Intragenomic Conflict between Knob Heterochromatin and B Chromosomes Is the Key to Understand Genome Size Variation along Altitudinal Clines in Maize
Abstract
1. Introduction
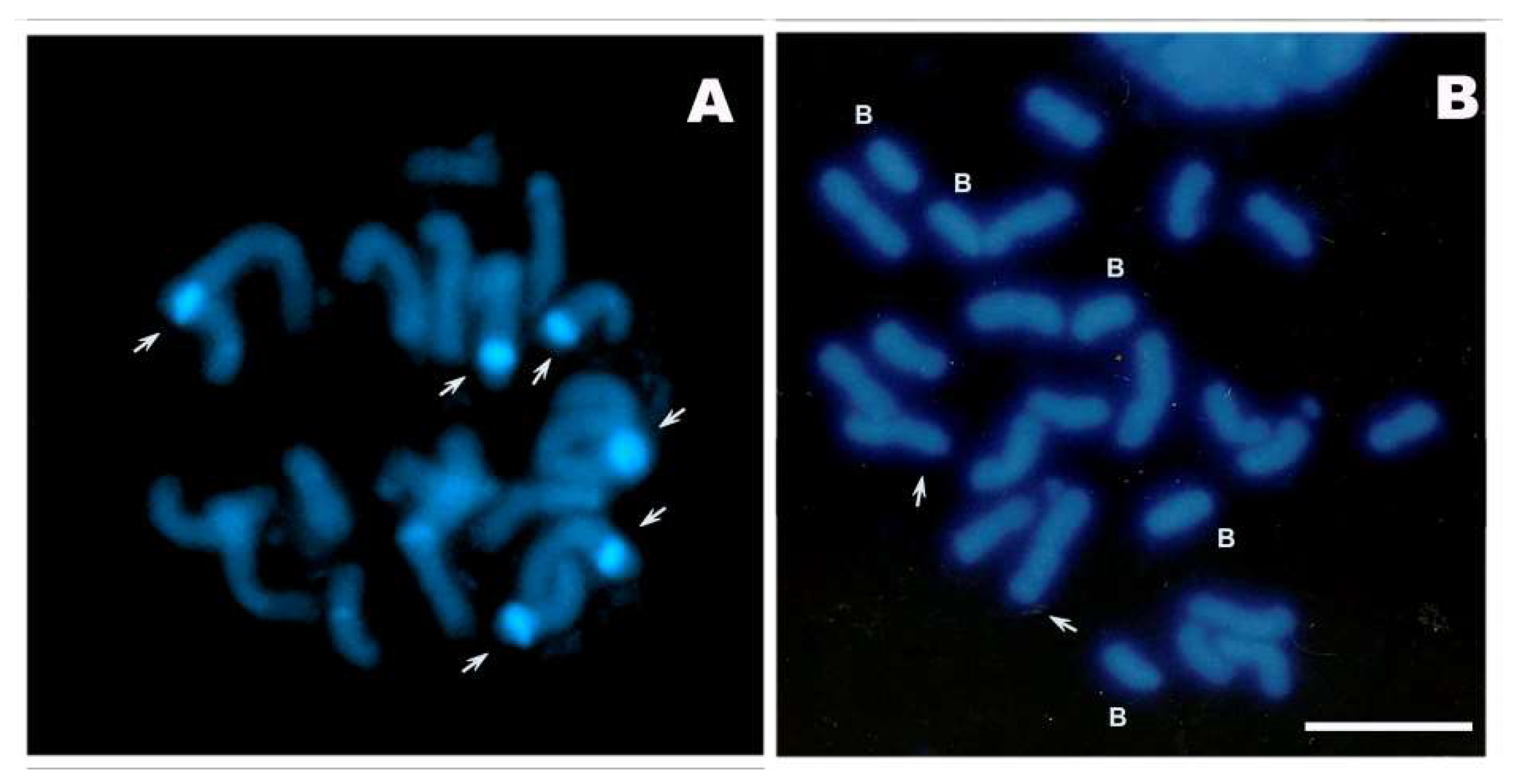
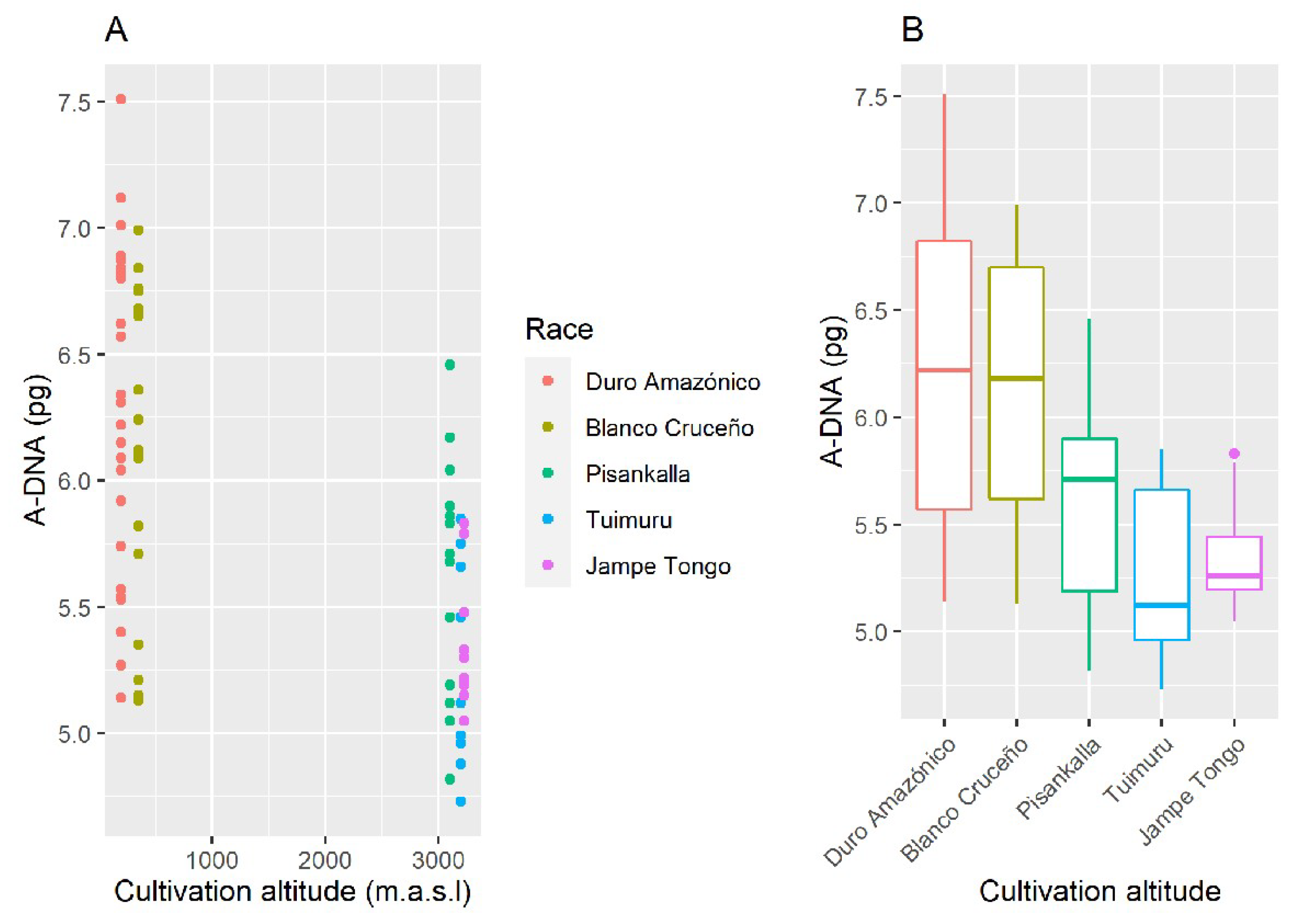
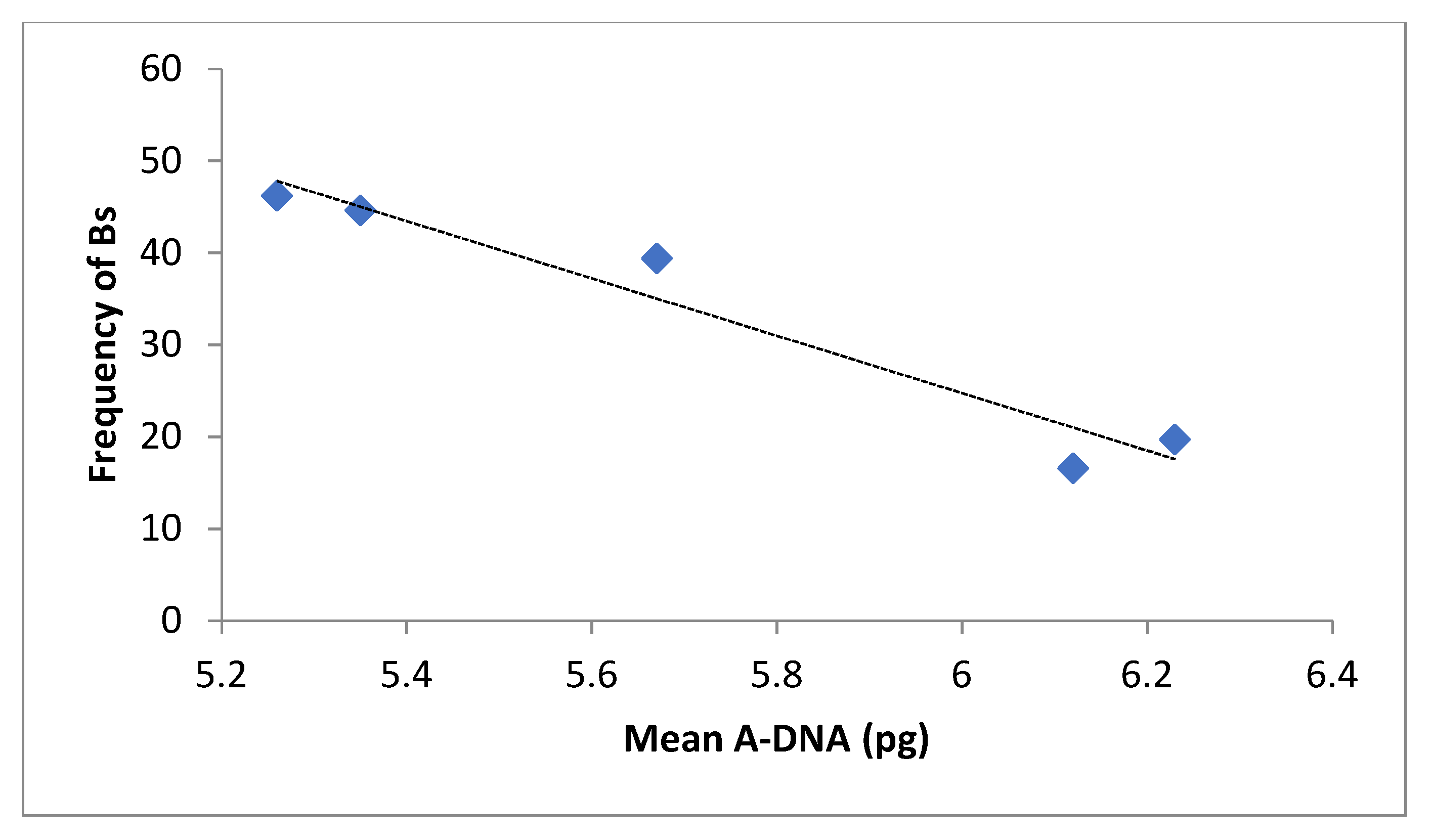
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Cytological Studies
4.3. Feulgen Staining and Cytophotometry
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kato, T.A.; Mapes, S.; Mera, O.; Serratos, H.; Bye, B. Origen y diversificación del maíz: Una revisión analítica. In Comisión Nacional Para el Conocimiento y Uso de la Biodiversidad; Universidad Nacional Autónoma de México: Ciudad de México, Mexico, 2009; p. 116. [Google Scholar]
- McClintock, B.; Kato, T.A.; Blumenschein, A. Constitución Cromosómica de las Razas de Maíz. Su Significado en la Interpretación de Relaciones Entre las Razas y Variedades en las Américas; Colegio de Postgraduados: Chapingo, Mexico, 1981; p. 517. [Google Scholar]
- Cámara Hernández, J.; Miante Alzogaray, A.M.; Bellón, R.; Galmarini, A.J. Razas de Maíz Nativas de la Argentina; Pascale, A.J., Ed.; Facultad de Agronomía, Universidad de Buenos Aires Press: Buenos Aires, Argentina, 2011; p. 192. [Google Scholar]
- Rodríguez, A.; Romero, M.; Quiroga, J.; Avila, G.; Brandolini, A. Maíces Bolivianos; Organización de las Naciones Unidas Para la Agricultura y la Alimentación, FAO: Roma, Italy, 1968; p. 243. [Google Scholar]
- Avila, G.; Brandolini, A.G. I mais boliviani. In Documenti per la Cooperazione Allo Sviluppo; Rivista di Agricoltura Subtropicale e Tropicale: Firenze, Italy, 1992; p. 105. [Google Scholar]
- Rayburn, A.L.; Auger, J.A. Genome size variation in Zea mays ssp. mays adapted to different altitudes. Theor. Appl. Genet. 1990, 79, 470–474. [Google Scholar] [CrossRef]
- Tito, C.; Poggio, L.; Naranjo, C.A. Cytogenetic studies in the genus Zea: DNA content and heterochromatin in species and hybrids. Theor. Appl. Genet. 1991, 83, 58–64. [Google Scholar] [CrossRef]
- Poggio, L.; Rosato, M.; Chiavarino, A.M.; Naranjo, C.A. Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Ann. Bot. 1998, 82, 107–115. [Google Scholar] [CrossRef]
- Rosato, M.; Chiavarino, A.M.; Naranjo, C.A.; Hernández, J.C.; Poggio, L. Genome size and numerical polymorphism for B-chromosome races of maize (Zea mays ssp. mays, Poaceae). Am. J. Bot. 1998, 85, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Realini, M.F.; Poggio, L.; Cámara-Hernández, J.; González, G.E. Intra-specific variation in genome size in maize: Cytological and phenotypic correlates. AoB Plants 2016, 8, plv138. [Google Scholar] [CrossRef] [PubMed]
- Fourastié, M.F.; Gottlieb, A.M.; Poggio, L.; González, G.E. Are cytological parameters of maize landraces (Zea mays ssp. mays) adapted along an altitudinal cline? J. Plant. Res. 2017, 131, 285–296. [Google Scholar] [CrossRef] [PubMed]
- SanMiguel, P.; Bennetzen, J.L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 1998, 82, 37–44. [Google Scholar] [CrossRef]
- Meyers, B.C.; Tingey, S.V.; Morgante, M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001, 11, 1660–1676. [Google Scholar] [CrossRef]
- Fourastié, M.F. Estudios Citogenéticos en Razas de Maíz del NOA: Caracterización Cariotípica, Evaluación del Tamaño del Genoma y Frecuencia de Cromosomas B. Ph.D. Thesis, Universidad de Buenos Aires, Buenos Aires, Argentina, 2015. [Google Scholar]
- Bilinski, P.; Albert, P.S.; Berg, J.J.; Birchler, J.A.; Grote, M.; Lorant, A.; Quezada, J.; Swarts, K.; Yang, J.; Ross-Ibarra, J. Parallel altitudinal clines reveal adaptive evolution of genome size in Zea mays. PLoS Genet. 2018, 14, e1007162. [Google Scholar] [CrossRef]
- Greilhuber, J.; Leitch, I.J. Genome Size and the Phenotype. Plant. Genome Diversity; Springer: Viena, Austria, 2013; Volume 2, pp. 323–340. [Google Scholar]
- Comertpay, G. Assessment of nuclear DNA contents variation and their relationship with flowering in corn genotypes. Turk. J. Field Crop. 2019, 24, 39–45. [Google Scholar] [CrossRef]
- Laurie, D.A.; Bennett, M.D. Nuclear DNA content in the genera Zea and Sorghum. Intergeneric, Interespecific and Intraspecific Variation. Heredity 1985, 55, 307–313. [Google Scholar] [CrossRef]
- Bennett, M.D.; Leitch, I.J. Nuclear DNA amounts in angiosperms: Progress, problems and prospects. Ann. Bot. 2005, 95, 45–90. [Google Scholar] [CrossRef]
- Realini, M.F.; Poggio, L.; Cámara-Hernández, J.; González, G.E. Genome size and repetitive sequences are driven by artificial selection on the length of the vegetative cycle in maize landraces from Northeastern Argentina. Rodriguésia 2021, 72, e03542018. [Google Scholar] [CrossRef]
- Tenaillon, M.; Manicacci, D.; Nicolas, S.; Tardieu, F.; Welcker, C. Testing the link between genome size and growth rate in maize. Peer J. 2016, 4, e2408. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Xu, C.; Guo, Z.; Wang, S.; Xu, Y.; Zou, C. Maize (Zea mays L.) genome size indicated by 180-bp knob abundance is associated with flowering time. Sci. Rep. 2017, 7, 5954. [Google Scholar] [CrossRef]
- Díez, C.M.; Gaut, B.S.; Meca, E.; Scheinvar, E.; Montes-Hernández, S.; Eguiarte, L.E.; Tenallion, M.I. Genome size variation in wild and cultivated maize along altitudinal gradients. New Phytol. 2013, 199, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Chiavarino, A.M.; Rosato, M.; Rossi, P.; Poggio, L.; Naranjo, C.A. Localization of the genes controlling B chromosome transmission rate in maize (Zea mays ssp. mays, Poaceae). Am. J. Bot. 1998, 85, 1581–1585. [Google Scholar] [CrossRef] [PubMed]
- Puertas, M.J.; Jiménez, G.; Manzanero, S.; Chiavarino, A.M.; Rosato, M.; Naranjo, C.A.; Poggio, L. Genetic Control of B Chromosome Transmission in Maize and rye. Chromosomes Today; Birkhäuser Verlag: Basel, Switzerland, 2000; Volume 13, pp. 79–91. [Google Scholar]
- Jones, N.; Houben, A. B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant. Sci. 2003, 8, 217–423. [Google Scholar] [CrossRef]
- Houben, A.; Banaei-Moghaddam, A.M.; Klemme, S.; Timmis, J.N. Evolution and biology of supernumerary B chromosomes. Cell Mol. Life Sci. 2014, 71, 467–478. [Google Scholar] [CrossRef]
- Huang, W.; Du, Y.; Zhao, X.; Jin, W. B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L.). BMC Plant. Biol. 2016, 16, 88. [Google Scholar] [CrossRef]
- Huang, Y.H.; Peng, S.F.; Lin, Y.P.; Cheng, Y.M. The maize B chromosome is capable of expressing microRNAs and altering the expression of microRNAs derived from A chromosomes. Chromosom. Res. 2020, 28, 129–138. [Google Scholar] [CrossRef]
- Bennett, M.D. Nuclear DNA content and minimum generation time in herbaceous plants. Proc. R. Soc. Lond. 1972, 181, 109–135. [Google Scholar] [PubMed]
- Bennett, M.D. Variation in genomic form in plants and its ecological implications. New Phytol. 1987, 106, 177–200. [Google Scholar] [CrossRef]
- Realini, M.F.; Poggio, L.; Cámara Hernández, J.; González, G.E. Exploring karyotype diversity of Argentinian Guaraní maize landraces: Relationship among South American maize. PLoS ONE 2018, 13, e0198398. [Google Scholar] [CrossRef] [PubMed]
- González, G.E.; Poggio, L. Karyotype of Zea luxurians and Z. mays subsp. mays using FISH/DAPI, and analysis of meiotic behavior of hybrids. Genome 2011, 54, 26–32. [Google Scholar] [PubMed]
- González, G.E.; Fourastié, M.F.; Poggio, L. Relevancia del número y composición de secuencias de los nudos cromosómicos en la caracterización de maíz y teocintle. Rev. Fitotec. Mex. 2013, 36, 127–135. [Google Scholar] [CrossRef]
- Ayonoadu, U.W.; Rees, H. The effects of B-chromosomes on the nuclear phenotype in root meristems of maize. Heredity 1971, 27, 365–383. [Google Scholar] [CrossRef]
- Lia, V.V.; Confalonieri, V.A.; Poggio, L. B chromosome polymorphism in maize landraces: Adaptive vs demographic hypothesis of clinal variation. Genetics 2007, 107, 896–904. [Google Scholar] [CrossRef]
- Ahmad, S.; Martins, C. The modern view of B chromosomes under the impact of high scale omics analyses. Cells 2019, 8, 156. [Google Scholar] [CrossRef]
- Blavet, N.; Yang, H.; Su, H.; Solanský, P.; Douglas, R.N.; Karafiátová, M.; Šimková, L.; Zhang, J.; Liu, Y.; Hou, J.; et al. Sequence of the supernumerary B chromosome of maize provides insight into its drive mechanism and evolution. Proc. Natl. Acad. Sci. USA 2021, 118, e2104254118. [Google Scholar] [CrossRef]
- Hewitt, G.M. Orthoptera: Grasshoppers and Crickets. Animal Cytogenetics; Gebrüder Borntraeger: Berlin-Stuttgart, Germany, 1979; Volume 3, p. 170. [Google Scholar]
- Barbujani, G.; Pigliucci, M. Geographical patterns of karyotype polymorphism in Italian populations of Ornithogalum montanum (Liliaceae). Heredity 1989, 62, 67–75. [Google Scholar] [CrossRef]
- López-León, M.D.; Pardo, M.C.; Cabrero, J.; Camacho, J.P.M. Undertransmission of the supernumerary segment through heterozygous females possessing B chromosomes in the grasshopper Eyprepocnemis plorans. Genome 1994, 37, 705–709. [Google Scholar] [CrossRef]
- Clemente, M.; Remis, M.I.; Vilardi, J.C.; Alberti, A. Supernumerary heterochromatin, chiasma conditions and abnormal sperm formation in Dichroplus elongatus (Orthoptera): Intra and interpopulation analysis. Caryologia 1994, 46, 321–335. [Google Scholar]
- Remis, M.I.; Clemente, M.; Pensel, S.; Vilardi, J.C. Non-random distribution patterns of supernumerary segments and B chromosomes in Diclzuuplus elongatus (Orthoptera). Hereditas 1998, 129, 207–213. [Google Scholar] [CrossRef]
- Tardif, B.; Morisset, P. Relation between numbers of B-chromosomes and C-bands in Allium schoenoprasum L. Cytologia 1992, 57, 349–352. [Google Scholar] [CrossRef][Green Version]
- Rhoades, M.M.; Dempsey, E. Chromatin elimination induced by the B chromosome of maize. Heredity 1973, 64, 13–18. [Google Scholar] [CrossRef]
- Kanizay, L.B.; Albert, P.S.; Birchler, J.B.; Dawe, R.K. Intragenomic conflict between the two major knob repeats of maize. Genetics 2013, 194, 81–89. [Google Scholar] [CrossRef]
- Bennet, M.D.; Smith, J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. 1976, 274, 227–274. [Google Scholar] [CrossRef]
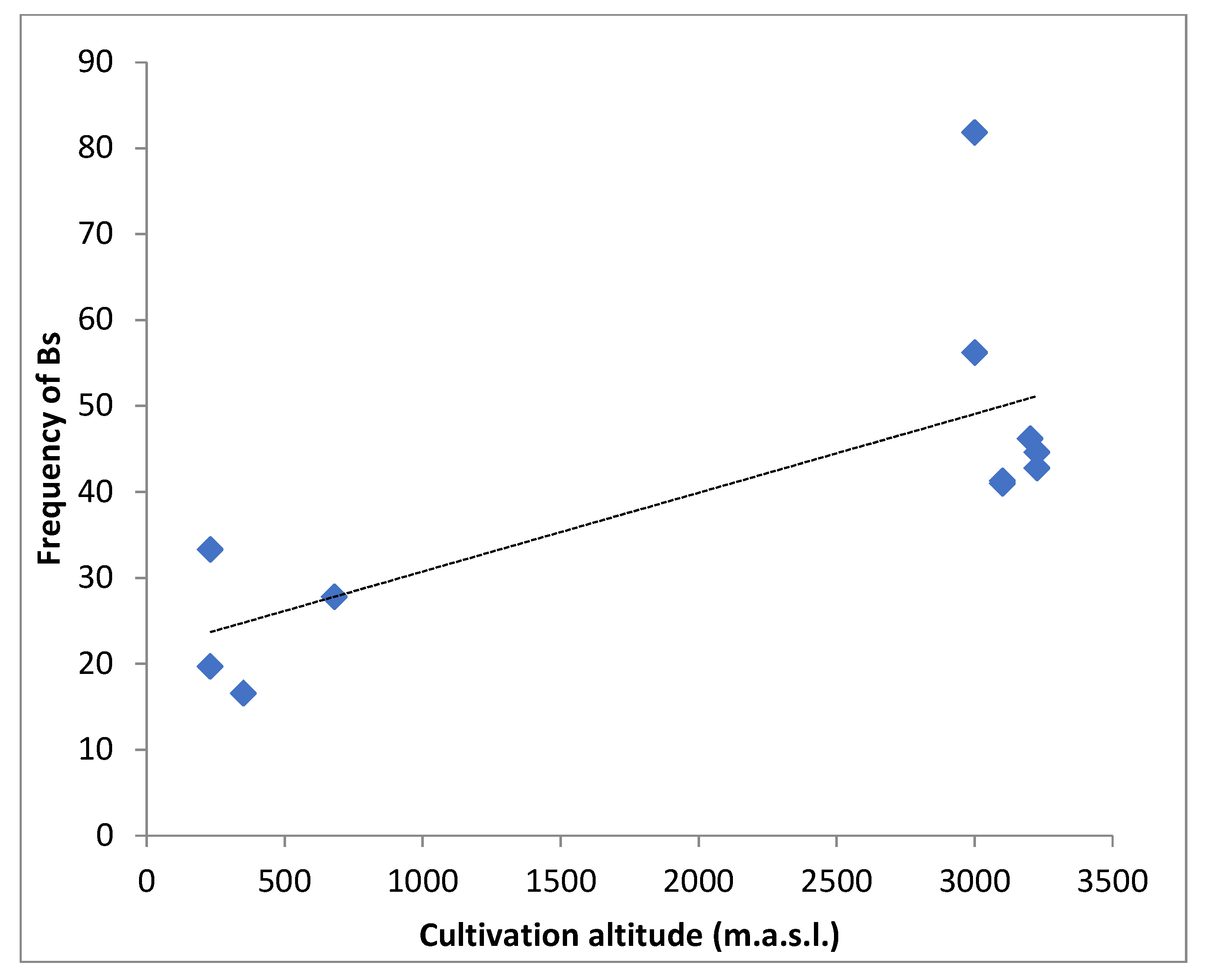

| Landrace | Population Voucher | Cultivation Altitude (m.a.s.l.) | 0B | 1B | 2B | 3B | 4B | 5B | Total indiv. | Mean of Bs | Frequency of Bs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Duro Amazónico | BOZM0724 | 200 | 23 | 7 | 6 | 2 | 0 | 0 | 38 | 0.65 | 39.40 |
| Duro Amazónico | BOZM0723 | 230 | 73 | 16 | 2 | 0 | 0 | 0 | 91 | 0.22 | 19.70 |
| Blando Amazónico | BOZM1050 | 230 | 20 | 8 | 1 | 0 | 1 | 0 | 30 | 0.46 | 33.30 |
| Blando Cruceño | BOZM0715 | 350 | 70 | 14 | 0 | 0 | 0 | 0 | 84 | 0.16 | 16.60 |
| Blando Cruceño | BOZM1421 | 680 | 13 | 4 | 0 | 1 | 0 | 0 | 18 | 0.39 | 27.80 |
| Pisankalla | BOZM0038 | 3000 | 4 | 9 | 7 | 2 | 0 | 0 | 22 | 1.32 | 81.80 |
| Tuimuru | BOZM0678 | 3000 | 7 | 7 | 2 | 0 | 0 | 0 | 16 | 0.69 | 56.20 |
| Pisankalla | BOZM0962 | 3100 | 23 | 10 | 4 | 1 | 0 | 1 | 39 | 0.66 | 41.02 |
| Pisankalla | BOZM0684 | 3100 | 37 | 20 | 5 | 1 | 0 | 0 | 63 | 0.52 | 41.30 |
| Tuimuru | BOZM0672 | 3200 | 29 | 15 | 6 | 4 | 0 | 0 | 54 | 0.72 | 46.29 |
| Jampe Tongo | BOZM0192 | 3225 | 31 | 14 | 10 | 1 | 0 | 0 | 56 | 0.66 | 44.60 |
| Jampe Tongo | BOZM0786 | 3250 | 12 | 3 | 5 | 1 | 0 | 0 | 21 | 0.76 | 42.85 |
| Total | 353 | 129 | 50 | 13 | 1 | 1 | 547 | 0.49 |
| Landrace | Cult. Altitude (m.a.s.l.) | 0B (±S/D) | 1B (±S/D) | 2B (±S/D) | 3B (±S/D) |
|---|---|---|---|---|---|
| Jampe Tongo-BOZM0192 | 3250 | 5.35 ± 0.25 | 5.88 ± 0.07 | 6.30 ± 0.07 | 6.45 ± 0.11 |
| Tuimuru-BOZM0672 | 3200 | 5.26 ± 0.39 | 5.08 ± 0.22 | 5.68 ± 0.69 | 6.43 ± 0.05 |
| Pisankalla-BOZM0684 | 3100 | 5.67 ± 0.49 | 5.58 ± 0.41 | 6.14 ± 0.30 | 6.63 ± 0.93 |
| Blando Cruceño-BOZM0715 | 350 | 6.12 ± 0.63 | 6.59 ± 0.79 | --- | --- |
| Duro Amazónico-BOZM0724 | 200 | 6.23 ± 0.64 | 6.57 ± 0.38 | --- | --- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, G.E.; Poggio, L. Intragenomic Conflict between Knob Heterochromatin and B Chromosomes Is the Key to Understand Genome Size Variation along Altitudinal Clines in Maize. Plants 2021, 10, 1859. https://doi.org/10.3390/plants10091859
González GE, Poggio L. Intragenomic Conflict between Knob Heterochromatin and B Chromosomes Is the Key to Understand Genome Size Variation along Altitudinal Clines in Maize. Plants. 2021; 10(9):1859. https://doi.org/10.3390/plants10091859
Chicago/Turabian StyleGonzález, Graciela Esther, and Lidia Poggio. 2021. "Intragenomic Conflict between Knob Heterochromatin and B Chromosomes Is the Key to Understand Genome Size Variation along Altitudinal Clines in Maize" Plants 10, no. 9: 1859. https://doi.org/10.3390/plants10091859
APA StyleGonzález, G. E., & Poggio, L. (2021). Intragenomic Conflict between Knob Heterochromatin and B Chromosomes Is the Key to Understand Genome Size Variation along Altitudinal Clines in Maize. Plants, 10(9), 1859. https://doi.org/10.3390/plants10091859





