Morphological Formation, Fatty Acid Profile, and Molecular Identification of Some Landraces of Ethiopian Brassica as a Promising Crop to Support Breeding Programs
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Vegetative Growth Characteristics
2.3. Extraction of Crude Oil from Seeds
2.4. Gas Chromatography Analysis of Oils
2.5. Morphological Data Analysis
2.6. Path Analysis
2.7. Molecular Characterization
3. Results
3.1. Agromorphological Traits
3.2. Total Oil Content of Rapeseeds Genotypes
3.3. Fatty Acid Composition
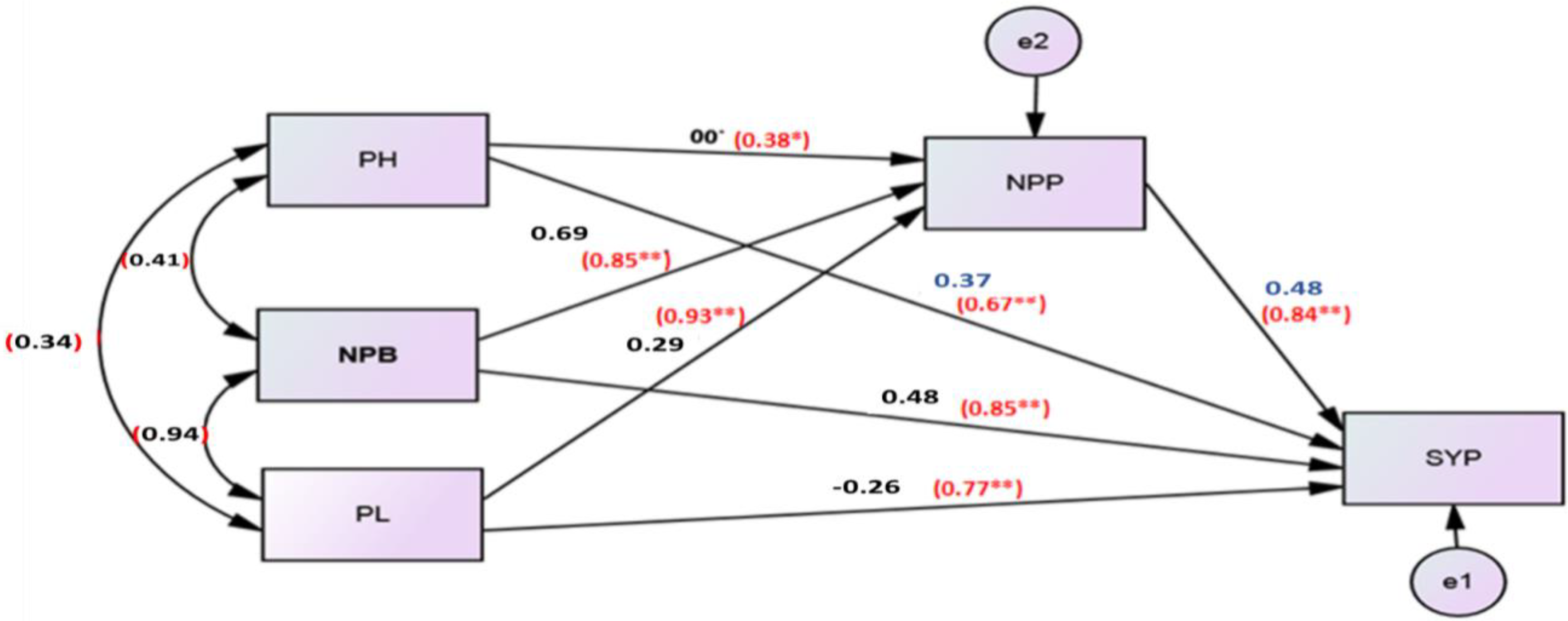
3.4. Path Analysis
3.5. Molecular Characterization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Balkaya, A.; Yanmaz, R.; Apaydin, A.; Kar, H. Morphological characterisation of white head cabbage (Brassica oleracea var. capitata subvar. alba) genotypes in Turkey. N. Z. J. Crop Hortic. Sci. 2005, 33, 333–341. [Google Scholar] [CrossRef]
- El-esawi, M.A.; Bourke, P.; Germaine, K.; Malone, R. Assessment of Morphological Variation in Irish Brassica oleracea Species. J. Agric. Sci. 2012, 4, 20–34. [Google Scholar] [CrossRef]
- Kİbar, B.; Karaağaç, O.; Kar, H. Determination of Morphological Variability among Cabbage (Brassica oleracea var. capitata L.) Hybrids and Their Parents. J. Inst. Sci. Technol. 2016, 6, 31–44. [Google Scholar] [CrossRef]
- Seifert, K.A.; Samson, R.A.; Dewaard, J.R.; Houbraken, J.; Lévesque, C.A.; Moncalvo, J.-M.; Louis-Seize, G.; Hebert, P. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc. Natl. Acad. Sci. USA 2007, 104, 3901–3906. [Google Scholar] [CrossRef]
- Cardone, M.; Mazzoncini, M.; Menini, S.; Rocco, V.; Senatore, A.; Seggiani, M.; Vitolo, S. Brassica carinata as an alternative oil crop for the production of biodiesel in Italy: Agronomic evaluation, fuel production by transesterification and characterization. Biomass Bioenergy 2003, 25, 623–636. [Google Scholar] [CrossRef]
- Thakur, A.K.; Singh, K.H.; Sharma, D.; Parmar, N.; Nanjundan, J. Breeding and genomics interventions in Ethiopian mustard (Brassica carinata A. Braun) improvement—A mini review. S. Afr. J. Bot. 2019, 125, 457–465. [Google Scholar] [CrossRef]
- Hagos, R.; Shaibu, A.S.; Zhang, L.; Cai, X.; Liang, J.; Wu, J.; Lin, R.; Wang, X. Ethiopian Mustard (Brassica carinata A. Braun) as an Alternative Energy Source and Sustainable Crop. Sustainability 2020, 12, 7492. [Google Scholar] [CrossRef]
- Ali, W.M.; Pant, D.P. Genetic analysis of oil content in Ethiopian mustard (Brassica carinata A. Braun). Pantnagar J. Res. 2013, 11, 208–213. [Google Scholar]
- Martínez-Valdivieso, D.; Font, R.; Del Río-Celestino, M. Prediction of agro-morphological and nutritional traits in Ethiopian mustard leaves (Brassica carinata A. Braun) by visible-near-infrared spectroscopy. Foods 2019, 8, 6. [Google Scholar] [CrossRef]
- Tiwari, M.; Babu, V.B.; Kotwaliwale, N.; Hamad, R.; Singh, K. Identification of Indian mustard (Brassica juncea L.) varieties by DUS testing using morphological characters Identification of Indian mustard (Brassica juncea L.) varieties by DUS testing using morphological characters. J. Oilseed Brassica 2017, 8, 57–61. [Google Scholar]
- Weerakoon, S.R.; Iqbal, M.C.M.; Somaratne, S.; Peiris, P.K.D.; Wimalasuriya, W.S.R. Delimitation of local mustard (Brassica juncea) germplasm in Sri Lanka and improvement of their nutritive quality. In Proceedings of the 12th International Rapeseed Congress, Wuhan, China, 26–30 March 2007; pp. 75–78. [Google Scholar]
- Weerakoon, S.; Somaratne, S. Agro-morphological characterization and relationships amoung mustard germplasm (Brassica juncea [ L.] Czern & Coss ) in Sri Lanka: A classification tree approach. J. Agric. Sci. 2010, 5, 89–97. [Google Scholar]
- Timpano, E.K.; Scheible, M.K.R.; Meiklejohn, K.A. Optimization of the second internal transcribed spacer (ITS2) for characterizing land plants from soil. PLoS ONE 2020, 15, e0231436. [Google Scholar] [CrossRef] [PubMed]
- Ziffer-berger, J.; Keren-keiserman, A.; Doron-faigenboim, A. Notes on the generic position of Brassica deserti (Brassicaceae). Isr. J. Plant Sci. 2019, 66, 89–93. [Google Scholar] [CrossRef]
- Takamiya, T.; Wongsawad, P.; Tajima, N.; Shioda, N.; Lu, A.J.F.; Wen, C.L.; Bin Wu, J.; Handa, T.; Iijima, H.; Kitanaka, S.; et al. Identification of Dendrobium Species Used for Herbal Medicines Based on Ribosomal DNA Internal Transcribed Spacer Sequence. Biol. Pharm. Bull. 2011, 34, 779–782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Panyal, N.; Panwar, S.; Sonah, H.; Singh, K.P.; Sharma, T.R. Genetic diversity analysis of marigold (Tagetes sp) genotypes using RAPD and ISSR markers. Indian J. Agric. Sci. 2013, 83, 484–490. [Google Scholar]
- Lee, S.; Mohamed, R.; Faridah-Hanum, I.; Lamasudin, D. Utilization of the internal transcribed spacer (ITS) DNA sequence to trace the geographical sources of Aquilaria malaccensis Lam. populations. Plant Genet. Resour. 2018, 16, 103–111. [Google Scholar] [CrossRef]
- Rush, T.A.; Golan, J.; McTaggart, A.; Kane, C.; Schneider, R.W.; Aime, M.C. Variation in the Internal Transcribed Spacer Region of Phakopsora pachyrhizi and Implications for Molecular Diagnostic Assays. Plant Dis. 2019, 103, 2237–2245. [Google Scholar] [CrossRef]
- Bhandari, H.R.; Bhanu, A.N.; Srivastava, K.; Singh, M.N.; Hemantaranjan, A. Assessment of genetic diversity in crop plants—An overview. Adv. Plants Agric. Res. 2017, 7, 279–286. [Google Scholar]
- Zeng, X.; Li, H.; Li, K.; Yuan, R.; Zhao, S.; Li, J.; Luo, J.; Li, X.; Ma, H.; Wu, G.; et al. Evolution of the Brassicaceae-specific MS5-Like family and neofunctionalization of the novel MALE STERILITY 5 gene essential for male fertility in Brassica napus. New Phytol. 2021, 229, 2339–2356. [Google Scholar] [CrossRef]
- Crevelari, J.A.; Durães, N.N.L.; Bendia, L.C.R.; Vettorazzi, J.C.F.; Entringer, G.C.; Junior, J.A.F.; Pereira, M.G. Correlations between agronomic traits and path analysis for silage production in maize hybrids. Bragantia 2018, 77, 243–252. [Google Scholar] [CrossRef]
- Granlund, M.; Zimmerman, D.C. Oil content of safflower seeds as determined by wide-line nuclear magnetic resonance (NMR). Proc. Natl. Acad. Sci. USA 1975, 27, 128–133. [Google Scholar]
- Velioglu, S.D.; Temiz, H.T.; Ercioglu, E.; Velioglu, H.M.; Topcu, A.; Boyaci, I.H. Use of Raman spectroscopy for determining erucic acid content in canola oil. Food Chem. 2017, 221, 87–90. [Google Scholar] [CrossRef]
- Mohdaly, A.A.; Mahmoud, A.A.; Housain, M.H.; Iryna, S. Chemical composition, physicochemical properties and fatty acid profile of Tiger Nut (Cyperus esculentus L) seed oil as affected by different preparation methods. Int. Food Res. J. 2015, 22, 1931–1938. [Google Scholar]
- Abdi, H. Bonferroni and Sidak Corrections for Mutliple Comparisons; Salkind, N.J., Ed.; Encyclopedia of Measurement and Statistics: Thousand Okas, CA, USA, 2007; pp. 103–107. [Google Scholar]
- Dewey, D.R.; Lu, K.H. A correlation and path coefficient analysis of components of crested wheat grass seed production. Agron. J. 1959, 51, 515–518. [Google Scholar] [CrossRef]
- Arbuckle, J.L. IBM SPSS Amos 24 User’s Guide; Amos Development Corporation, SPSS Inc.: Crawfordville, FL, USA, 2017. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- AbdAl-Aziz, S.A.; El-Metwally, M.M.; Saber, W.I. Molecular identification of a novel inulinolytic fungus isolated from and grown on tubers of Helianthus tuberosus and statistical screening of medium components. World J. Microbiol. Biotechnol. 2012, 28, 3245–3254. [Google Scholar] [CrossRef]
- Hewavitharana, G.G.; Perera, D.N.; Navaratne, S.B.; Wickramasinghe, I. Extraction methods of fat from food samples and preparation of fatty acid methyl esters for gas Chromatography: A Review. Arab. J. Chem. 2020. [Google Scholar] [CrossRef]
- Froyen, E.; Burns-Whitmore, B. The Effects of Linoleic Acid Consumption on Lipid Risk Markers for Cardiovascular Disease in Healthy Individuals: A Review of Human Intervention Trials. Nutrients 2020, 12, 2329. [Google Scholar] [CrossRef] [PubMed]
- Babaoglu, M.; Guel, M. Safflower (Carthamus tinctorius L.) breeding activities at Trakya Agricultural. J. Crop Breed. Genet. 2015, 1, 20–25. [Google Scholar]
- Kose, A.; Onder, O.; Bilir, O.; Kosar, F. Application of multivariate statistical analysis for breeding strategies of spring safflower (Carthamus tinctorius L.). Turkish J. Field Crops 2018, 23, 12–19. [Google Scholar] [CrossRef]
- Teklehaymanot, T.; Wang, H.; Liang, J.; Wu, J.; Lin, R.; Zhou, Z.; Cai, X.; Wang, X. Variation in Plant Morphology and Sinigrin Content in Ethiopian Mustard (Brassica carinata L.). Hortic. Plant J. 2019, 5, 205–212. [Google Scholar] [CrossRef]
- Hossain, Z.; Johnson, E.N.; Wang, L.; Blackshaw, E.R.; Gan, Y. Comparative analysis of oil and protein content and seed yield of five Brassicaceae oilseeds on the Canadian prairie. Ind. Crop Prod. 2019, 136, 77–86. [Google Scholar] [CrossRef]
- Stamigna, C.; Chiaretti, D.; Chiaretti, E.; Prosini, P.P. Oil and furfural recovery from Brassica carinata. Biomass Bioenerg. 2012, 39, 478–483. [Google Scholar] [CrossRef]
- Aghdam, A.M.; Sayfzadeh, S.; Shirani Rad, A.H.; Valadabadi, S.A.; Zakerin, H.R. The assessment of water stress and delay cropping on quantitative and qualitative traits of rapeseed genotypes. Ind. Crop Prod. 2019, 131, 160–165. [Google Scholar] [CrossRef]
- Cartea, E.; De Haro-Bailón, A.; Padilla, G.; Obregón-Cano, S.; Del Rio-Celestino, M.; Ordás, A. Seed Oil Quality of Brassica napus and Brassica rapa Germplasm from Northwestern Spain. Foods 2019, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Zahran, H.A.; Abd-Elsaber, A.; Tawfeuk, H.Z. Genetic diversity, chemical composition and oil characteristics of six sesame genotypes. OCL 2020, 27, 39. [Google Scholar] [CrossRef]
- Kumar, S.; Seepaul, R.; Mulvaney, M.J.; Colvin, B.; George, S.; Marois, J.J.; Bennett, R.; Leon, R.; Wright, D.L.; Small, I.M. Brassica carinata genotypes demonstrate potential as a winter biofuel crop in South East United States. Ind. Crop Prod. 2020, 150, 112353. [Google Scholar] [CrossRef]
- Sokoła-Wysoczańska, E.; Wysoczański, T.; Wagner, J.; Czyż, K.; Bodkowski, R.; Lochyński, S.; Patkowska-Sokoła, B. Polyunsaturated fatty acids and their potential therapeutic role in cardiovascular system disorders—A review. Nutrients 2018, 10, 1561. [Google Scholar] [CrossRef]
- Kirkegaard, J.A.; Lilley, J.M.; Brill, R.D.; Ware, A.H.; Walela, C.K. The critical period for yield and quality determination in canola (Brassica napus L.). Field Crop. Res. 2018, 222, 180–188. [Google Scholar] [CrossRef]
- Abebe, D. Genetic Variability and Association among Seed Yield and Yield Related Traits in Ethiopian Mustard (Brassica carinata A. Braun) at Kulumsa, Arsi. Master’s Thesis, School of Graduate Studies of Alemaya University, Dire Dawa, Ethiopia, 2006. [Google Scholar]
- Rondanini, D.P.; Vilarino, M.d.P.; Roberts, M.E.; Polosa, M.A.; Botto, J. Physiological responses of spring rapeseed (Brassica napus) to red/far-red ratios and irradiance during pre- and post-flowering stages. Physiol. Plant. 2014, 152, 784–794. [Google Scholar] [CrossRef]
- Paul, M.; Panda, G.; Kumar, P.; Mohapatra, D.; Thatoi, H. Study of structural and molecular interaction for the catalytic activity of cellulases: An insight in cellulose hydrolysis for higher bioethanol yield. J. Mol. Struct. 2020, 1204, 127547. [Google Scholar] [CrossRef]
- Ton, L.B.; Neik, T.X.; Batley, J. The Use of Genetic and Gene Technologies in Shaping Modern Rapeseed Cultivars (Brassica napus L.). Genes 2020, 11, 1161. [Google Scholar] [CrossRef] [PubMed]
- Zuppa, A.; Costantini, S.; Costantini, M. Comparative sequence analysis of bacterial symbionts from the marine sponges Geodia cydonium and Ircinia muscarum. Bioinformation 2014, 10, 196–200. [Google Scholar] [CrossRef][Green Version]
- Kaya, I.; Demir, I.; Usta, M.; Sipahioglu, M.H. Molecular phylogeny based on its sequences of nrDNA of some species belonging to dodder (Cuscuta L.) genus from various ecological sites of Turkey. Not. Bot. Horti Agrobot Cluj-Napoca 2020, 48, 1332–1340. [Google Scholar] [CrossRef]
- Elshafei, A.A.; Afiah, S.A.; Amer, M.A.; El-enany, M.A.M. Validation of molecular markers linked with salinity tolerance in wheat (Triticum aestivum L.) grown on saline soil. Biosci. Res. 2019, 16, 963–978. [Google Scholar]
- Paul, M.; Islam, T.; Hoque, M.I.; Sarker, R.H. Analysis of genetic diversity in oilseed brassica germplasm through issr markers and isozyme profiling. Bangladesh J. Bot. 2020, 49, 147–158. [Google Scholar] [CrossRef]
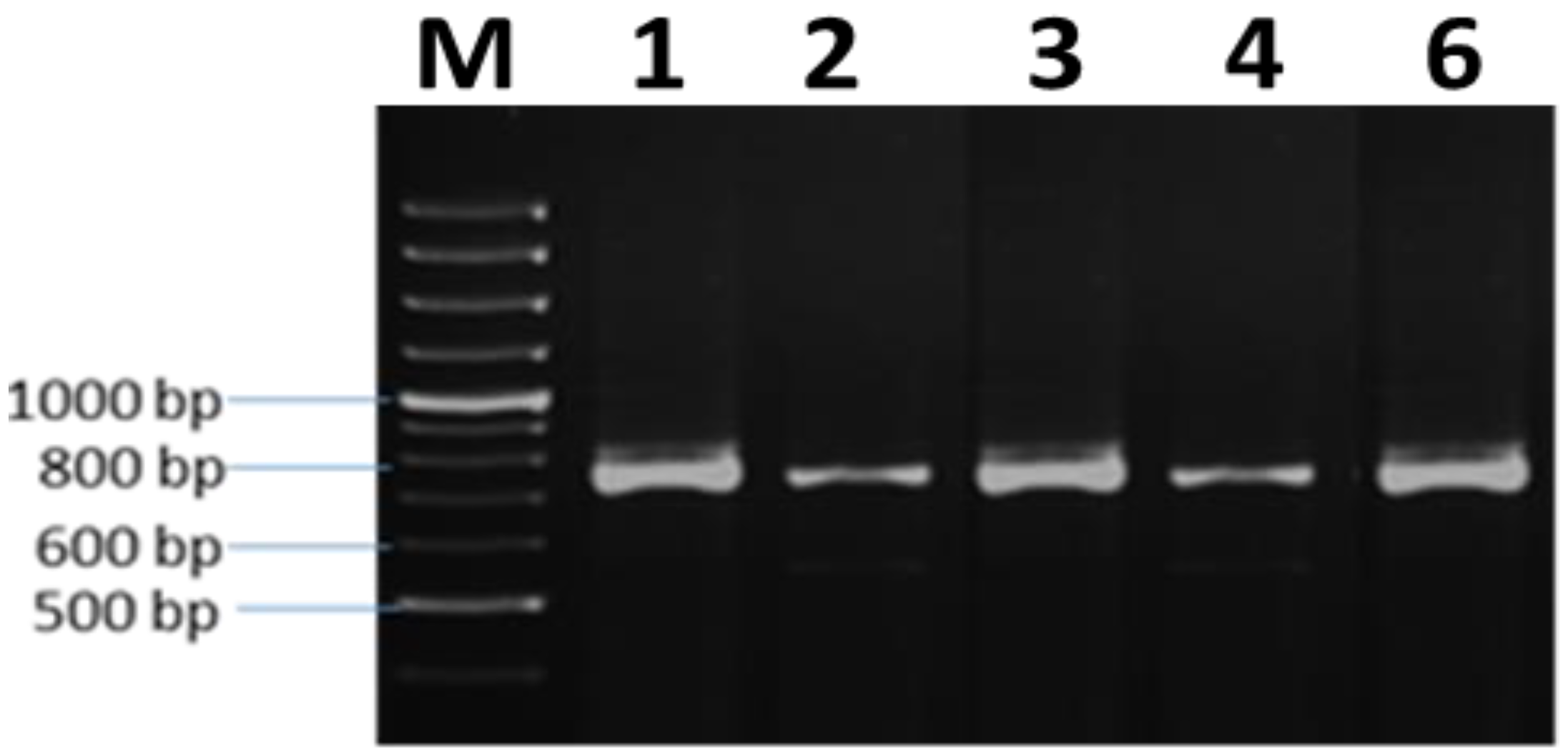

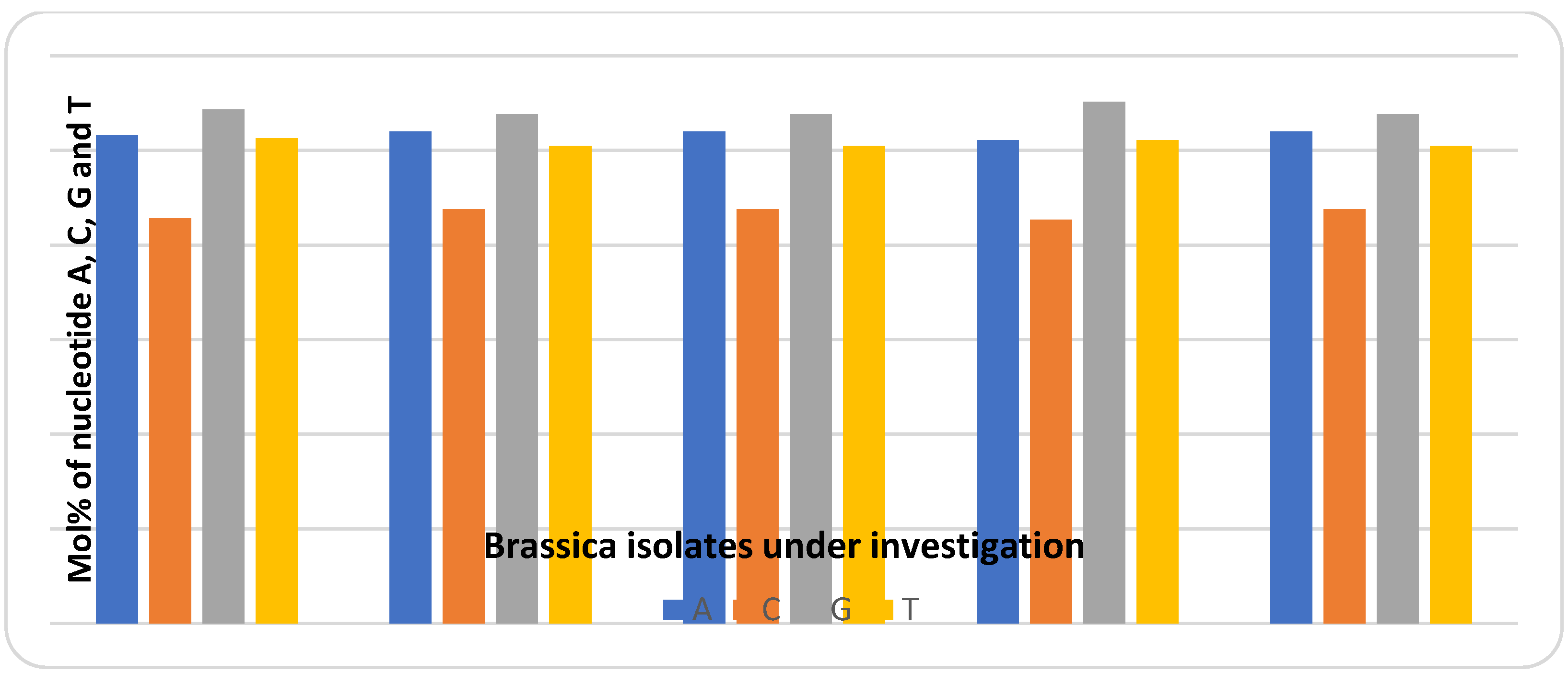
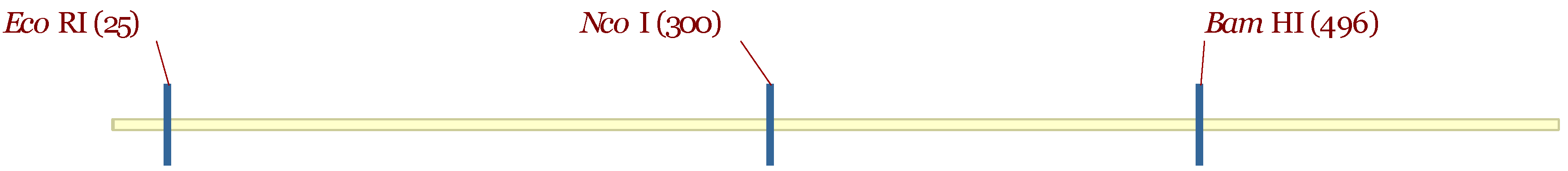

| Accession | Fay1 | Fay2 | Fay3 | Fay4 | Fay6 |
| 2017/2018 | |||||
| Plant height | 175.3 a ± 4.9 | 145.9 b ± 4.1 | 141.8 b ± 5.1 | 141.2 b ± 5.8 | 142.0 b ± 1.3 |
| Height of first branch | 14.0 e ± 0.4 | 32.9 b ± 0.5 | 43.5 a ± 0.8 | 17.3 d ± 0.7 | 23.3 c ± 0.7 |
| No. of branches per plant | 22.2 a ± 0.8 | 13.7 c ± 0.6 | 8.5 d ± 0.4 | 20.7 a ± 0.5 | 17.8 b ± 0.4 |
| No. of pods per plant | 2968.7 a ± 5.3 | 1992.9 d ± 3.4 | 1147.8 e ± 13.3 | 2764.3 b ± 9.4 | 2536.5 c ± 18.2 |
| Seed yield per plant | 87.8 a ± 2.6 | 49.6 d ± 0.4 | 37.9 e ± 0.6 | 60.2 b ± 2.22 | 56.6 c ± 0.5 |
| Average pod length | 5.2 a ± 0.05 | 3.3 c ± 0.1 | 2.5 d ± 0.2 | 5.1 a ± 0.1 | 4.7 b ± 0.1 |
| Number of seeds per pod | 21.3 a ± 0.3 | 13.0 d ± 0.7 | 7.8 e ± 0.3 | 19.8 b ± 0.5 | 17.3 c ± 0.3 |
| Protein content | 31.2 a ± 0.2 | 23.0 c ± 0.5 | 26.3 b ± 1.4 | 26.8 b ± 0.2 | 26.5 b ± 0.7 |
| Oil content | 41.2 c ± 0.4 | 44.8 b ± 0.2 | 49.2 a ± 0.5 | 44.8 b ± 0.9 | 42.4 c ± 0.5 |
| 2018/2019 | |||||
| Plant height | 165.2 a ± 4.3 | 160.0 a ± 2.1 | 141.0 b ± 5.1 | 136.5 b ± 5.8 | 134.3 b ± 5.1 |
| Height of first branch | 14.3 e ± 0.3 | 33.1 b ± 0.6 | 43.8 a ± 0.9 | 19.3 d ± 0.5 | 23.4 c ± 0.4 |
| No. of branches per plant | 21.8 a ± 0.6 | 15.9 c ± 0.5 | 7.3 d ± 0.4 | 20.3 b ± 0.5 | 19.7 b ± 0.5 |
| No. of pods per plant | 2970.3 a ± 5.6 | 1979.5 d ± 3.7 | 1130.7 e ± 7.3 | 2741.3 b ± 14.2 | 2565.4 c ± 11.4 |
| Seed yield per plant | 90.3 a ± 1.1 | 51.2 c ± 1.1 | 37.0 d ± 0.7 | 58.2 b ± 0.8 | 55.3 b ± 0.1 |
| Average pod length | 5.3 a ± 0.1 | 3.7 d ± 0.1 | 2.7 e ± 0.1 | 4.6 b ± 0.1 | 4.1 c ± 0.1 |
| Number of seeds per pod | 20.8 a ± 0.3 | 13.9 c ± 0.4 | 11.3 d ± 0.6 | 19.7 a ± 0.7 | 16.9 b ± 0.5 |
| Protein content | 31.8 a ± 0.2 | 24.3 c ± 0.4 | 26.7 b ± 1.1 | 26.8 b ± 0.3 | 28.3 b ± 0.4 |
| Oil content | 41.1 d ± 0.4 | 44.8 b ± 0.2 | 49.3 a ± 0.2 | 45.1 b ± 0.3 | 43.4 c ± 0.2 |
| Compounds | Fay1 | Fay2 | Fay3 | Fay4 | Fay6 |
|---|---|---|---|---|---|
| C14 Myristic | 0.04 * ± 0.01 | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.04 ± 0.01 | 0.04 ± 0.01 |
| C16 Palmitic | 3.16 ± 0.41 | 3.08 ± 0.40 | 3.28 ± 0.43 | 3.31 ± 0.43 | 3.37 ± 0.44 |
| C16:1 Palmitoleic | 0.08 ± 0.01 | 0.13 ± 0.02 | 0.22 ± 0.03 | 0.08 ± 0.01 | 0.12 ± 0.02 |
| C17 Margaric | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.03 ± 0.00 | 0.07 ± 0.01 | 0.06 ± 0.01 |
| C18 Stearic | 0.88 ± 0.12 | 0.77 ± 0.10 | 1.12 ± 0.15 | 0.82 ± 0.11 | 0.93 ± 0.12 |
| C18:1 n9 cis Oleic | 7.73 ± 1.12 | 7.02 ± 0.92 | 22.98 ± 3.00 | 8.30 ± 1.08 | 9.90 ± 1.29 |
| C18:1 n9 t Elaidic | 0.11 ± 0.01 | 0.06 ± 0.01 | 0.03 ± 0.00 | 0.11 ± 0.01 | 0.08 ± 0.01 |
| C18:2 n6 cis Linoleic | 15.52 ± 2.03 | 15.65 ± 2.05 | 21.69 ± 2.84 | 16.44 ± 2.15 | 16.34 ± 2.14 |
| C18:3 n3 Linolenic | 14.14 ± 1.85 | 14.27 ± 1.87 | 14.02 ± 1.83 | 12.29 ± 1.61 | 12.11 ± 1.58 |
| C20 Arachidic | 0.73 ± 0.10 | 0.56 ± 0.07 | 0.71 ± 0.09 | 0.68 ± 0.09 | 0.74 ± 0.10 |
| C20:1n11c Paullinic | 6.45 ± 0.84 | 5.78 ± 0.76 | 11.01 ± 1.44 | 6.80 ± 0.89 | 6.54 ± 0.85 |
| C20:1 n-9 Gondoic | 0.97 ± 0.13 | 0.87 ± 0.11 | 0.81 ± 0.11 | 0.83 ± 0.11 | 0.80 ± 0.10 |
| C22 Behenic acid | 0.48 ± 0.06 | 0.49 ± 0.06 | 0.30 ± 0.04 | 0.47 ± 0.06 | 0.56 ± 0.07 |
| C22 1 n9 Erucic | 45.87 ± 6.0 | 47.78 ± 6.25 | 20.83 ± 2.72 | 45.88 ± 6.00 | 44.25 ± 5.78 |
| C20 5n3 Eicosapentaenoic | 1.23 ± 0.16 | 1.28 ± 0.17 | 0.29 ± 0.04 | 1.15 ± 0.15 | 1.12 ± 0.15 |
| C22 4n6 Docosatetraenoic | 1.67 ± 0.22 | 1.71 ± 0.22 | 0.91 ± 0.1 | 1.66 ± 0.22 | 1.98 ± 0.26 |
| Unknown compound | 0.89 ± 0.12 | 0.44 ± 0.06 | 1.71 ± 0.22 | 1.07 ± 0.14 | 1.06 ± 0.14 |
| Total SFA | 5.34 ± 0.70 | 5.01 ± 0.65 | 5.50 ± 0.72 | 5.39 ± 0.70 | 5.70 ± 0.75 |
| Total unsaturated | 93.77 ± 12.26 | 94.55 ± 12.36 | 92.79 ± 12.13 | 93.54 ± 12.23 | 93.24 ± 12.19 |
| Total MUFA | 61.21 ± 8.0 | 61.64 ± 8.08 | 55.88 ± 7.30 | 62.00 ± 8.10 | 61.69 ± 8.06 |
| Total PUFA | 32.56 ± 4.26 | 32.91 ± 4.03 | 36.91 ± 4.82 | 31.54 ± 4.12 | 31.55 ± 4.12 |
| ω-6/ω-3 | 1.12 ± 0.15 | 1.12 ± 0.15 | 1.58 ± 0.21 | 1.35 ± 0.18 | 1.38 ± 0.18 |
| PUFA/SFA | 6.10 ± 0.80 | 6.57 ± 0.86 | 6.71 ± 0.88 | 5.85 ± 0.76 | 5.54 ± 0.72 |
| SFA/UnSFA | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.06 ± 0.01 | 0.06 ± 0.01 |
| SFA/PUFA | 0.16 ± 0.02 | 0.15 ± 0.02 | 0.15 ± 0.02 | 0.17 ± 0.02 | 0.18 ± 0.02 |
| Oleic/linoleic | 0.50 ± 0.07 | 0.45 ± 0.06 | 1.06 ± 0.14 | 0.50 ± 0.07 | 0.61 ± 0.08 |
| Characters | Direct Effect | Indirect Effect Via | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PH | HFB | NPB | NPP | SYP | Protein | PL | NSP | TIE | ||
| PH | −0.16 | 0.82 | −0.08 | −0.62 | −0.32 | 0.13 | −0.11 | −0.11 | −0.29 | |
| HFB | −1.95 | 0.07 | 0.19 | 1.58 | 0.42 | −0.11 | 0.31 | 0.27 | 2.73 | |
| NPB | −0.20 | −0.07 | 1.87 | −1.54 | −0.41 | 0.12 | −0.31 | −0.26 | −0.6 | |
| NPP | −0.19 | −0.06 | 1.92 | −1.61 | −0.41 | 0.10 | −0.31 | −0.27 | −0.64 | |
| SYP | 0.48 | −0.11 | 1.69 | −0.17 | −1.36 | 0.15 | −0.25 | −0.23 | −0.28 | |
| Protein | −0.23 | −0.10 | 0.97 | −0.10 | −0.73 | −0.32 | −0.18 | −0.14 | −0.6 | |
| PL | −0.33 | −0.06 | 1.84 | −0.18 | −1.50 | −0.37 | 0.13 | −0.26 | −0.4 | |
| NSP | −0.28 | −0.06 | 1.90 | −0.18 | −1.56 | −0.40 | 0.11 | −0.30 | −0.49 | |
| Person correlation with oil percent (r) | −0.46 | 0.77 | −0.79 | −0.82 | −0.76 | −0.37 | −0.74 | −0.78 | ||
| coefficient of determination (R2) | 21.16% | 59.29% | 62.41% | 67.24% | 57.76% | 13.69% | 54.76% | 60.84% | ||
| p−value | 0.00 ** | 0.00 ** | 0.00 ** | 0.00 ** | 0.00 ** | 0.04 * | 0.00 ** | 0.00 ** | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalaf, A.E.A.; Abd Al-Aziz, S.A.; Ali, S.M.; Mohdaly, A.A.; Rady, M.M.; Majrashi, A.; Ali, E.F.; Yassein, A.A.M. Morphological Formation, Fatty Acid Profile, and Molecular Identification of Some Landraces of Ethiopian Brassica as a Promising Crop to Support Breeding Programs. Plants 2021, 10, 1431. https://doi.org/10.3390/plants10071431
Khalaf AEA, Abd Al-Aziz SA, Ali SM, Mohdaly AA, Rady MM, Majrashi A, Ali EF, Yassein AAM. Morphological Formation, Fatty Acid Profile, and Molecular Identification of Some Landraces of Ethiopian Brassica as a Promising Crop to Support Breeding Programs. Plants. 2021; 10(7):1431. https://doi.org/10.3390/plants10071431
Chicago/Turabian StyleKhalaf, Ahmed E. A., Samia A. Abd Al-Aziz, Safaa M. Ali, Adel A. Mohdaly, Mostafa M. Rady, Ali Majrashi, Esmat F. Ali, and Ahmed A. M. Yassein. 2021. "Morphological Formation, Fatty Acid Profile, and Molecular Identification of Some Landraces of Ethiopian Brassica as a Promising Crop to Support Breeding Programs" Plants 10, no. 7: 1431. https://doi.org/10.3390/plants10071431
APA StyleKhalaf, A. E. A., Abd Al-Aziz, S. A., Ali, S. M., Mohdaly, A. A., Rady, M. M., Majrashi, A., Ali, E. F., & Yassein, A. A. M. (2021). Morphological Formation, Fatty Acid Profile, and Molecular Identification of Some Landraces of Ethiopian Brassica as a Promising Crop to Support Breeding Programs. Plants, 10(7), 1431. https://doi.org/10.3390/plants10071431








