 |
| 2 | 178.4 | 178.4 | 2 | ||||||
| 3 | 146.2 | 146.1 | 3 | 6.97 | s | 6.69 | s | ||
| 4 | 117.1 | 117.1 | 4 | ||||||
| 5 | 127.9 | 127.7 | 5 | ||||||
| 6 | 133.3 | 133.6 | 6 | 7.01 | dd | 7.0, 1,0 | 7.03 | dd | 7, 1 |
| 7 | 118.3 | 118.2 | 7 | 6.36 | d | 7.0 | 6.37 | d | 7.0 |
| 8 | 168.7 | 168.7 | 8 | ||||||
| 9 | 42.9 | 42.7 | 9 | ||||||
| 10 | 164.2 | 164.7 | 10 | ||||||
| 11 | 33.2 | 32 | 11α | 1.95 | td | 14.0, 7.0 | 1.95 | m | |
| 11β | 2.21 | ddd | 14.0, 5.0, 3.0 | 2.26 | ddd | 14.0, 5.0, 3.0 | |||
| 12 | 29.9 | 29.9 | 12α | 1.76 | ddd | 14.0, 7.0, 3.0 | 1.76 | m | |
| 12β | 1.82 | td | 14.0, 5.0 | 1,79 | m | ||||
| 13 | 40 | 40.6 | 13 | ||||||
| 14 | 44.2 | 44.6 | 14 | ||||||
| 15 | 29.4 | 29.5 | 15α | 1.84 | ddd | 12.5, 10.5, 4.5 | 1.83 | m | |
| 15β | 1.73 | m | 1.74 | m | |||||
| 16 | 35.7 | 34 | 16α | 1.65 | ddd | 13.5, 4.5, 2.5 | 1.67 | m | |
| 16β | 1.91 | ddd | 13.5, 10.5, 5.5 | 1.86 | m | ||||
| 17 | 35.9 | 35.5 | 17 | ||||||
| 18 | 43.3 | 43.5 | 18 | 1.93 | dd | 9.5 | 1.88 | m | |
| 19 | 36.9 | 38.2 | 19α | 2.28 | dd | 15.0, 9.0 | 2.21 | dd | 15.0, 9.0 |
| 19β | 2.2 | dd | 15.0, 5.0 | 2.19 | dd | 15.0, 7.0 | |||
| 20 | 73.7 | - ¥ | 20 | 3.24 | s | 3.5 | d | 5.0 | |
| 21 | 214.9 | 213.6 | 21 | ||||||
| 22 | 50.5 | 50.9 | 22α | 2.99 | d | 14.0 | 2.93 | d | 14.0 |
| 22β | 1.95 | d | 14.0 | 1.99 | m | ||||
| 23 | 10.3 | 10.3 | 23 | 2.22 | s | 2.23 | s | ||
| 24 | - | - | 24 | ||||||
| 25 | 38.5 | 39 | 25 | 1.48 | s | 1.51 | s | ||
| 26 | 23.3 | 21.5 | 26 | 1.36 | s | 1.35 | s | ||
| 27 | 19.4 | 19.7 | 27 | 0.89 | s | 0.87 | s | ||
| 28 | 33.2 | 32 | 28 | 1.13 | s | 1.26 | s | ||
| 29 | - | - | 29 | - | - | - | |||
| 30 | 29 | 28.5 | 30 | 1.36 | s | 1.35 | s | ||
| Group | Description | Mean | SD | SE |
|---|---|---|---|---|
| 1 | 5 mg/L IBA + 0 mg/L NAA | 39.60 | 49.40 | 7.13 |
| 2 | 3 mg/L IBA + 1 mg/L NAA | 72.70 | 46.70 | 14.08 |
| 3 | 2 mg/L IBA + 2 mg/L NAA | 26.70 | 45.80 | 11.82 |
| 4 | 1 mg/L IBA + 5 mg/L NAA | 82.40 | 39.30 | 9.53 |
| 5 | 0 mg/L IBA + 5 mg/L NAA | 20.00 | 41.00 | 9.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitakbut, T.; Spiteller, M.; Kayser, O. In Vitro Production and Exudation of 20-Hydroxymaytenin from Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. Cell Culture. Plants 2021, 10, 1493. https://doi.org/10.3390/plants10081493
Pitakbut T, Spiteller M, Kayser O. In Vitro Production and Exudation of 20-Hydroxymaytenin from Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. Cell Culture. Plants. 2021; 10(8):1493. https://doi.org/10.3390/plants10081493
Chicago/Turabian StylePitakbut, Thanet, Michael Spiteller, and Oliver Kayser. 2021. "In Vitro Production and Exudation of 20-Hydroxymaytenin from Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. Cell Culture" Plants 10, no. 8: 1493. https://doi.org/10.3390/plants10081493
APA StylePitakbut, T., Spiteller, M., & Kayser, O. (2021). In Vitro Production and Exudation of 20-Hydroxymaytenin from Gymnosporia heterophylla (Eckl. and Zeyh.) Loes. Cell Culture. Plants, 10(8), 1493. https://doi.org/10.3390/plants10081493







