Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and FISH Mapping of CON/COM Satellite DNA Families
Abstract
1. Introduction
2. Results
2.1. PCR and BLAST Analysis
2.2. Chromosomal Structural Variations in the Studied Accessions
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Genomic DNA Extraction, PCR, and Sequencing
4.3. Chromosome Spread Preparation
4.4. Fluorescence In Situ Hybridization
- In FISH assays, the following probes were used:
- pTa71 containing a 9 kb long DNA sequence of common wheat enclosing 18S-5.8S-26S (45S) rDNA [49];
- pTa794 containing a 420 bp long DNA sequence of wheat containing 5S rDNA [50];
- CON1, CON2, and COM2 sequences obtained by the PCR-based technique.
4.5. Chromosomal Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tzvelev, N.N. Poaceae of the USSR; Nauka: Leningrad, Russia, 1976. [Google Scholar]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Zuloaga, F.O.; Judziewicz, E.J.; Filgueiras, T.S.; Davis, J.I.; Morrone, O. A worldwide phylogenetic classification of the Poaceae (Gramineae). J. Syst. Evol. 2015, 53, 117–137. [Google Scholar] [CrossRef]
- Kellogg, E.A. Flowering plants. Monocots: Poaceae. In The Families and Genera of Vascular Plants; Kubitzki, K., Ed.; Springer: Cham, Switzerland, 2015; Volume XIII. [Google Scholar] [CrossRef]
- Soreng, R.J.; Peterson, P.M.; Romaschenko, K.; Davidse, G.; Teisher, J.K.; Clark, L.G.; Barber, P.; Gillespie, L.J.; Zuloaga, F.O. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: An update and a comparison of two 2015 classifications. J. Syst. Evol. 2017, 55, 259–290. [Google Scholar] [CrossRef]
- Tzvelev, N.N. Arctic Flora of the USSR; Nauka: Leningrad, Russia, 1964; Volume 2. [Google Scholar]
- Hulten, E. Flora of Alaska and Neighboring Territories; Stanford University Press: Stanford, UK, 1968. [Google Scholar]
- Alberdi, M.; Bravo, L.A.; Gutierrez, A.; Gidekel, M.; Corcuera, L.J. Ecophysiology of Antarctic vascular plants. Physiol. Plant. 2002, 115, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.M. Chromosome numbers of Falkland Islands angiosperms. BAS Bull. 1967, 14, 69–82. [Google Scholar]
- Moore, D.M. Studies in Colobanthus Quitensis (Kunth) Bartl. and Deschampsia antarctica Desv. II. Taxonomy, distribution and relationships. BAS Bull. 1970, 23, 63–80. [Google Scholar]
- Chiapella, J.; Zuloaga, F.O. A Revision of Deschampsia, Avenella, and Vahlodea (Poaceae, Poeae, Airinae) in South America. Ann. Mo. Bot. Gard. 2010, 97, 141–162. [Google Scholar] [CrossRef]
- Lee, J.; Noh, E.K.; Choi, H.S.; Shin, S.C.; Park, H.; Lee, H. Transcriptome sequencing of the Antarctic vascular plant Deschampsia antarctica Desv. under abiotic stress. Planta 2013, 237, 823–836. [Google Scholar] [CrossRef]
- Long, Q.; Rabanal, F.A.; Meng, D.; Huber, C.D.; Farlow, A.; Platzer, A.; Zhang, Q.; Vilhjalmsson, B.J.; Korte, A.; Nizhynska, V.; et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 2013, 45, 884–890. [Google Scholar] [CrossRef]
- Byun, M.Y.; Lee, J.; Cui, L.H.; Kang, Y.; Oh, T.K.; Park, H.; Lee, H.; Kim, W.T. Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 2015, 236, 61–74. [Google Scholar] [CrossRef]
- Waminal, N.E.; Perumal, S.; Liu, S.; Chalhoub, B.; Kim, H.H.; Yang, T.J.; Liu, S.; Snowdon, R.; Chalhoub, B. Quantity, distribution, and evolution of major repeats in Brassica napus. In The Brassica Napus Genome; Liu, S., Snowdon, R., Chalhoub, B., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 111–129. [Google Scholar]
- Roser, M.; Winterfeld, G.; Doring, E.; Schneider, J. Chromosome evolution in grass tribes Aveneae/Poeae (Poaceae): Insights from karyotype structure and molecular phylogeny. Schlechtendalia 2014, 28, 1–21. [Google Scholar]
- Amosova, A.V.; Bolsheva, N.L.; Zoshchuk, S.A.; Twardovska, M.O.; Yurkevich, O.Y.; Andreev, I.O.; Samatadze, T.E.; Badaeva, E.D.; Kunakh, V.A.; Muravenko, O.V. Comparative molecular cytogenetic characterization of seven Deschampsia (Poaceae) species. PLoS ONE 2017, 12, e0175760. [Google Scholar] [CrossRef]
- Soreng, R.J.; Davis, J.I. Phylogenetic structure in Poaceae subfamily Pooideae as inferred from molecular and morphological characters: Misclassification versus reticulation. In Grasses: Systematics and Evolution; Jacobs, S.W.L., Everett, J., Eds.; CSIRO Publishing: Collingwood, UK, 2000; pp. 61–74. [Google Scholar]
- Saarela, J.M.; Bull, R.D.; Paradis, M.J.; Ebata, S.N.; Peterson, P.M.; Soreng, R.J.; Paszko, B. Molecular phylogenetics of cool-season grasses in the subtribes Agrostidinae, Anthoxanthinae, Aveninae, Brizinae, Calothecinae, Koeleriinae and Phalaridinae (Poaceae, Pooideae, Poeae, Poeae chloroplast group 1). PhytoKeys 2017, 87, 1–139. [Google Scholar] [CrossRef]
- Amosova, A.V.; Bolsheva, N.L.; Samatadze, T.E.; Twardovska, M.O.; Zoshchuk, S.A.; Andreev, I.O.; Badaeva, E.D.; Kunakh, V.A.; Muravenko, O.V. Molecular cytogenetic analysis of Deschampsia antarctica Desv. (Poaceae), Maritime Antarctic. PLoS ONE 2015, 10, e0138878. [Google Scholar] [CrossRef]
- Androsiuk, P.; Chwedorzewska, K.J.; Dulska, J.; Milarska, S.; Giełwanowska, I. Retrotransposon based genetic diversity of Deschampsia antarctica Desv. from King George Island (Maritime Antarctic). Ecol. Evol. 2021, 11, 648–663. [Google Scholar] [CrossRef]
- Flavell, R.B.; O’Dell, M.; Hutchinson, J. Nucleotide sequence organization in plant chromosomes and evidence for sequence translocation during evolution. Cold Spring Harb. Symp. Quant. Biol. 1981, 45, 501–508. [Google Scholar] [CrossRef]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Tomas, D.; Rodrigues, J.; Varela, A.; Veloso, M.M.; Viegas, W.; Silva, M. Use of repetitive sequences for molecular and cytogenetic characterization of Avena species from Portugal. Int. J. Mol. Sci. 2016, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- González, M.L.; Chiapella, J.O.; Urdampilleta, J.D. Characterization of some satellite DNA families in Deschampsia antarctica (Poaceae). Polar Biol. 2018, 41, 457–468. [Google Scholar] [CrossRef]
- González, M.L.; Chiapella, J.O.; Urdampilleta, J.D. Genomic differentiation of Deschampsia antarctica and D. cespitosa (Poaceae) based on satellite DNA. Bot. J. Linn. Soc. 2020, 194, 326–341. [Google Scholar] [CrossRef]
- Grebenstein, B.; Grebenstein, O.; Sauer, W.; Hemleben, V. Characterization of a highly repeated DNA component of perennial oats (Helictotrichon, Poaceae) with sequence similarity to a A-genome-specific satellite DNA of rice (Oryza). Theor. Appl. Genet. 1995, 90, 1101–1105. [Google Scholar] [CrossRef]
- Grebenstein, B.; Grebenstein, O.; Sauer, W.; Hemleben, V. Distribution and complex organization of satellite DNA sequences in Aveneae species. Genome 1996, 39, 1045–1050. [Google Scholar] [CrossRef]
- Winterfeld, G.; Röser, M. Chromosomal localization and evolution of satellite DNAs and heterochromatin in grasses (Poaceae), especially tribe Aveneae. Plant Syst. Evol. 2007, 264, 75–100. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar]
- Flavell, R.B. Repetitive DNA and chromosome evolution in plants. Philos. Trans. R. Soc. Lond. B 1986, 312, 227–242. [Google Scholar]
- Carrido-Ramos, M. Satellite DNA in plants: More than just rubbish. Cytogenet Genome Res 2015, 146, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Ta, T.D.; Waminal, N.E.; Nguyen, T.H.; Pellerin, R.J.; Kim, H.H. Comparative FISH analysis of Senna tora tandem repeats revealed insights into the chromosome dynamics in Senna. Genes Genom. 2021, 43, 237–249. [Google Scholar] [CrossRef]
- Volkov, R.A.; Medina, F.J.; Zentgraf, U.; Hemleben, V. Molecular cell biology: Organization and molecular evolution of rDNA, nucleolar dominance, and nucleolus structure. Prog. Bot. 2004, 65, 106–146. [Google Scholar]
- Volkov, R.A.; Panchuk, I.I.; Borisjuk, N.V.; Hosiawa-Baranska, M.; Maluszynska, J.; Hemleben, V. Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biol. 2017, 17, 21. [Google Scholar] [CrossRef]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the Plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Ugarković, Đ. Satellite DNA libraries and centromere evolution. Open Evol. J. 2008, 2, 1–6. [Google Scholar] [CrossRef][Green Version]
- Kawano, S. Cytogeography and evolution of the Deschampsia caespitosa complex. Can. J. Bot. 1963, 41, 719–742. [Google Scholar] [CrossRef]
- Garsia-Suarez, R.; Alonso-Blanco, C.; Fernandez-Carvajal, M.C.; Fernandez-Prieto, J.A.; Roca, F.; Giraldez, R. Diversity and systematics of Deschampsia sensu lato (Poaceae), inferred from karyotypes, protein electrophoresis, total genomic DNA hybridization and chloroplast DNA analysis. Plant Syst. Evol. 1997, 205, 99–110. [Google Scholar] [CrossRef]
- Torres, G.A.; Gong, Z.; Iovene, M.; Hirsch, C.D.; Buell, C.R.; Bryan, G.J.; Novák, P.; Macas, J.; Jiang, J. Organization and evolution of subtelomeric satellite repeats in the potato genome. G3 Genes Genomes Genet. 2011, 1, 85–92. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Chen, N.W.G.; Thareau, V.; Pflieger, E.; Blanchet, S.; Bryan, G.J.; Novák, P.; Macas, J.; Jiang, J. The subtelomeric khipu satellite repeat from Phaseolus vulgaris: Lessons learned from the genome analysis of the Andean genotype G19833. Front Plant. Sci. 2013, 4, 109. [Google Scholar] [CrossRef]
- Vershinin, A.V.; Schwarzacher, T.; Heslop-Harrison, J.S. The largescale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 1995, 7, 1823–1833. [Google Scholar] [PubMed]
- Sharma, S.; Raina, S.N. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet Genome Res. 2005, 109, 15–26. [Google Scholar] [CrossRef]
- Kubis, S.; Schmidt, T.; Heslop-Harrison, J.S. Repetitive DNA elements as a major component of plant genomes. Ann. Bot. 1998, 82, 45–55. [Google Scholar] [CrossRef]
- Macas, J.; Mészáros, T.; Nouzová, M. PlantSat: A specialized database for plant satellite repeats. Bioinformatics 2002, 18, 28–35. [Google Scholar] [CrossRef] [PubMed]
- González, M.L.; Urdampilleta, J.D.; Fasanella, M.; Premoli, A.C.; Chiapella, J.O. Distribution of rDNA and polyploidy in Deschampsia antarctica E. Desv. in Antarctic and Patagonic populations. Polar Biol. 2016, 39, 1663–1677. [Google Scholar] [CrossRef]
- Trifonov, E.N. The multiple codes of nucleotide sequences. Bull. Math. Biol. 1989, 51, 417–432. [Google Scholar] [CrossRef]
- Sýkorová, E.; Fajkus, J.; Ito, M.; Fukui, K. Transition between two forms of heterochromatin at plant subtelomeres. Chromosome Res. 2001, 9, 309–323. [Google Scholar] [CrossRef]
- Chiapella, J. A molecular phylogenetic study of Deschampsia (Poaceae: Aveneae) inferred from nuclear ITS and plastid trnL sequence data: Support for the recognition of Avenella and Vahlodea. Taxon 2007, 56, 55–64. [Google Scholar]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, W.L.; Dyer, T.A. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980, 8, 4851–4855. [Google Scholar] [CrossRef] [PubMed]
- Muravenko, O.V.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Nosova, I.V.; Zelenina, D.A.; Volkov, A.A.; Popov, K.V.; Zelenin, A.V. Comparison of genomes of eight species of sections Linum and Adenolinum from the genus Linum based on chromosome banding, molecular markers and RAPD analysis. Genetica 2009, 135, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Zoshchuk, S.A.; Rodionov, A.V.; Ghukasyan, L.; Samatadze, T.E.; Punina, E.O.; Loskutov, I.G.; Yurkevich, O.Y.; Muravenko, O.V. Molecular cytogenetics of valuable Arctic and sub-Arctic pasture grass species from the Aveneae/Poeae tribe complex (Poaceae). BMC Genet. 2019, 20, 92. [Google Scholar] [CrossRef] [PubMed]


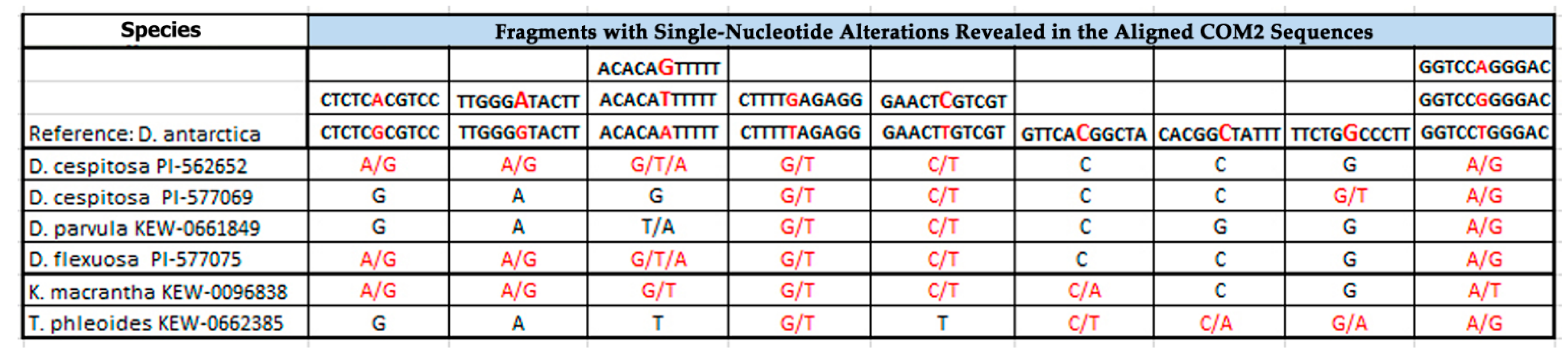
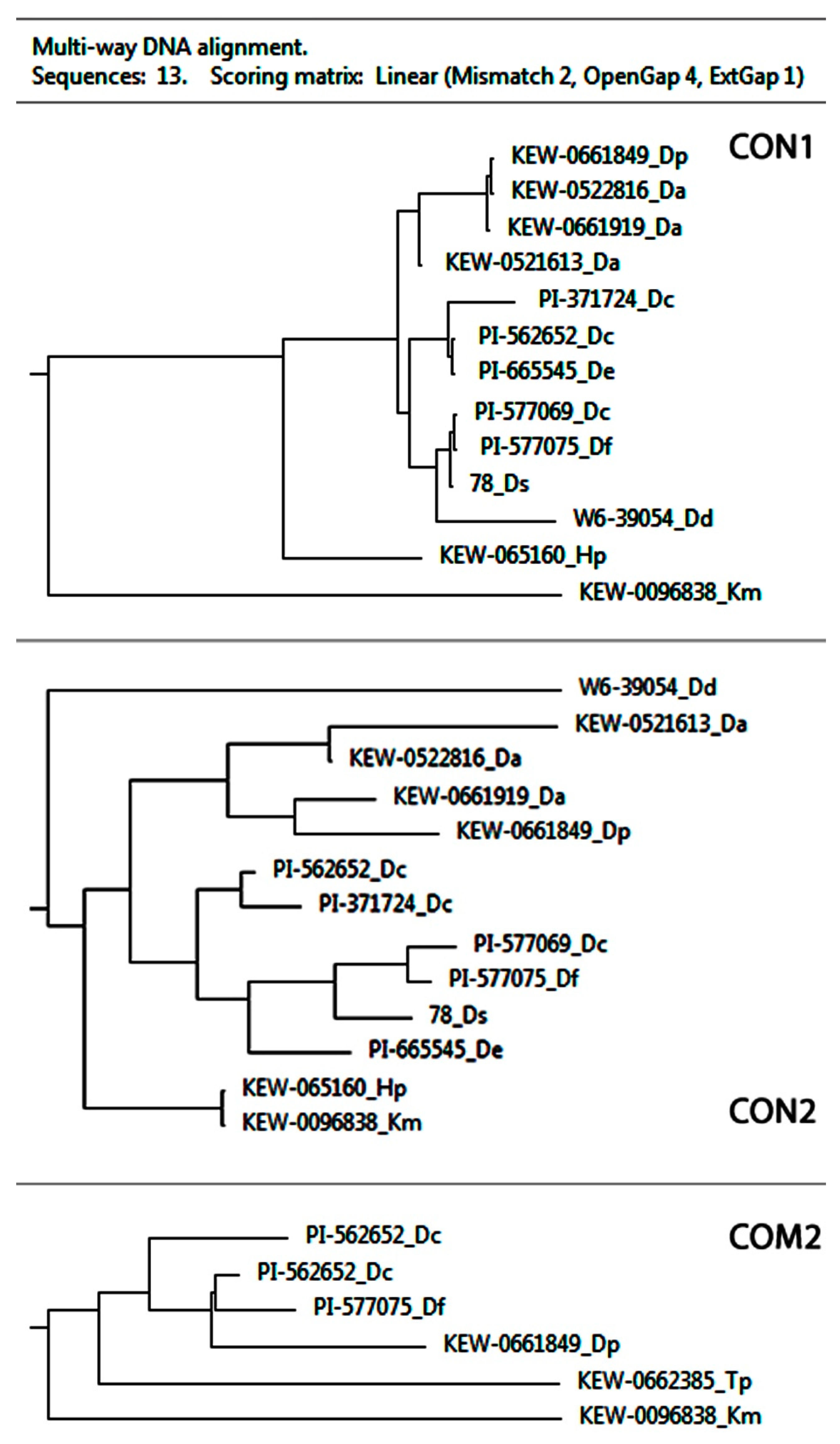

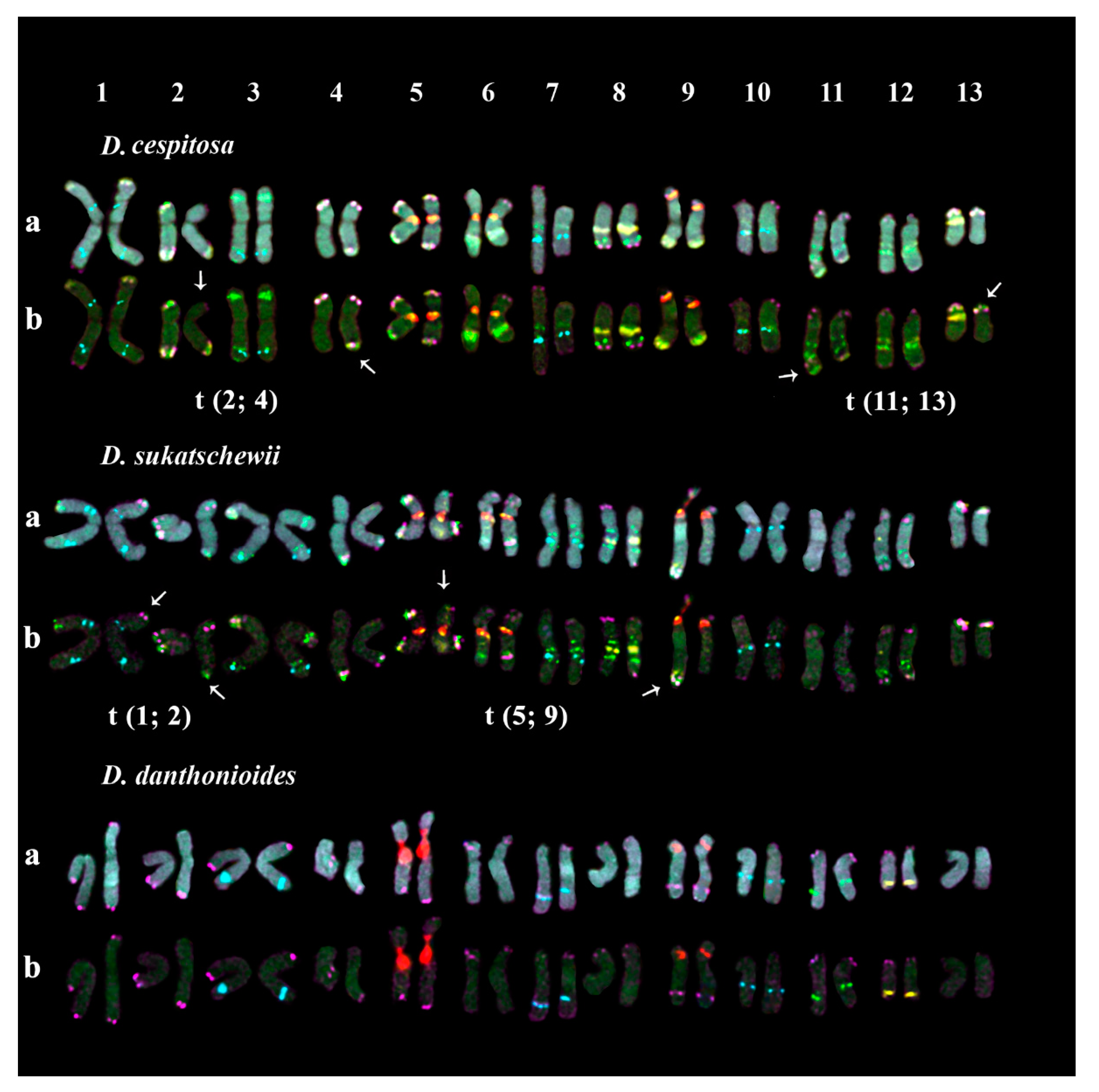
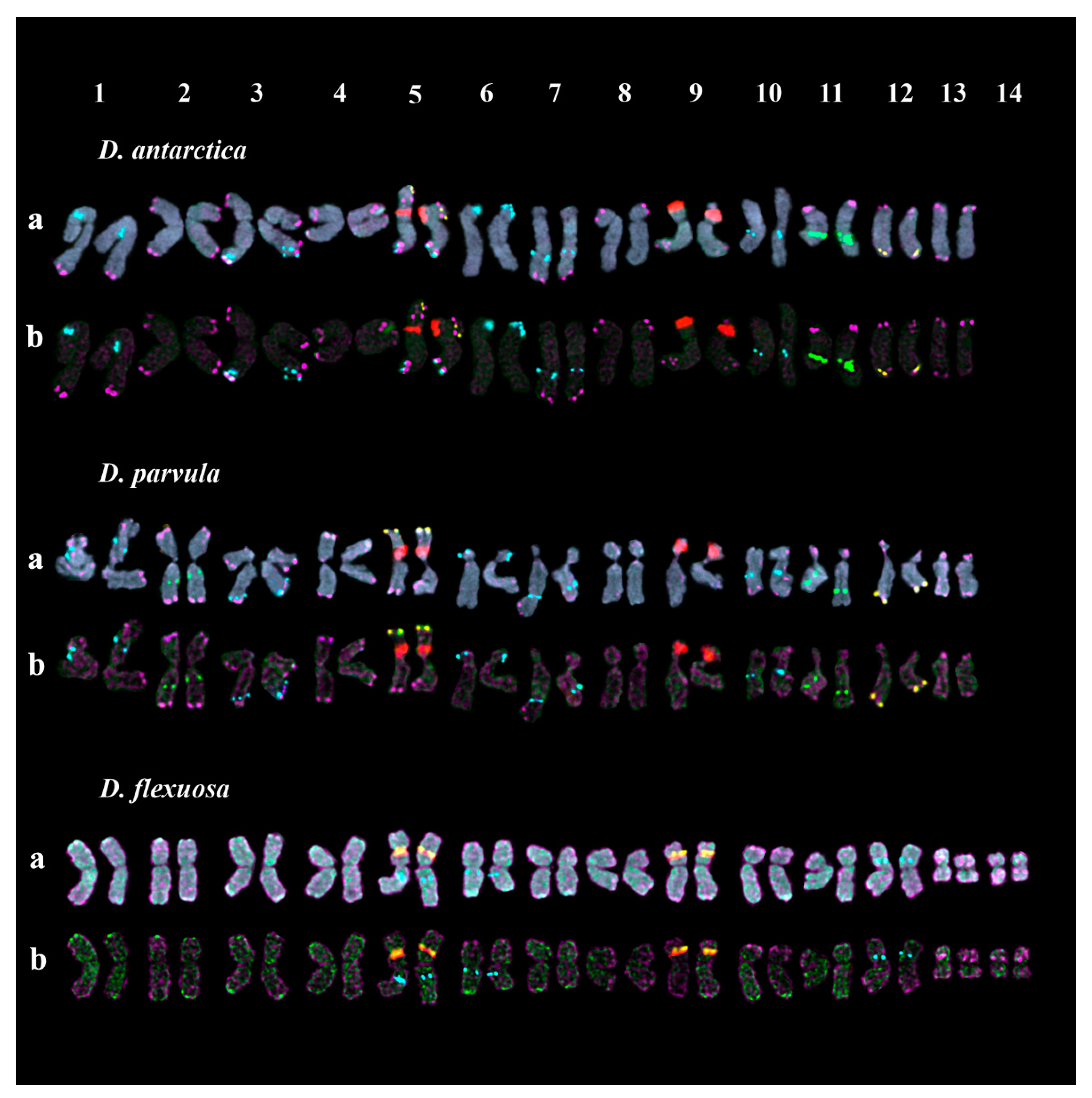
| Species | Catalog Number/Origin | Seed Source |
|---|---|---|
| Deschampsiaantarctica E. Desv. | KEW-0522816/Falkland Is., St. Georgia | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
| Deschampsiaantarctica E. Desv. | KEW-0661919/Falkland Is., Weddle | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
| Deschampsiaantarctica E. Desv. | KEW-0521613/Falkland Is., St. Georgia | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
| Deschampsia cespitosa ssp. beringensis (Hultén) W.E. Lawr. | PI-562652/Alaska, USA | Western Regional Plant Introduction Station, USDA ARS NPGS, Pullman, WA, USA |
| Deschampsia cespitosa (L.) P. Beauv. | PI-371724/Alaska, Valdez. USA | Western Regional Plant Introduction Station, USDA ARS NPGS, Pullman, WA, USA |
| Deschampsia cespitosa (L.) P. Beauv. | PI-577069/Great Britain | Western Regional Plant Introduction Station, USDA ARS NPGS, Pullman, WA, USA |
| Deschampsia danthonioides (Trin.) Munro | W6-39054/Washington, USA | Western Regional Plant Introduction Station, USDA ARS NPGS, Pullman, WA, USA |
| Deschampsia elongata (Hook.) | Munro PI-665545/Oregon, USA | Western Regional Plant Introduction Station, USDA ARS NPGS, Pullman, WA, USA |
| Deschampsia. flexuosa (L.) Trin. (= Avenella flexuosa (L.) Drejer) | PI-577075/Wales, UK | Western Regional Plant Introduction Station, USDA ARS NPGS, Pullman, WA, USA |
| Deschampsia parvula (Hook.f.) E. Desv. | KEW-0661849/Falkland Is. | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
| Deschampsia sukatschewii (Popl.) Roshev | 78/Altai Mountains, RF | Laboratory of genetic resources of fodder plants, Federal Williams Research Center of Forage Production and Agroecology (FWRC FPA) |
| Helictotrichon pubescens (Huds.) Pilg. (syn. Aenula pubescens (Hud.) Dumort.) | KEW-065160/England, UK | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
| Kolleria macrantha (Ledeb.) Schult. (syn. Kolleria cristata (L.) Pers.) | KEW-0096838/Greece | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
| Tricetum phleoides (d’Urv.) Kunth. | KEW-0662385/Falkland Is. | Seed Conservation Department, Royal Botanic Gardens, Kew, UK |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amosova, A.V.; Ghukasyan, L.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and FISH Mapping of CON/COM Satellite DNA Families. Plants 2021, 10, 1105. https://doi.org/10.3390/plants10061105
Amosova AV, Ghukasyan L, Yurkevich OY, Bolsheva NL, Samatadze TE, Zoshchuk SA, Muravenko OV. Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and FISH Mapping of CON/COM Satellite DNA Families. Plants. 2021; 10(6):1105. https://doi.org/10.3390/plants10061105
Chicago/Turabian StyleAmosova, Alexandra V., Lilit Ghukasyan, Olga Yu. Yurkevich, Nadezhda L. Bolsheva, Tatiana E. Samatadze, Svyatoslav A. Zoshchuk, and Olga V. Muravenko. 2021. "Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and FISH Mapping of CON/COM Satellite DNA Families" Plants 10, no. 6: 1105. https://doi.org/10.3390/plants10061105
APA StyleAmosova, A. V., Ghukasyan, L., Yurkevich, O. Y., Bolsheva, N. L., Samatadze, T. E., Zoshchuk, S. A., & Muravenko, O. V. (2021). Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and FISH Mapping of CON/COM Satellite DNA Families. Plants, 10(6), 1105. https://doi.org/10.3390/plants10061105






