Early Growth Stage Characterization and the Biochemical Responses for Salinity Stress in Tomato
Abstract
1. Introduction
2. Results
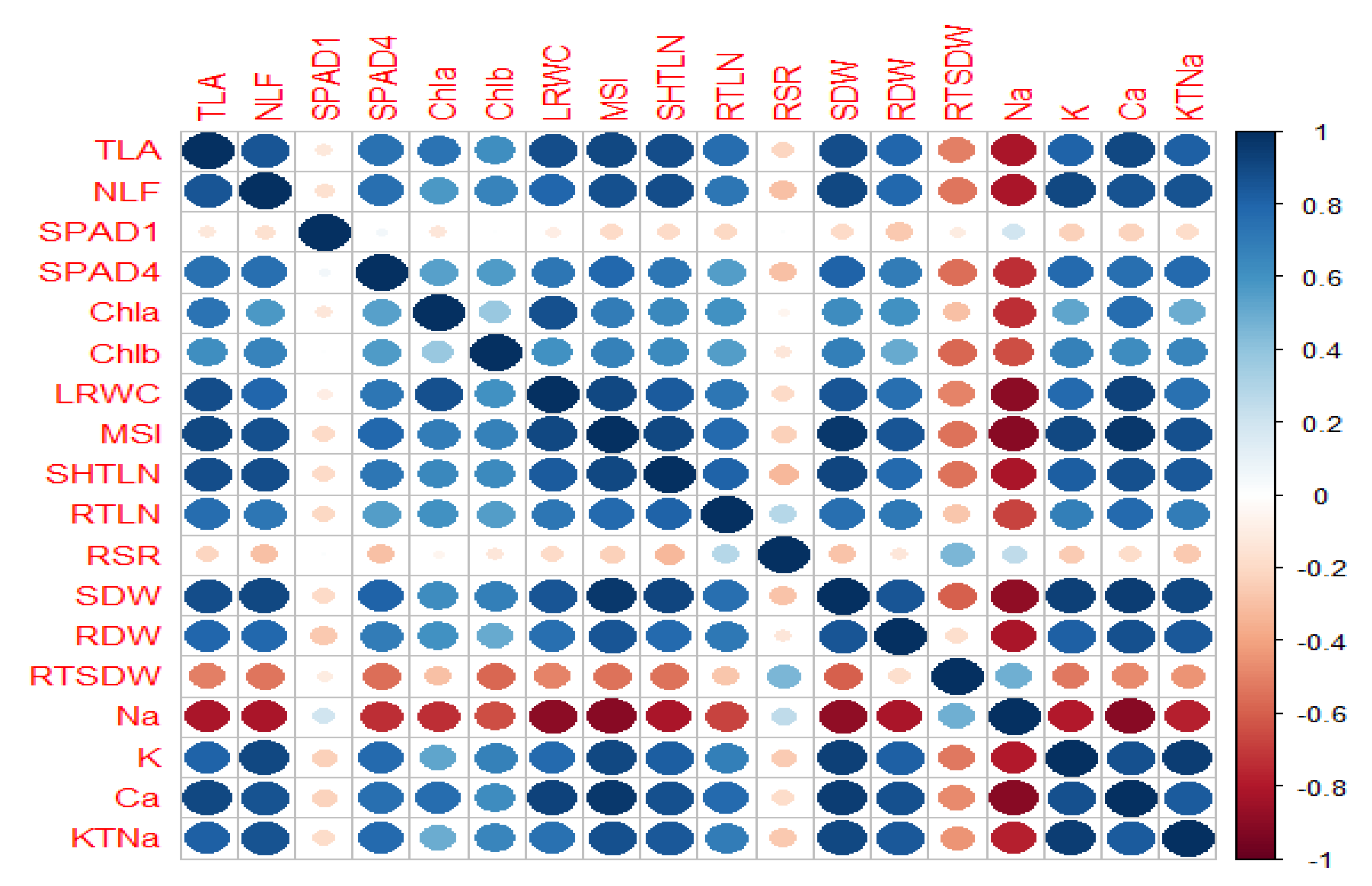
2.1. Variability and Correlation Analysis
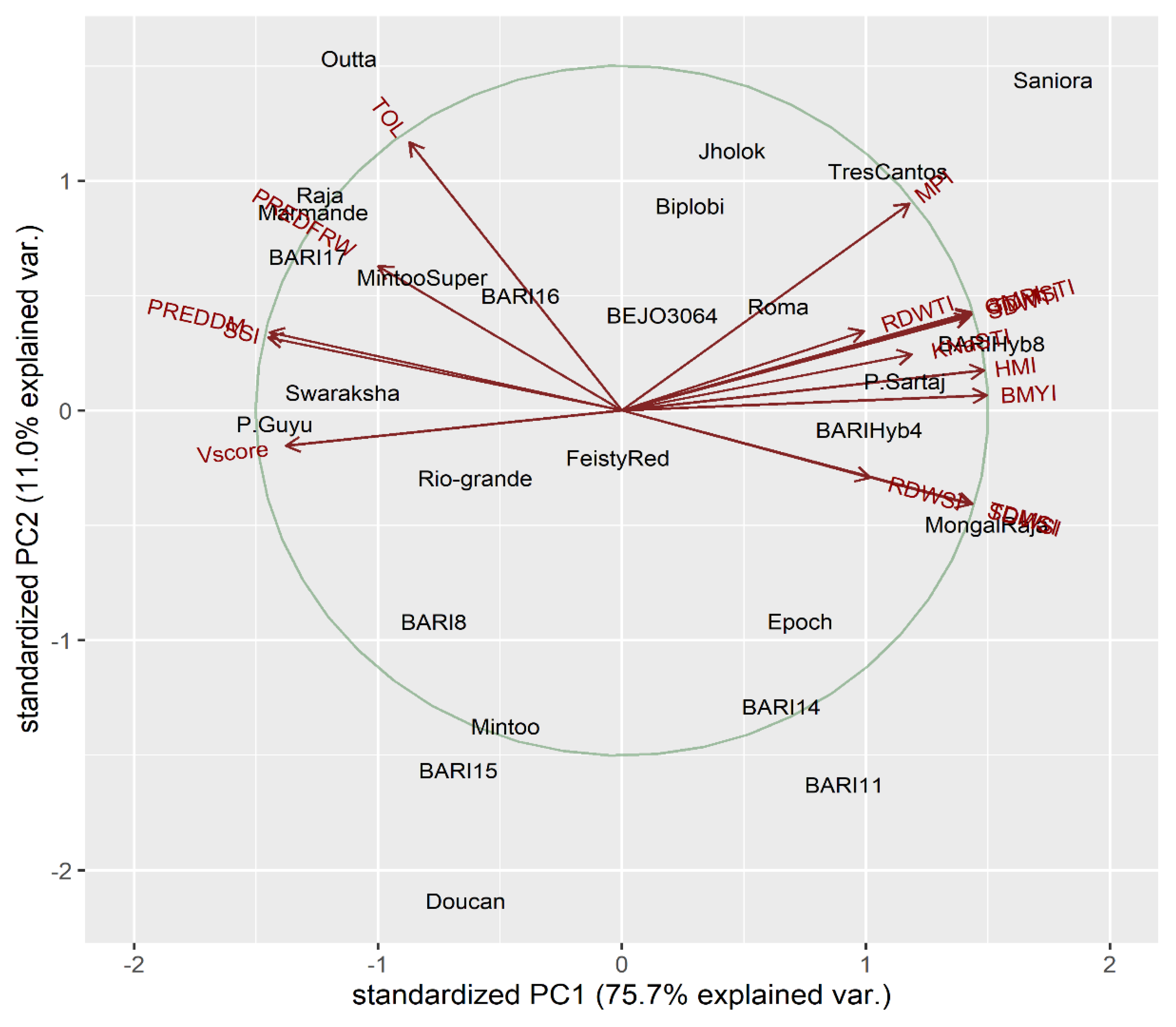
2.2. PCA for Physio-Morphological Traits
2.3. PCA for the Salt Tolerance Indices
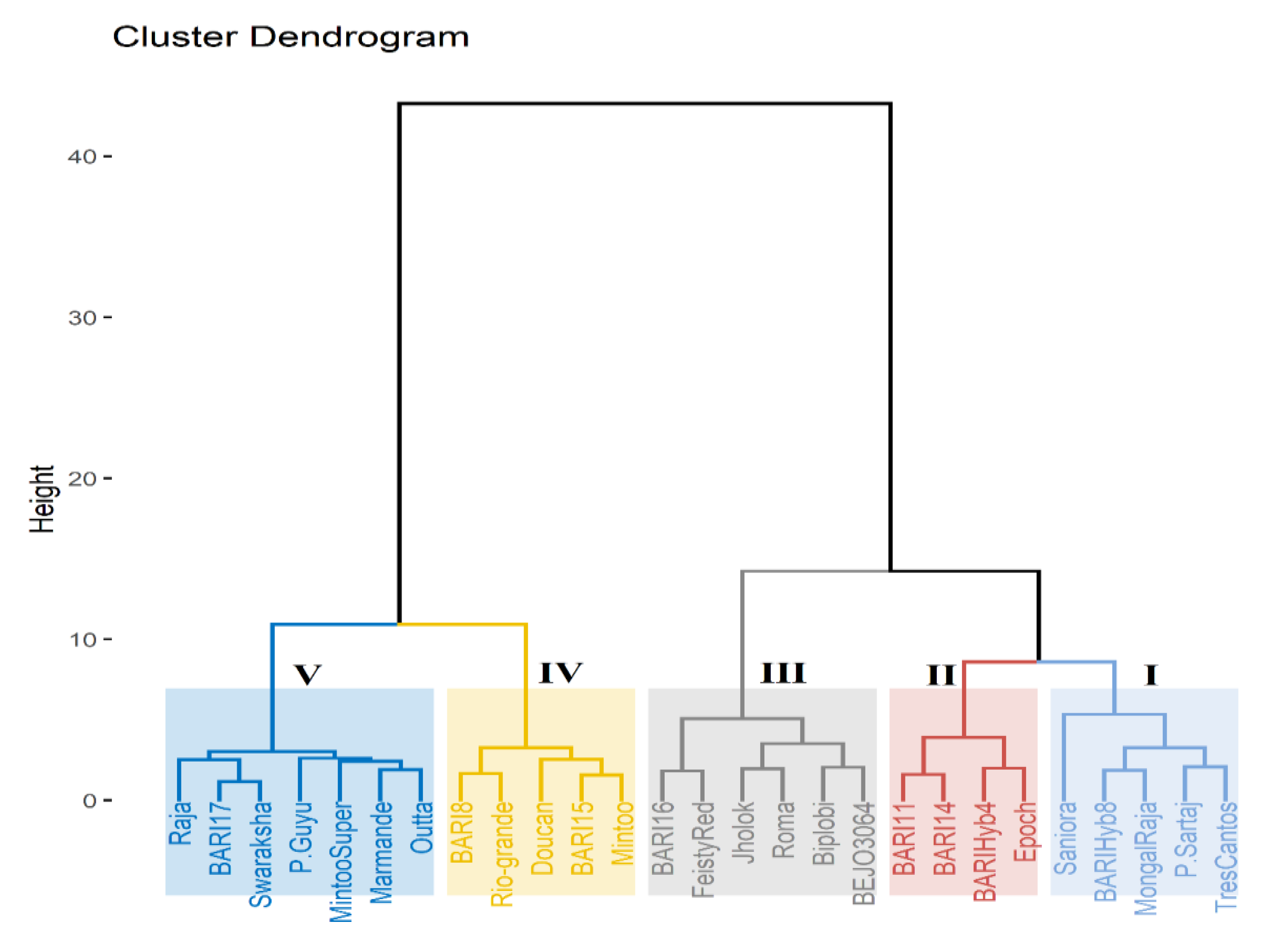
2.4. Hierarchical Cluster Analysis for Salt Tolerance Indices
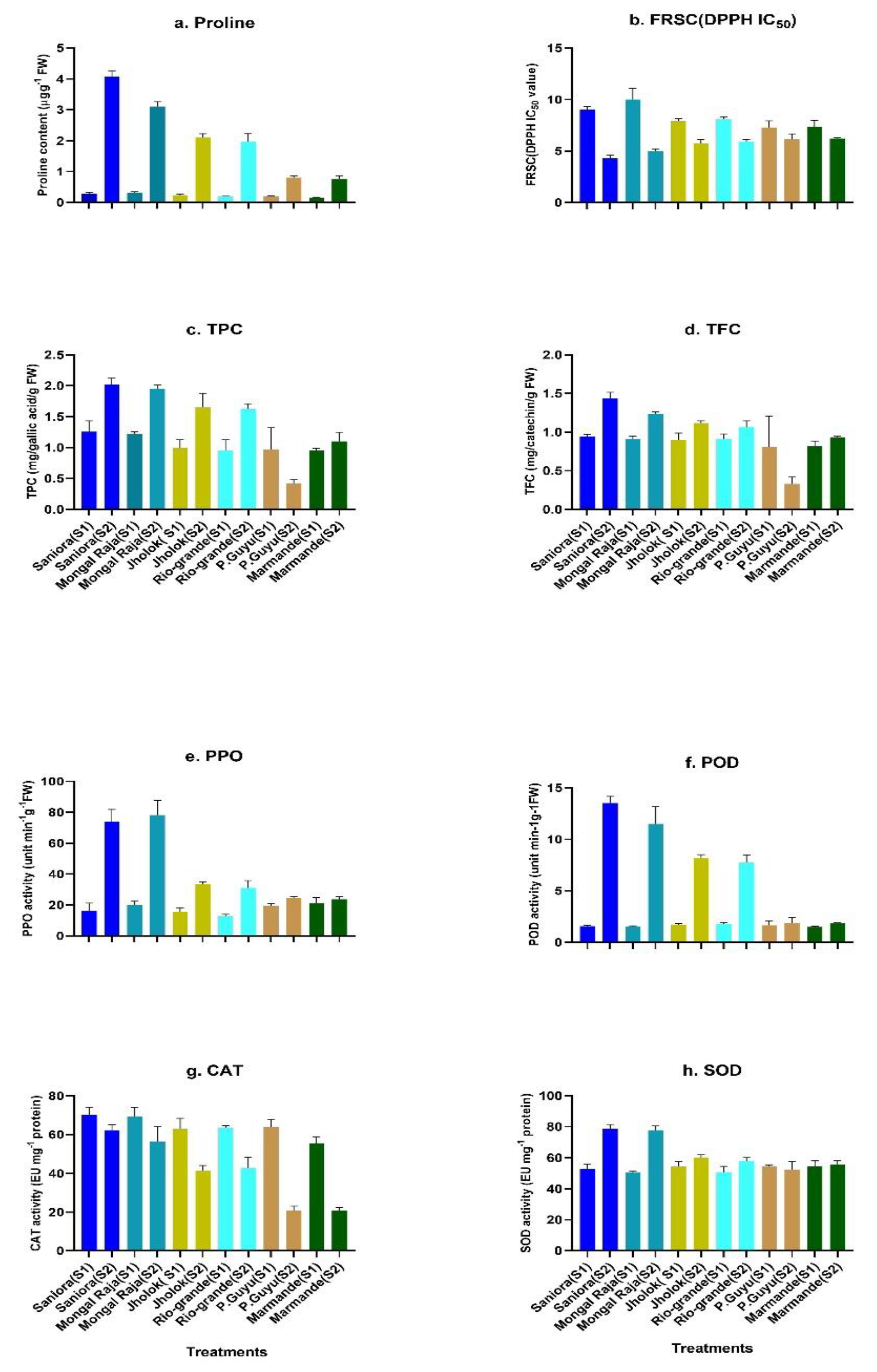
2.5. Salt Tolerance and Characterization of Biochemical Traits
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growing Conditions
4.2. Experimental Design, Salt Stress Application, and Nutrient Management
4.3. Measurements
4.3.1. Growth Parameters
4.3.2. Relative Leaf Water Content (RLWC)
4.3.3. Estimation of Percent Yield Reduction
4.3.4. Membrane Stability Index (MSI)
4.3.5. Measurement of Stress Tolerance Indices
4.3.6. Photosynthetic Pigment Measurement
4.3.7. Mineral Analysis
4.3.8. Determination of Leaf Proline Content
4.3.9. Measurement of Total Phenols and Flavonoids
4.3.10. Free Radical Scavenging Capacity (FRSC) Assessment
4.3.11. Enzyme Measurements
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010, 37, 613–620. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Bacha, H.; Tekaya, M.; Drine, S.; Guasmi, F.; Touil, L.; Enneb, H.; Triki, T.; Cheour, F.; Ferchichi, A. Impact of salt stress on morpho-physiological and biochemical parameters of Solanum lycopersicum cv. Microtom leaves. South Afr. J. Bot. 2017, 108, 364–369. [Google Scholar] [CrossRef]
- Foolad, M.R. Recent Advances in Genetics of Salt Tolerance in Tomato. Plant Cell Tissue Organ Cult. (PCTOC) 2004, 76, 101–119. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.; Foster, K.J.; et al. Energy costs of salt tolerance in crop plants. New Phytol. 2019, 225, 1072–1090. [Google Scholar] [CrossRef]
- Tang, R.-J.; Zhao, F.-G.; Garcia, V.J.; Kleist, T.J.; Yang, L.; Zhang, H.-X.; Luan, S. Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3134–3139. [Google Scholar] [CrossRef]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.-G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef]
- Tavakkoli, E.; Rengasamy, P.; McDonald, G.K. High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010, 61, 4449–4459. [Google Scholar] [CrossRef] [PubMed]
- Mbarki, S.; Skalicky, M.; Vachova, P.; Hajihashemi, S.; Jouini, L.; Zivcak, M.; Tlustos, P.; Brestic, M.; Hejnak, V.; Khelil, A.Z. Comparing Salt Tolerance at Seedling and Germination Stages in Local Populations of Medicago ciliaris L. to Medicago intertexta L. and Medicago scutellata L. Plants 2020, 9, 526. [Google Scholar] [CrossRef] [PubMed]
- Sdouga, D.; Ben Amor, F.; Ghribi, S.; Kabtni, S.; Tebini, M.; Branca, F.; Trifi-Farah, N.; Marghali, S. An insight from tolerance to salinity stress in halophyte Portulaca oleracea L.: Physio-morphological, biochemical and molecular responses. Ecotoxicol. Environ. Saf. 2019, 172, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Durazzo, A.; Azzini, E.; Foddai, M.S.; Nobili, F.; Garaguso, I.; Raguzzini, A.; Finotti, E.; Tisselli, V.; Del Vecchio, S.; Piazza, C.; et al. Influence of different crop management practices on the nutritional properties and benefits of tomato -Lycopersicon esculentum cv Perfectpeel-. Int. J. Food Sci. Technol. 2010, 45, 2637–2644. [Google Scholar] [CrossRef]
- Saito, T.; Matsukura, C. Effect of Salt Stress on the Growth and Fruit Quality of Tomato Plants. In Abiotic Stress Biology in Horticultural Plants; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–16. [Google Scholar]
- Zhai, Y.; Yang, Q.; Hou, M. The Effects of Saline Water Drip Irrigation on Tomato Yield, Quality, and Blossom-End Rot Incidence—A 3a Case Study in the South of China. PLoS ONE 2015, 10, e0142204. [Google Scholar] [CrossRef]
- Singh, J.; Sastry, E.V.D.; Singh, V. Effect of salinity on tomato (Lycopersicon esculentum Mill.) during seed germination stage. Physiol. Mol. Biol. Plants 2011, 18, 45–50. [Google Scholar] [CrossRef]
- Ibrahimova, U.F.; Mammadov, A.C.; Feyziyev, Y.M. The effect of NaCl on some physiological and biochemical parameters in Triticum aestivum L. genotypes. Plant Physiol. Rep. 2019, 24, 370–375. [Google Scholar] [CrossRef]
- Pailles, Y.; Awlia, M.; Julkowska, M.M.; Passone, L.; Zemmouri, K.; Negrão, S.; Schmöckel, S.M.; Tester, M. Diverse Traits Contribute to Salinity Tolerance of Wild Tomato Seedlings from the Galapagos Islands. Plant Physiol. 2020, 182, 534–546. [Google Scholar] [CrossRef]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel—Dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Saade, S.; Maurer, A.; Shahid, M.; Oakey, H.; Schmöckel, S.M.; Negrão, S.; Pillen, K.; Tester, M. Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dasgan, H.; Aktas, H.; Abak, K.; Cakmak, I. Determination of screening techniques to salinity tolerance in tomatoes and investigation of genotype responses. Plant Sci. 2002, 163, 695–703. [Google Scholar] [CrossRef]
- Turhan, A.; Seniz, V. Salt tolerance during vegetative growth in cross of tomato and effect of cytoplasm in response to salt tolerance. Bulg. J. Agric. Sci. 2012, 18, 207–218. [Google Scholar]
- Joshi, R.; Gupta, P.; Singla-Pareek, S.L.; Pareek, A. Biomass production and salinity response in plants: Role of MicroRNAs. Indian J. Plant Physiol. 2017, 22, 448–457. [Google Scholar] [CrossRef]
- Foolad, M.R. Genetic analysis of salt tolerance during vegetative growth in tomato, Lycopersicon esculentum Mill. Plant Breed. 1996, 115, 245–250. [Google Scholar] [CrossRef]
- Cuartero, J.; Yeo, A.R.; Flowers, T.J. Selection of donors for salt-tolerance in tomato using physiological traits. New Phytol. 1992, 121, 63–69. [Google Scholar] [CrossRef]
- Sivakumar, J.; Prashanth, J.E.P.; Rajesh, N.; Reddy, S.M.; Pinjari, O.B. Principal component analysis approach for comprehensive screening of salt stress-tolerant tomato germplasm at the seedling stage. J. Biosci. 2020, 45, 1–11. [Google Scholar] [CrossRef]
- Raza, M.A.; Saeed, A.; Munir, H.; Ziaf, K.; Shakeel, A.; Saeed, N.; Munawar, A.; Rehman, F. Screening of tomato genotypes for salinity tolerance based on early growth attributes and leaf inorganic osmolytes. Arch. Agron. Soil Sci. 2016, 63, 501–512. [Google Scholar] [CrossRef]
- Rehman, F.; Saeed, A.; Yaseen, M.; Shakeel, A.; Ziaf, K.; Munir, H.; Tariq, S.A.; Raza, M.A.; Riaz, A. Genetic evaluation and characterization using cluster heat map to assess NaCl tolerance in tomato germplasm at the seedling stage. Chil. J. Agric. Res. 2019, 79, 56–65. [Google Scholar] [CrossRef]
- Derbali, W.; Goussi, R.; Koyro, H.-W.; Abdelly, C.; Manaa, A. Physiological and biochemical markers for screening salt tolerant quinoa genotypes at early seedling stage. J. Plant Interact. 2020, 15, 27–38. [Google Scholar] [CrossRef]
- Gholinezhad, E.; Darvishzadeh, R.; Bernousi, I. Evaluation of Drought Tolerance Indices for Selection of Confectionery Sunflower (Helianthus anuus L.) Landraces under Various Environmental Conditions. Not. Bot. Horti Agrobot. Cluj-Napoca 2014, 42, 187–201. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High Salinity Induces Different Oxidative Stress and Antioxidant Responses in Maize Seedlings Organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef]
- Chawla, S.; Jain, S.; Jain, V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2013, 22, 27–34. [Google Scholar] [CrossRef]
- AbdAllah, A.M.; Burkey, K.O.; Mashaheet, A.M. Reduction of plant water consumption through anti-transpirants foliar application in tomato plants ( Solanum lycopersicum L.). Sci. Hortic. 2018, 235, 373–381. [Google Scholar] [CrossRef]
- Martinez, V.; Nieves-Cordones, M.; Lopez-Delacalle, M.; Rodenas, R.; Mestre, T.C.; Garcia-Sanchez, F.; Rubio, F.; Nortes, P.A.; Mittler, R.; Rivero, R.M. Tolerance to Stress Combination in Tomato Plants: New Insights in the Protective Role of Melatonin. Molecules 2018, 23, 535. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Mousa, M.A.; Al-Qurashi, A.D.; Bakhashwain, A.A. Response of tomato genotypes at early growing stages to irrigation water salinity. J. Food Agric. Env. 2013, 11, 501–507. [Google Scholar]
- Ben Chikha, M.; Hessini, K.; Ourteni, R.N.; Ghorbel, A.; Zoghlami, N. Identification of barley landrace genotypes with contrasting salinity tolerance at vegetative growth stage. Plant Biotechnol. 2016, 33, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Salam, A.; Hollington, P.A.; Gorham, J.; Jones, R.G.W.; Gliddon, C. Physiological Genetics of Salt Tolerance in Wheat (Triticum aestivum L.): Performance of Wheat Varieties, Inbred Lines and Reciprocal F1 Hybrids under Saline Conditions. J. Agron. Crop. Sci. 1999, 183, 145–156. [Google Scholar] [CrossRef]
- Ashraf, M.; McNeilly, T. Effect of salinity on some cultivars of maize. Maydica 1989, 34, 179–189. [Google Scholar]
- Azhar, F.; Ahmad, M. Inheritance Pattern of Cotton Seed Oil In Diverse Germplasm of Gossypium hirsutum L. Pak. J. Biol. Sci. 2000, 3, 1250–1252. [Google Scholar] [CrossRef][Green Version]
- Al-Amin, M.; Islam, M.M.; Begum, S.N.; Alam, M.; Moniruzzaman, M.; Patwary, M. Evaluation of Rice Germplasm under Salt Stress at the Seedling Stage through SSR Markers. Int. J. Agric. Res. Innov. Technol. 2013, 3, 52–59. [Google Scholar] [CrossRef]
- Magudeeswari, P.; Sastry, E.V.D.; Devi, T.R. Principal component (PCA) and cluster analyses for plant nutrient traits in baby corn (Zea mays L.). Indian J. Agric. Res. 2019, 53, 353–357. [Google Scholar] [CrossRef]
- Alam, M.S.; Hossain, S.; Ali, M.A.; Hossain, M.G.; Islam, M.F. Assessment of Genetic Divergence in Tomato (Solanum lycopersicum L.) through Clustering and Principal Component Analysis. J. Agric. Sci. Eng. Innov. 2020, 1, 10–14. [Google Scholar] [CrossRef]
- Ali, Z.; Salam, A.; Azhar, F.M.; Khan, I.A.; Khan, A.A.; Bahadur, S.; Mahmood, T.; Ahmad, A.; Trethowan, R. The response of genetically distinct bread wheat genotypes to salinity stress. Plant Breed. 2012, 131, 707–715. [Google Scholar] [CrossRef]
- Maas, E.V.; Hoffman, G.J. Crop salt tolerance–current assessment. J. Irrig. Drain. Div. 1977, 103, 115–134. [Google Scholar] [CrossRef]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of salt stress on ion concentration, proline content, antioxidant enzyme activities and gene expression in tomato cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef] [PubMed]
- Cruz, V.; Cuartero, J. Effects of salinity at several developmental stages of six genotypes of tomato (Lycopersicon spp.). In Proceedings of the XIth Eucarpia Meeting on Tomato Genetics and Breeding, Malaga, Spain, 6–8 March 1990; pp. 81–86. [Google Scholar]
- Zobel, R.L. The genetics of root development. In The Development and Function of Roots; Academic Press: New York, NY, USA, 1975; pp. 261–275. [Google Scholar]
- An, P.; Inanaga, S.; Li, X.J.; Eneji, A.E.; Zhu, N.W. Interactive Effects of Salinity and Air Humidity on Two Tomato Cultivars Differing in Salt Tolerance. J. Plant Nutr. 2005, 28, 459–473. [Google Scholar] [CrossRef]
- Amjad, M.; Akhtar, J.; Anwar-Ul-Haq, M.; Yang, A.; Akhtar, S.S.; Jacobsen, S.-E. Integrating role of ethylene and ABA in tomato plants adaptation to salt stress. Sci. Hortic. 2014, 172, 109–116. [Google Scholar] [CrossRef]
- Murillo-Amador, B.; Reyes-Pérez, J.J.; Hernández-Montiel, L.G.; Rueda-Puente, E.O.; De Lucia, B.; Beltrán-Morales, F.A.; Ruiz-Espinoza, F.H. Physiological responses to salinity in Solanum lycopersicum L. varieties. Pak. J. Bot. 2017, 49, 809–818. [Google Scholar]
- Chen, Z.; Newman, I.; Zhou, M.; Mendham, N.; Zhang, G.; Shabala, S. Screening plants for salt tolerance by measuring K+ flux: A case study for barley. Plant Cell Environ. 2005, 28, 1230–1246. [Google Scholar] [CrossRef]
- Khan, N.; Syeed, S.; Masood, A.; Nazar, R.; Iqbal, N. Application of salicylic acid increases contents of nutrients and antioxidative metabolism in mungbean and alleviates adverse effects of salinity stress. Int. J. Plant Biol. 2010, 4, 1. [Google Scholar] [CrossRef]
- Sucre, B.; Suárez, N. Effect of salinity and PEG-induced water stress on water status, gas exchange, solute accumulation, and leaf growth in Ipomoea pes-caprae. Environ. Exp. Bot. 2011, 70, 192–203. [Google Scholar] [CrossRef]
- Dekov, I.; Tsonev, T.; Yordanov, I. Effects of Water Stress and High-Temperature Stress on the Structure and Activity of Photosynthetic Apparatus of Zea Mays and Helianthus Annuus. Photosynthetica 2000, 38, 361–366. [Google Scholar] [CrossRef]
- Kwon, O.K.; Mekapogu, M.; Kim, K.S. Effect of salinity stress on photosynthesis and related physiological responses in carnation (Dianthus caryophyllus). Hortic. Environ. Biotechnol. 2019, 60, 831–839. [Google Scholar] [CrossRef]
- Zaharieva, M.; Gaulin, E.; Havaux, M.; Acevedo, E.; Monneveux, P. Drought and heat responses in the wild wheat relative Aegilops geniculata Roth: Potential interest for wheat improvement. Crop Sci. 2001, 41, 1–25. [Google Scholar] [CrossRef]
- Sairam, R.; Chandrasekhar, V.; Srivastava, G. Comparison of Hexaploid and Tetraploid Wheat Cultivars in their Responses to Water Stress. Biol. Plant. 2001, 44, 89–94. [Google Scholar] [CrossRef]
- Shannon, M.; Grieve, C. Tolerance of vegetable crops to salinity. Sci. Hortic. 1998, 78, 5–38. [Google Scholar] [CrossRef]
- Rasel, M.; Arif, M.T.-U.; Hossain, M.A.; Hassan, L.; Farzana, S.; Brestic, M. Screening of Salt-Tolerant Rice Landraces by Seedling Stage Phenotyping and Dissecting Biochemical Determinants of Tolerance Mechanism. J. Plant Growth Regul. 2020, 1–16. [Google Scholar] [CrossRef]
- Narimani, T.; Toorchi, M.; Tarinejad, A.R.; Mohammadi, S.A.; Mohammadi, H. Physiological and Biochemical Evaluation of Barley (Hordeum vulgare L.) under Salinity Stress. J. Agric. Sci. Technol. 2020, 22, 1009–1021. [Google Scholar]
- Tavakoli, M.; Poustini, K.; Besharati, H.; Ali, S. Variable Salinity Responses of 25 Alfalfa Genotypes and Comparative Salt-Response Ion Distribution. Russ. J. Plant Physiol. 2019, 66, 231–239. [Google Scholar] [CrossRef]
- Grzesiak, S.; Hordyńska, N.; Szczyrek, P.; Grzesiak, M.T.; Noga, A.; Szechyńska-Hebda, M. Variation among wheat (Triticum easativum L.) genotypes in response to the drought stress: I—Selection approaches. J. Plant Interact. 2018, 14, 30–44. [Google Scholar] [CrossRef]
- Mohsin, S.M.; Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Fujita, M. Exogenous Tebuconazole and Trifloxystrobin Regulates Reactive Oxygen Species Metabolism Toward Mitigating Salt-Induced Damages in Cucumber Seedling. Plants 2019, 8, 428. [Google Scholar] [CrossRef]
- Rady, M.M.; Taha, R.; Mahdi, A.H. Proline enhances growth, productivity and anatomy of two varieties of Lupinus termis L. grown under salt stress. South Afr. J. Bot. 2016, 102, 221–227. [Google Scholar] [CrossRef]
- Shahbaz, M.; Mushtaq, Z.; Andaz, F.; Masood, A. Does proline application ameliorate adverse effects of salt stress on growth, ions and photosynthetic ability of eggplant (Solanum melongena L.)? Sci. Hortic. 2013, 164, 507–511. [Google Scholar] [CrossRef]
- Taïbi, K.; Taïbi, F.; Abderrahim, L.A.; Ennajah, A.; Belkhodja, M.; Mulet, J.M. Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defence systems in Phaseolus vulgaris L. South Afr. J. Bot. 2016, 105, 306–312. [Google Scholar] [CrossRef]
- Di Ferdinando, M.; Brunetti, C.; Fini, A.; Tattini, M. Flavonoids as Antioxidants in Plants Under Abiotic Stresses. In Abiotic Stress Responses in Plants; Springer: Berlin/Heidelberg, Germany, 2011; pp. 159–179. [Google Scholar]
- Hamooh, B.T.; Sattar, F.A.; Wellman, G.; Mousa, M.A.A. Metabolomic and Biochemical Analysis of Two Potato (Solanum tuberosum L.) Cultivars Exposed to In Vitro Osmotic and Salt Stresses. Plants 2021, 10, 98. [Google Scholar] [CrossRef]
- Willekens, H.; Chamnongpol, S.; Davey, M.; Schraudner, M.; Langebartels, C.; Van Montagu, M.; Inzé, D.; Van Camp, W. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 1997, 16, 4806–4816. [Google Scholar] [CrossRef] [PubMed]
- Murshed, R.; Lopez-Lauri, F.; Sallanon, H. Effect of salt stress on tomato fruit antioxidant systems depends on fruit development stage. Physiol. Mol. Biol. Plants 2013, 20, 15–29. [Google Scholar] [CrossRef]
- Parvin, K.; Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Mohsin, S.M.; Fujita, A.M. Quercetin Mediated Salt Tolerance in Tomato through the Enhancement of Plant Antioxidant Defense and Glyoxalase Systems. Plants 2019, 8, 247. [Google Scholar] [CrossRef]
- Soleimani, Z.; Afshar, A.S.; Nematpour, F.S. Responses of antioxidant gene and enzymes to salinity stress in the Cuminum cyminum L. Russ. J. Plant Physiol. 2017, 64, 361–367. [Google Scholar] [CrossRef]
- Estaji, A.; Roosta, H.R.; Rezaei, S.A.; Hosseini, S.S.; Niknam, F. Morphological, physiological and phytochemical response of different Satureja hortensis L. accessions to salinity in a greenhouse experiment. J. Appl. Res. Med. Aromat. Plants 2018, 10, 25–33. [Google Scholar] [CrossRef]
- Asadi, M.; Mirvaghefei, A.; Nematollahi, M.; Banaee, M.; Ahmadi, K. Effects of Watercress (Nasturtium nasturtium) extract on selected immunological parameters of rainbow trout (Oncorhynchus mykiss). Open Veter J. 2012, 2, 32–39. [Google Scholar]
- Ekbic, E.; Cagıran, C.; Kose, M.A.; Aras, V.; Korkmaz, K. Assessment of watermelon accessions for salt tolerance using stress tolerance indices. Ciênc. Agrotecnol. 2017, 41, 616–625. [Google Scholar] [CrossRef]
- Easlon, H.M.; Bloom, A.J. Easy Leaf Area: Automated Digital Image Analysis for Rapid and Accurate Measurement of Leaf Area. Appl. Plant Sci. 2014, 2, 1400033. [Google Scholar] [CrossRef]
- Yamasaki, S.; Dillenburg, L.R. Measurements of leaf relative water content in Araucaria angustifolia. Rev. Bras. Fisiol. Veg. 1999, 11, 69–75. [Google Scholar]
- El Goumi, Y.; Fakiri, M.; Lamsaouri, O.; Benchekroun, M. Salt stress effect on seed germination and some physiological traits in three Moroccan barley (Hordeum vulgare L.) cultivars. J. Mater. Environ. Sci. 2014, 5, 625–632. [Google Scholar]
- Sairam, R.K.; Srivastava, G.C.; Agarwal, S.; Meena, R.C. Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol. Plant. 2005, 49, 85–91. [Google Scholar] [CrossRef]
- Fischer, R.; Maurer, R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978, 29, 897–912. [Google Scholar] [CrossRef]
- Clarke, J.M.; DePauw, R.M.; Townley-Smith, T.F. Evaluation of methods for quantification of drought tolerance in wheat. Crop Sci. 1992, 32, 723–728. [Google Scholar] [CrossRef]
- Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and other Food Crops in Temperature and Water Stress, Shanhua, Taiwan, 13–16 August 1992; pp. 257–270. [Google Scholar]
- Hossain, A.B.S.; Sears, R.G.; Cox, T.S.; Paulsen, G.M. Desiccation Tolerance and Its Relationship to Assimilate Partitioning in Winter Wheat. Crop. Sci. 1990, 30, 622–627. [Google Scholar] [CrossRef]
- Schneider, K.A.; Rosales-Serna, R.; Ibarra-Perez, F.; Cazares-Enriquez, B.; Acosta-Gallegos, J.A.; Ramirez-Vallejo, P.; Wassimi, N.; Kelly, J.D. Improving common bean performance under drought stress. Crop Sci. 1997, 37, 43–50. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- McKinney, R.H.; Bailey, A.E. Some notes on the keeping quality of fats in baked goods. J. Am. Oil Chem. Soc. 1941, 18, 147–148. [Google Scholar] [CrossRef]
- Saida, C.; Houria, B.; Mébarek, B. Interactive effects of salinity and potassium on physio-morphological traits of tomato (Lycopersicon esculentum Mill.; var: Heintz). Agric. Biol. J. N. Am. 2014, 5, 135–143. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Velioglu, Y.S.; Mazza, G.; Gao, L.; Oomah, B.D. Antioxidant activity and total phenolics in selected fruits, vegetables, and grain products. J. Agric. Food Chem. 1998, 46, 4113–4117. [Google Scholar] [CrossRef]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Ao, C.; Li, A.; Elzaawely, A.A.; Xuan, T.D.; Tawata, S. Evaluation of antioxidant and antibacterial activities of Ficus microcarpa L. fil. extract. Food Control. 2008, 19, 940–948. [Google Scholar] [CrossRef]
- Antoniou, C.; Xenofontos, R.; Chatzimichail, G.; Christou, A.; Kashfi, K.; Fotopoulos, V. Exploring the Potential of Nitric Oxide and Hydrogen Sulfide (NOSH)-Releasing Synthetic Compounds as Novel Priming Agents against Drought Stress in Medicago sativa Plants. Biomolecules 2020, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, L.; Chen, S.; Sun, H.; Ma, S. Transcription profile analysis of Lycopersicum esculentum leaves, unravels volatile emissions and gene expression under salinity stress. Plant Physiol. Biochem. 2018, 126, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Chance, B.; Maehly, A. Assay of catalases and peroxidases. Vitam. Coenzymes Part F 1955, 2, 764–775. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Hossain, M.A.; Fujita, M. Nitric oxide modulates antioxidant defense and the methylglyoxal detoxification system and reduces salinity-induced damage of wheat seedlings. Plant Biotechnol. Rep. 2011, 5, 353–365. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [CrossRef]
- Analytical Software. Statistix 8.1; Maurice/Thomas text 2008; Analytical Software: Tallahassee, FL, USA, 2008. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- GraphPad Software. Prism, G. Version 8.0.0; GraphPad Software: La Joalla, CA, USA, 2018. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, M.S.; Tester, M.; Fiene, G.; Mousa, M.A.A. Early Growth Stage Characterization and the Biochemical Responses for Salinity Stress in Tomato. Plants 2021, 10, 712. https://doi.org/10.3390/plants10040712
Alam MS, Tester M, Fiene G, Mousa MAA. Early Growth Stage Characterization and the Biochemical Responses for Salinity Stress in Tomato. Plants. 2021; 10(4):712. https://doi.org/10.3390/plants10040712
Chicago/Turabian StyleAlam, Md Sarowar, Mark Tester, Gabriele Fiene, and Magdi Ali Ahmed Mousa. 2021. "Early Growth Stage Characterization and the Biochemical Responses for Salinity Stress in Tomato" Plants 10, no. 4: 712. https://doi.org/10.3390/plants10040712
APA StyleAlam, M. S., Tester, M., Fiene, G., & Mousa, M. A. A. (2021). Early Growth Stage Characterization and the Biochemical Responses for Salinity Stress in Tomato. Plants, 10(4), 712. https://doi.org/10.3390/plants10040712






