Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing
Abstract
1. Introduction
2. Results
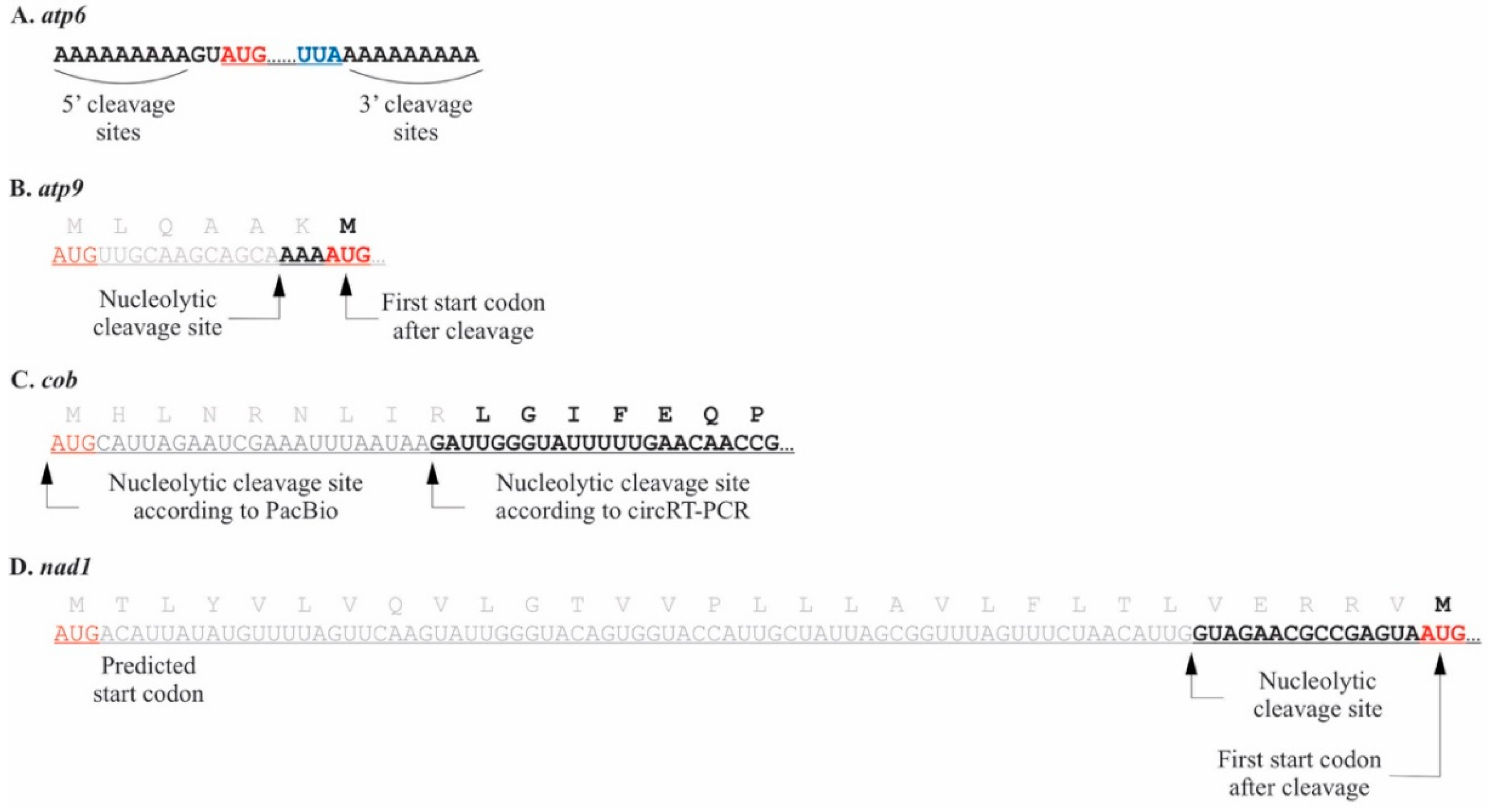
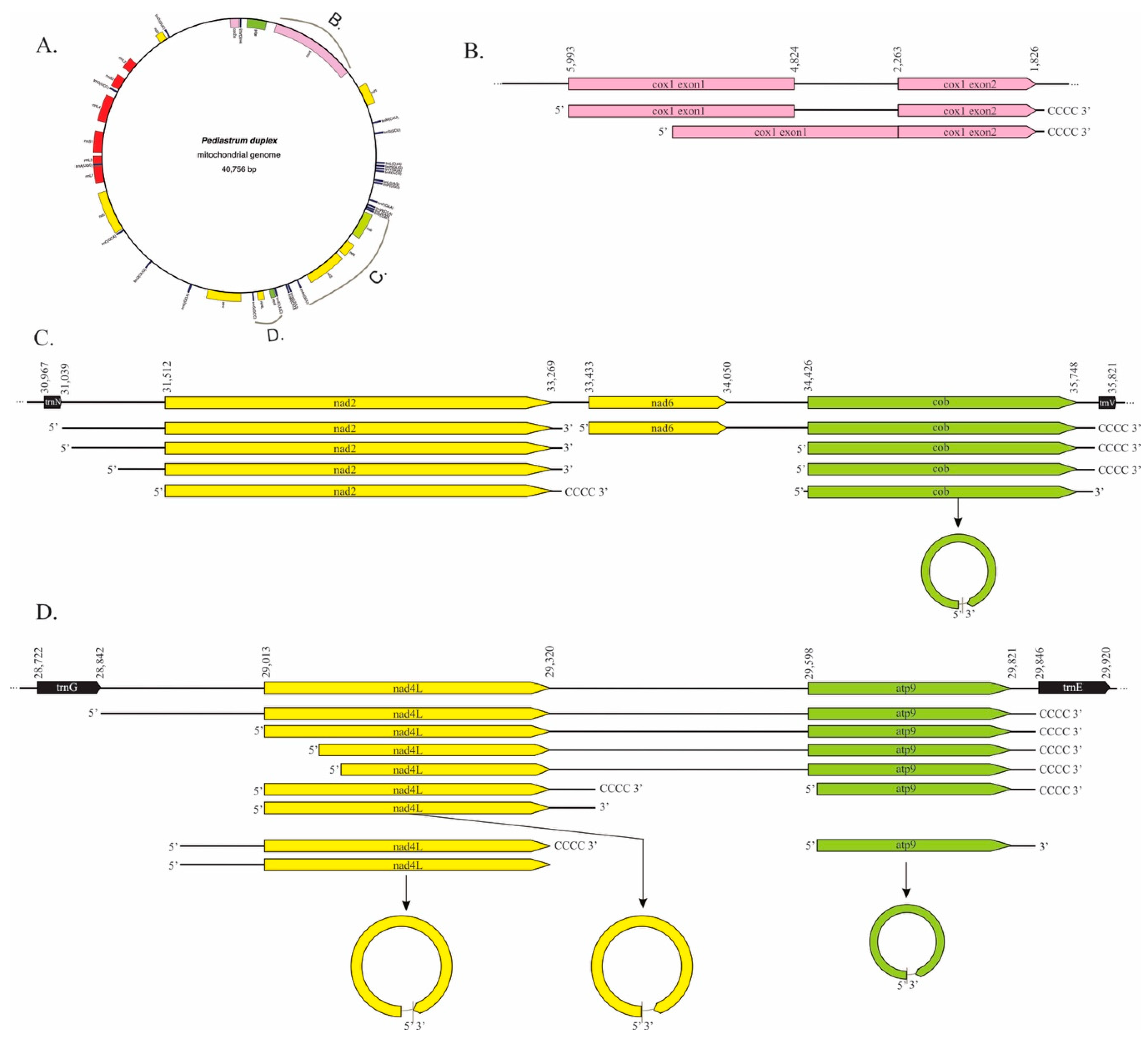
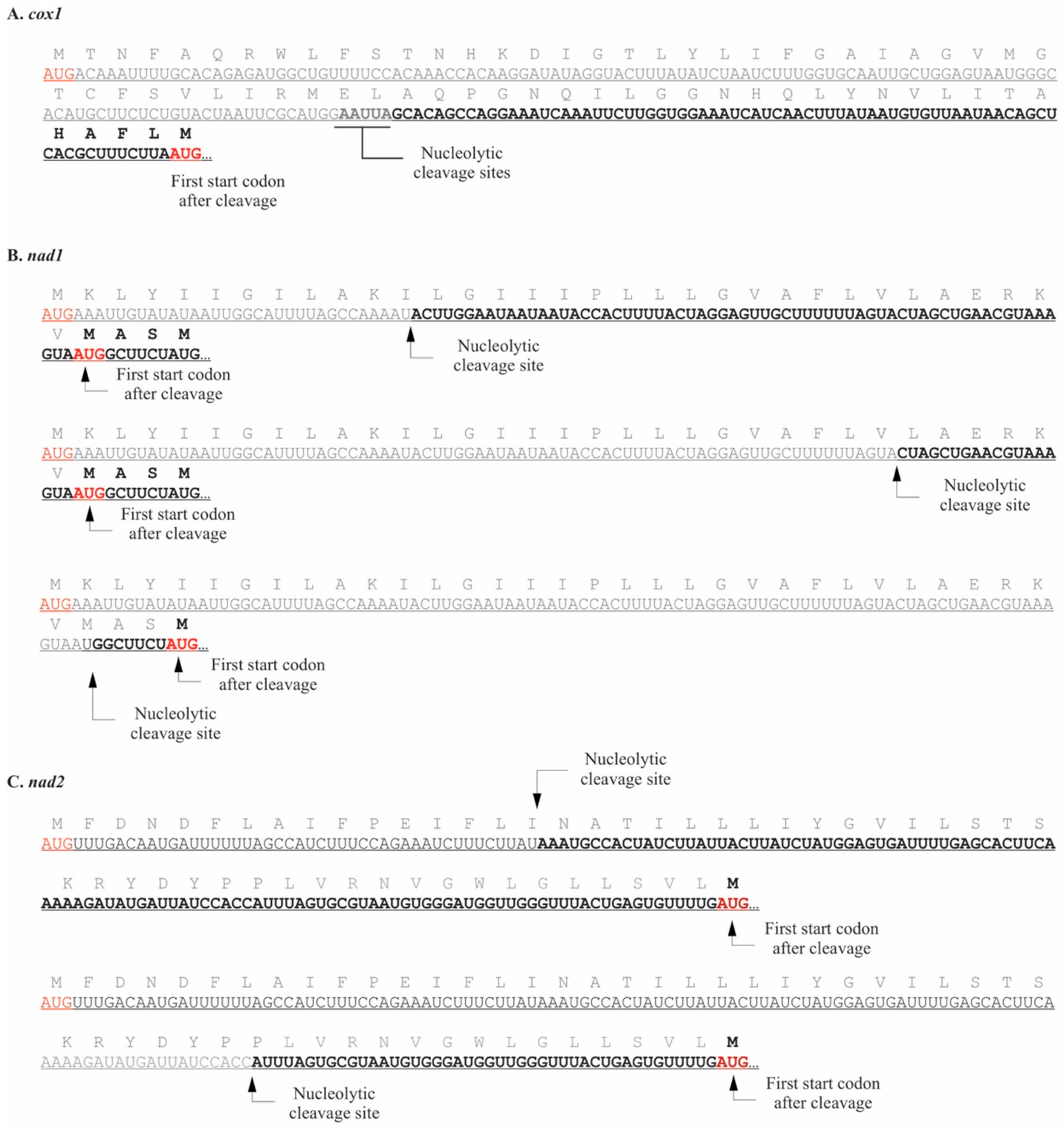
2.1. Pediastrum duplex
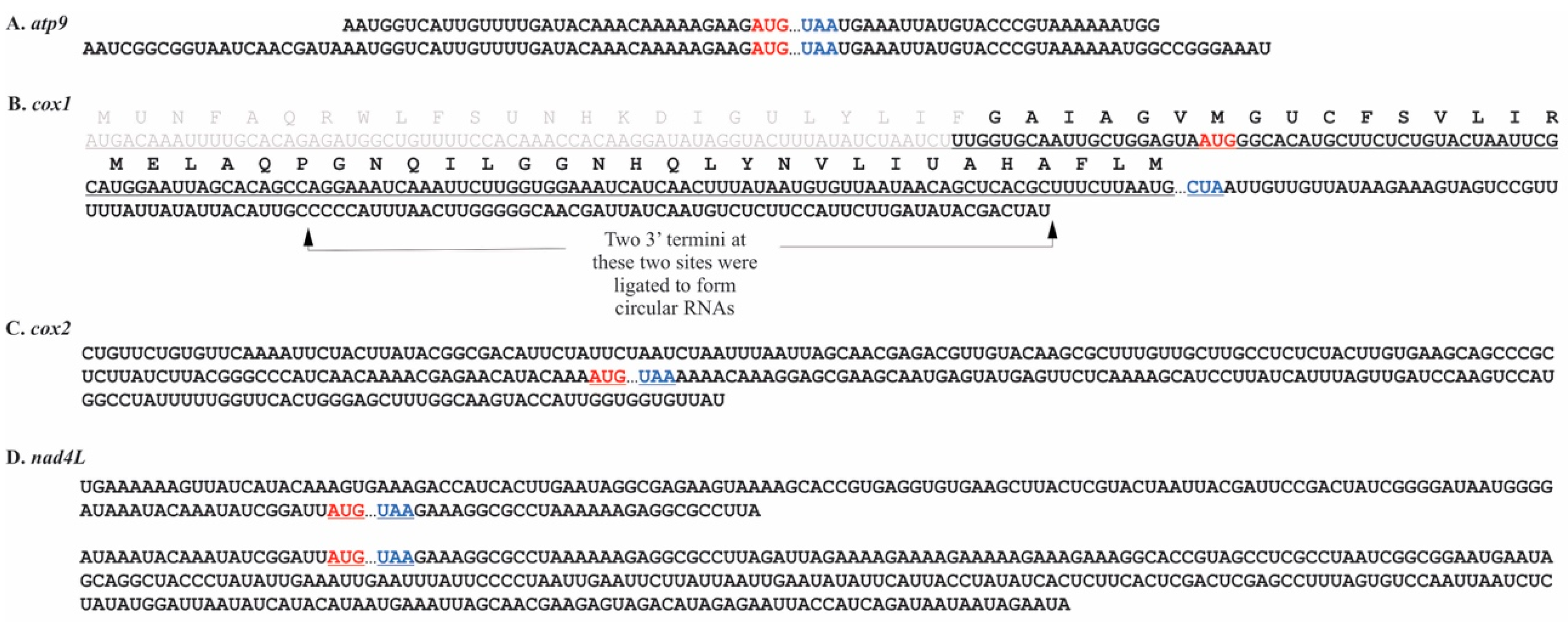
2.2. Chara vulgaris
3. Discussion
3.1. Mitochondrial Processing in P. duplex Resembles That Seen in C. reinhardtii
3.2. Mitochondrial Processing in C. vulgaris Resembles That Seen in Embryophytes.
4. Materials and Methods
4.1. Cultures
4.2. RNA Extraction and CircRT-PCR
4.3. Sequencing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rose, R.J.; Tiew, T.W.-Y.; Dashek, W. The mitochondrion. In Plant Cells and Their Organelles; Dashek, W.V., Miglani, G.S., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Archibald, J.M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015, 25, R911–R921. [Google Scholar] [CrossRef]
- López-García, P.; Eme, L.; Moreira, D. Symbiosis in eukaryotic evolution. J. Theor. Biol. 2017, 434, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 2015, 5, 7949. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.G.; Williams, B.P. Evolutionary inference across eukaryotes identifies specific pressures favoring mitochondrial gene retention. Cell Syst. 2016, 2, 101–111. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, A.R.; Minczuk, M. Mitochondrial transcription and translation: Overview. Essays Biochem. 2018, 62, 309–320. [Google Scholar] [CrossRef]
- Tiranti, V.; Savoia, A.; Forti, F.; D’Apolito, M.F.; Centra, M.; Rocchi, M.; Zeviani, M. Identification of the gene encoding the human mitochondrial RNApolymerase (h-mtRPOL) by cyberscreening of the expressed sequence tags database. Hum. Mol. Genet. 1997, 6, 615–625. [Google Scholar] [CrossRef]
- Falkenberg, M.; Gaspari, M.; Rantanen, A.; Trifunovic, A.; Larsson, N.G.; Gustafsson, C.M. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002, 31, 289–294. [Google Scholar] [CrossRef]
- Kanki, T.; Nakayama, H.; Sasaki, N.; Takio, K.; Alam, T.I.; Hamasaki, N.; Kang, D. Mitochondrial nucleoid and transcription factor A. Ann. N. Y. Acad. Sci. 2004, 1011, 61–68. [Google Scholar] [CrossRef]
- Montoya, J.; Christianson, T.; Levens, D.; Rabinowitz, M.; Attardi, G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1982, 79, 7195–7199. [Google Scholar] [CrossRef]
- Montoya, J.; Gaines, G.L.; Attardi, G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell 1983, 34, 151–159. [Google Scholar] [CrossRef]
- Chang, D.D.; Clayton, D.A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell 1984, 36, 635–643. [Google Scholar] [CrossRef]
- Gao, S.; Tian, X.; Chang, H.; Sun, Y.; Wu, Z.; Cheng, Z.; Dong, P.; Zhao, Q.; Ruan, J.; Bu, W. Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondria 2018, 38, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Minczuk, M.; He, J.; Duch, A.M.; Ettema, T.J.; Chlebowski, A.; Dzionek, K.; Nijtmans, L.G.J.; Huynen, M.A.; Holt, I.J. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011, 39, 4284–4299. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing inhuman mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Montoya, J.; Ojala, D.; Attardi, G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 1981, 290, 465–470. [Google Scholar] [CrossRef]
- Temperley, R.J.; Wydro, M.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M. Human mitochondrial mRNAs—Like members of all families, similar but different. Biochim. Biophys. Acta 2010, 1797, 1081–1085. [Google Scholar] [CrossRef]
- Kummer, E.; Leibundgut, M.; Rackham, O.; Lee, R.G.; Boehringer, D.; Filipovska, A.; Ban, N. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 2018, 560, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Nagaike, T.; Suzuki, T.; Katoh, T.; Ueda, T. Human mitochondrial mRNAs are stabilized with polyaenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005, 280, 19721–19727. [Google Scholar] [CrossRef]
- Slomovic, S.; Lauer, D.; Geiger, D.; Schuster, G. Polyadenylation and degradation of human mitochondrial RNA: The prokaryotic past leaves its mark. Mol. Cell. Biol. 2005, 25, 6427–6435. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, I.; Siira, S.J.; Shearwood, A.-M.J.; Ermer, J.A.; Filipovska, A.; Rackham, O. Simultaneous processing and degradation of mitochondrial RNAs revealed by circularized RNA sequencing. Nucleic Acids Res. 2017, 45, 5487–5500. [Google Scholar] [CrossRef] [PubMed]
- Mance, L.G.; Mawla, I.; Shell, S.M.; Cahoon, A.B. Mitochondrial mRNA fragments are circularized in a human HEK cell line. Mitochondrion 2020, 51, 1–6. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Hedtke, B.; Börner, T.; Weihe, A. One RNA polymerase serving two genomes. EMBO Rep. 2000, 1, 435–440. [Google Scholar] [CrossRef]
- Kühn, K.; Richter, U.; Meyer, E.H.; Delannoy, E.; de Longevialle, A.F.; O’Toole, N.; Börner, T.; Millar, A.H.; Small, I.D.; Whelan, J. Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 2009, 21, 2762–2779. [Google Scholar] [CrossRef]
- Roberti, M.; Polosa, P.L.; Bruni, F.; Manzari, C.; Deceglie, S.; Gadaleta, M.N.; Cantatore, P. The MTERF family proteins: Mitochondrial transcription regulators and beyond. Biochim. Biophys. Acta 2009, 1787, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.; Weihe, A.; Börner, T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005, 33, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Perrin, R.; Meyer, E.H.; Zaepfel, M.; Kim, Y.J.; Mache, R.; Grienenberger, J.M.; Gualberto, J.M.; Gagliardi, D. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 2004, 279, 25440–25446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, X.; Zou, J.; Liu, Y.; Lian, T.; Nian, H. Major contribution of transcription initiation to 5′-end formation of mitochondrial steady-state transcripts in maize. RNA Biol. 2019, 16, 104–117. [Google Scholar] [CrossRef]
- Binder, S.; Stoll, K.; Stoll, B. Maturation of 5′ ends of plant mitochondrial RNAs. Physiol. Plant 2016, 157, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Forner, J.; Weber, B.; Thuss, S.; Wildum, S.; Binder, S. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: T-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007, 35, 3676–3692. [Google Scholar] [CrossRef] [PubMed]
- Hanic-Joyce, P.J.; Spencer, D.F.; Gray, M.W. In vitro processing of transcripts containing novel tRNA-like sequences (‘t-elements’) encoded by wheat mitochondrial DNA. Plant Mol. Biol. 1990, 15, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, S.; Brennicke, A.; Binder, S. 3′-inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators. EMBO J. 1997, 16, 5069–5076. [Google Scholar] [CrossRef]
- Gobert, A.; Gutmann, B.; Taschner, A.; GöBringer, M.; Holzmann, J.; Hartmann, R.K.; Rossmanith, W.; Giegé, P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010, 17, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Gobert, A.; Giegé, P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012, 26, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.; Binder, S. Two NYN domain containing putative nucleases are involved in transcript maturation in Arabidopsis mitochondria. Plant J. 2016, 85, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Jonietz, C.; Forner, J.; Hölzle, A.; Thuss, S.; Binder, S. RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell 2010, 22, 443–453. [Google Scholar] [CrossRef]
- Hölzle, A.; Jonietz, C.; Törjek, O.; Altmann, T.; Binder, S.; Forner, J. A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 2011, 65, 65,737–744. [Google Scholar] [CrossRef]
- Hauler, A.; Jonietz, C.; Stoll, B.; Stoll, K.; Braun, H.-P.; Binder, S. RNA PROCESSING FACTOR 5 is required for efficient 5′ cleavage at a processing site conserved in RNAs of three different mitochondrial genes in Arabidopsis thaliana. Plant J. 2013, 74, 593–604. [Google Scholar] [CrossRef]
- Haili, N.; Arnal, N.; Quadrado, M.; Amiar, S.; Tcherkez, G.; Dahan, J.; Briozzo, P.; Colas des Francs-Small, C.; Vrielynck, N.; Mireau, H. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013, 41, 6650–6663. [Google Scholar] [CrossRef]
- Hammani, K.; Giegé, P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014, 19, 380–389. [Google Scholar] [CrossRef]
- Brown, G.G.; des Francs-Small, C.C.; Ostersetzer-Biran, O. Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 2014, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, D.; Leaver, C.J. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999, 18, 3757–3766. [Google Scholar] [CrossRef] [PubMed]
- Lupold, D.S.; Caoile, A.G.; Stern, D.B. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell 1999, 11, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Kühn, J.; Tengler, U.; Binder, S. Transcript lifetime is balanced between stabilizing stem-loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol. 2001, 21, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Schuster, G.; Stern, D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 2009, 85, 393–422. [Google Scholar] [CrossRef]
- Harris, E.H. The Chlamydomonas Sourcebook, 2nd ed.; Harris, E.H., Stern, D.B., Witman, G.B., Eds.; Academic Press: Oxford, UK, 2009. [Google Scholar]
- Gray, M.W.; Boer, P.H. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial DNA. Phil. Trans. R. Soc. Lon. B Biol. Sci. 1988, 319, 135–147. [Google Scholar] [CrossRef]
- Laflamme, M.; Lee, R.W. Mitochondrial genome conformation among CW-group chlorophycean algae. J. Phycol. 2003, 39, 213–220. [Google Scholar] [CrossRef]
- Mallet, M.A.; Lee, R.W. Identification of three distinct Polytomella lineages based on mitochondrial DNA features. J. Eukaryot. Microbiol. 2006, 53, 79–84. [Google Scholar] [CrossRef]
- Tracy, R.L.; Stern, D.B. Mitochondrial transcription initiation: Promoter structures and RNA poymerases. Curr. Genet. 1995, 28, 205–216. [Google Scholar] [CrossRef]
- Duby, F.; Cardol, P.; Matgne, R.F.; Remacle, C. Structure of the telomeric ends of mt DNA, transcriptional analysis and complex I assembly in the dum24 mitochondrial mutant of Chlamydomonas reinhardtii. Mol. Genet. Genom. 2001, 266, 109–114. [Google Scholar] [CrossRef]
- Zimmer, S.L.; Schein, A.; Zipor, G.; Stern, D.B.; Schuster, G. Polyadenylation in Arabidopsis and Chlamydomonas organelles: The input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase. Plant J. 2009, 59, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Giegé, T.; Cavaiuolol, M.; Cognat, V.; Ubrig, E.; Remacle, C.; Duchêne, A.-M.; Vallon, O.; Maréchal-Drouard, L. Polycytidylation of mitochondrial mRNAs in Chlamydomonas reinhardtii. Nucleic Acids Res. 2017, 45, 12963–12973. [Google Scholar] [CrossRef]
- Cahoon, A.B.; Qureshi, A.A. Leaderless mRNAs are circularized in Chlamydomonas reinhardtii mitochondria. Curr. Genet. 2018, 64, 1321–1333. [Google Scholar] [CrossRef]
- Gallaher, S.D.; Fitz-Gibbon, S.T.; Strenkert, D.; Purvine, S.O.; Pellegrini, M.; Merchant, S.S. High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant J. 2017, 93, 545–565. [Google Scholar] [CrossRef]
- Levy, S.; Schuster, G. Polyadenylation and degradation of RNA in the mitochondria. Biochem. Soc. Trans. 2016, 44, 1475–1482. [Google Scholar] [CrossRef]
- Cahoon, A.B.; Nauss, J.A.; Stanley, C.D.; Qureshi, A. Deep Transcriptome Sequencing of Two Green Algae, Chara vulgaris and Chlamydomonas reinhardtii, Provides no Evidence of Organellar RNA Editing. Genes 2017, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Farwagi, A.; Fučíková, K.; McManus, H.A. Phylogenetic patterns of gene rearrangements in four mitochondrial genomes from the green algal family Hydrodictyaceae (Sphaeropleales, Chlorophyceae). BMC Genom. 2015, 16, 826. [Google Scholar] [CrossRef] [PubMed]
- Proulex, G.C.R.; Lor, B.; Manoylov, K.M.; Cahoon, A.B. The chloroplast and mitochondrial genomes of the green alga Pediastrum duplex isolated from central Georgia (USA). Mitochondrial DNA Part B Resour. 2019, 4, 3070–3071. [Google Scholar] [CrossRef]
- Turmel, M.; Otis, C.; Lemieux, C. The mitochondrial genome of Chara vulgaris: Insights into the mitochondrial DNA architecture or the last common ancestor of green algae and land plants. Plant Cell 2003, 15, 1888–1903. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Otis, C.; Lemieux, C. Tracing the evolution of streptophyte algae and their mitochondrial genome. Genome Biol. Evol. 2013, 5, 1817–1835. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDraw (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Boer, P.H.; Bonen, L.; Lee, R.W.; Gray, M.W. Genes for respiratory chain proteins and ribosomal RNAs are present on a 16-kilobase-pair DNA species from Chlamydomonas reinhardtii mitochondria. Proc. Natl. Acad. Sci. USA 1985, 3340–3344. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Lemieux, C.; Burger, G.; Lang, B.F.; Otis, C.; Plante, I.; Gray, M.W. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor: Two radically different evolutoionary patterns within green algae. Plant Cell 1999, 11, 1717–1729. [Google Scholar] [CrossRef] [PubMed]
- Nedelcu, A.M.; Lee, R.W.; Lemieux, C.; Gray, M.W.; Burger, G. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 2000, 10, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Robbens, S.; Derelle, E.; Ferraz, C.; Wuyts, J.; Moreau, H.; Van de Peer, Y. The complete chloroplast and mitochondrial DNA sequences of Ostreococcus tauri: Organelle genomes of the smallest eukaryote are examples of compaction. Mol. Biol. Evol. 2007, 24, 956–968. [Google Scholar] [CrossRef] [PubMed]
- Brouard, J.-S.; Otis, C.; Lemieux, C.; Turmel, M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): Unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genom. 2008, 9, 290. [Google Scholar] [CrossRef]
- Pombert, J.-F.; Otis, C.; Lemieux, C.; Turmel, M. The complete mitochondrial DNA sequence of the green alga Pseudenclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the Chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol. Biol. Evol. 2004, 21, 922–935. [Google Scholar] [CrossRef][Green Version]
- Pombert, J.-F.; Otis, C.; Turmel, M.; Lemieux, C. The mitochondrial genome of the prasinophyte Prasinoderma colonial reveals two trans-spliced group I introns in the large subunit rRNA gene. PLoS ONE 2013, 8, e84325. [Google Scholar] [CrossRef]
- Smith, D.R.; Hamaji, T.; Olson, B.J.S.C.; Durand, P.M.; Ferris, P.; Michod, R.E.; Featherston, J.; Nozaki, H.; Keeling, P.J. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol. Biol. Evol. 2013, 30, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lim, J.-M.; Park, J.; Sim, Y.M.; Choi, H.-G.; Lee, J.; Jeong, W.-J. Plastid and mitochondrion genomic sequences from arctic Chlorella sp. ArM0029B. BMC Genom. 2014, 15, 286. [Google Scholar] [CrossRef] [PubMed]
- Fučíková, K.; Lewis, P.O.; González-Halphen, D.; Lewis, L.A. Gene arrangement convergence, diverse intron content, and genetic code modifications in mitochondrial genomes of Sphaeropleales (Chlorophyta). Genome Biol. Evol. 2014, 6, 2170–2180. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 2018, 174, 448–464. [Google Scholar] [CrossRef]
- Karol, K.G.; McCourt, R.M.; Cimino, M.T.; Delwiche, C.F. The closest living relatives of land plants. Science 2001, 294, 2351–2353. [Google Scholar] [CrossRef] [PubMed]
- Guria, A.; Sharma, P.; Natesan, S.; Pandi, G. Circular RNAs—The road less traveled. Front. Mol. Biosci. 2020, 6, 146. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schriner, S.; Hung, L.-H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Circular RNAs: Biogenesis, Function and a Role as Possible Cancer Biomarkers. Int. J. Genom. 2017, 2017, 6218353. [Google Scholar] [CrossRef]
- Tippery, N.P.; Fučiková, K.; Lewis, P.O.; Lewis, L.A. Probing the monophyly of the Sphaeropleales (Chlorophyceae) using data from five genes. J. Phycol. 2012, 48, 1482–1493. [Google Scholar] [CrossRef]
- Arnal, N.; Quadrado, M.; Simon, M.; Mireau, H. A restorer-of-fertility like pentatricopeptide repeat gene directs ribonucleolytic processing within the coding sequence of rps3-rpl16 and orf240a mitochondrial transcripts in Arabidopsis thaliana. Plant J. 2014, 78, 134–145. [Google Scholar] [CrossRef]
- Jonietz, C.; Forner, J.; Hildebrandt, T.; Binder, S. RNA PROCESSING FACTOR 3 is crucial for the accumulation of mature ccmC transcripts in mitochondria of Arabidopsis thaliana accession Columbia. Plant Physiol. 2011, 157, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.; Stoll, K.; Steinhilber, J.; Jonietz, C.; Binder, S. Mitochondrial transcript length polymorphisms are a widespread phenomenon in Arabidopsis thaliana. Plant Mol. Biol. 2012, 1, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.; Zendler, D.; Binder, S. RNA processing factor 7 and polynucleotide phosphorylase are necessary for processing and stability of nad2 mRNA in Arabidopsis mitochondria. RNA Biol. 2014, 11, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Stoll, K.; Jonietz, C.; Binder, S. In Arabidopsis thaliana two co-adapted cyto-nuclear systems correlate with distinct ccmC transcript sizes. Plant J. 2015, 81, 247–257. [Google Scholar] [CrossRef]
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsässer, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kühn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 2019, 101, 420–441. [Google Scholar] [CrossRef]
- Darbani, B.; Noeparvar, S.; Borg, S. Identification of circular RNAs from parental genes involved in multiple aspects of cellular metabolism in barley. Front. Plant Sci. 2016, 7, 776. [Google Scholar] [CrossRef]
- Sun, X.; Wang, X.; Ding, J.; Wang, Y.; Wang, J.; Zhang, X.; Che, Y.; Liu, Z.; Zhang, X.; Ye, J.; et al. Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett. 2016, 590, 3510–3516. [Google Scholar] [CrossRef]
- Bischoff, H.W.; Bold, H.C. Phycological studies. IV. Some soil algae from Enchanted Rock and related algal species. Univ. Tex. Publ. 1963, 6318, 1–95. [Google Scholar]
- Meade, M.J.; Proulex, G.C.R.; Manoylov, K.M.; Cahoon, A.B. Chloroplast MRNAs are 3′ Polyuridylylated in The Green Alga Pithophora roettleri (Cladophorales). J. Phycol. 2020, 56, 1124–1134. [Google Scholar] [CrossRef]



| Site of 5′ UTR Terminus Upstream from Start Codon (in Nucleotides) | ||
|---|---|---|
| Gene | Circular RT-PCR | PacBio |
| atp6 | −2–11 | nd |
| atp9 | −3 * | nd |
| cob | −2 * | 0 |
| cox1 | nd | 0 |
| cox2a | 0 | nd |
| cox3 | 0 | 0 |
| nad1 | −15 * | nd |
| nad2 | 0 | nd |
| nad4 | 0 | 0 |
| nad4L | 0 | 0 |
| −81–93 | nd | |
| nad5 | nd | nd |
| nad6 | nd | 0 |
| Circular RT-PCR | PacBio Long Reads | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Length of 3′ UTR from Stop Codon | UTRs with polyC Addition | UTRs with polyA + polyC Addition | UTR with polyA Addition | Length of 3′ UTR from Stop Codon | UTRs with polyC Addition | Genes with an Adjacent tRNA (Distance from Stop Codon in nt) |
| atp6 | 21 | + | - | - | nd | nd | |
| 0–9 | - | - | - | nd | nd | ||
| atp9 | 22 | + | - | - | 16–22 | + | 26 |
| 25 | - | - | - | nd | nd | ||
| cob | 110 | - | - | - | 104–110 | + | 110 |
| 100 | + | - | - | nd | nd | ||
| cox1 | 13 | + | - | - | 7–14 | + | |
| cox2a | 10 | + | - | - | nd | nd | 28 |
| cox3 | 13 | + | - | - | 10–13 | - | |
| nad1 | 18 | - | - | - | nd | nd | |
| nad2 | 9–21 | + | - | - | 5–23 | - | |
| nad4 | 8 | - | - | - | 7–8 | + | 786 |
| nad4L | −1 | - | - | - | 37 | - | |
| nad5 | 0–8 | - | - | + | 1–3 | - | 42 |
| 9–24 | + | + | |||||
| nad6 | nd | nd | nd | nd | nd | nd | |
| Site of 5′ UTR Terminus Upstream from Start Codon (in Nucleotides) | ||
|---|---|---|
| Gene | Circular RT-PCR | PacBio |
| atp6 | −76 | nd |
| −62 | ||
| atp9 | −56 | −93 |
| cob | −161 | nd |
| −46 | ||
| cox1 | −150 * | nd |
| cox2 | −318 | −52 |
| −18 | ||
| cox3 | −153 | nd |
| −31 | ||
| nad1 | −73 * | nd |
| −27 * | ||
| −8 * | ||
| nad2 | −112 * | nd |
| nad4 | −273 | nd |
| −107 | ||
| −99 | ||
| −82 | ||
| −18 | ||
| −6 | ||
| nad4L | −260 | nd |
| −18 | ||
| nad5 | nd | nd |
| nad6 | −36 | nd |
| Circular RT-PCR | PacBio | ||||
|---|---|---|---|---|---|
| Gene | Length of 3′ UTR from Stop Codon | UTRs with polyA Addition | Length of 3′ UTR from Stop Codon | Genes with an Adjacent tRNA (Distance from Stop Codon in nt) | Evidence of RNA Secondary Structure Immediately Downstream of 3′ Terminus |
| atp6 | 79 | + | nd | 459 | + |
| 81 | + | ||||
| 83 | + | ||||
| atp9 | 52–57 | + | 56 | 92 | - |
| 58 | - | ||||
| cob | 54 | - | 121 | - | |
| 68 | + | ||||
| cox1 | 73 | - | 73 | ||
| cox2 | 51 | - | nd | - | |
| 82 | - | ||||
| 107 | - | ||||
| 162 | - | ||||
| cox3 | 23 | + | 52 | 6056 | - |
| 51 | - | ||||
| 56 | - | ||||
| nad1 | 20 | - | nd | + | |
| 67 | - | ||||
| 70 | + | ||||
| 141 | - | ||||
| nad2 | 87 | + | nd | 1189 | + |
| nad4 | 10 | + | nd | - | |
| 13 | - | ||||
| 21 | - | ||||
| nad4L | 0 | - | nd | 2508 | - |
| 29 | + | ||||
| nad5 | nd | nd | nd | ||
| nad6 | nd | nd | nd | 5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proulex, G.C.R.; Meade, M.J.; Manoylov, K.M.; Cahoon, A.B. Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing. Plants 2021, 10, 576. https://doi.org/10.3390/plants10030576
Proulex GCR, Meade MJ, Manoylov KM, Cahoon AB. Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing. Plants. 2021; 10(3):576. https://doi.org/10.3390/plants10030576
Chicago/Turabian StyleProulex, Grayson C. R., Marcus J. Meade, Kalina M. Manoylov, and A. Bruce Cahoon. 2021. "Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing" Plants 10, no. 3: 576. https://doi.org/10.3390/plants10030576
APA StyleProulex, G. C. R., Meade, M. J., Manoylov, K. M., & Cahoon, A. B. (2021). Mitochondrial mRNA Processing in the Chlorophyte Alga Pediastrum duplex and Streptophyte Alga Chara vulgaris Reveals an Evolutionary Branch in Mitochondrial mRNA Processing. Plants, 10(3), 576. https://doi.org/10.3390/plants10030576






