A Core Module of Nuclear Genes Regulated by Biogenic Retrograde Signals from Plastids
Abstract
1. Introduction
2. Material and Methods
2.1. Sets of Retrograde Controlled Genes Used in This Study
2.2. Comparison of Gene Lists
2.3. Functional Annotation and Localization
3. Results and Discussion
3.1. Selection and Comparability of Microarray Data Sets
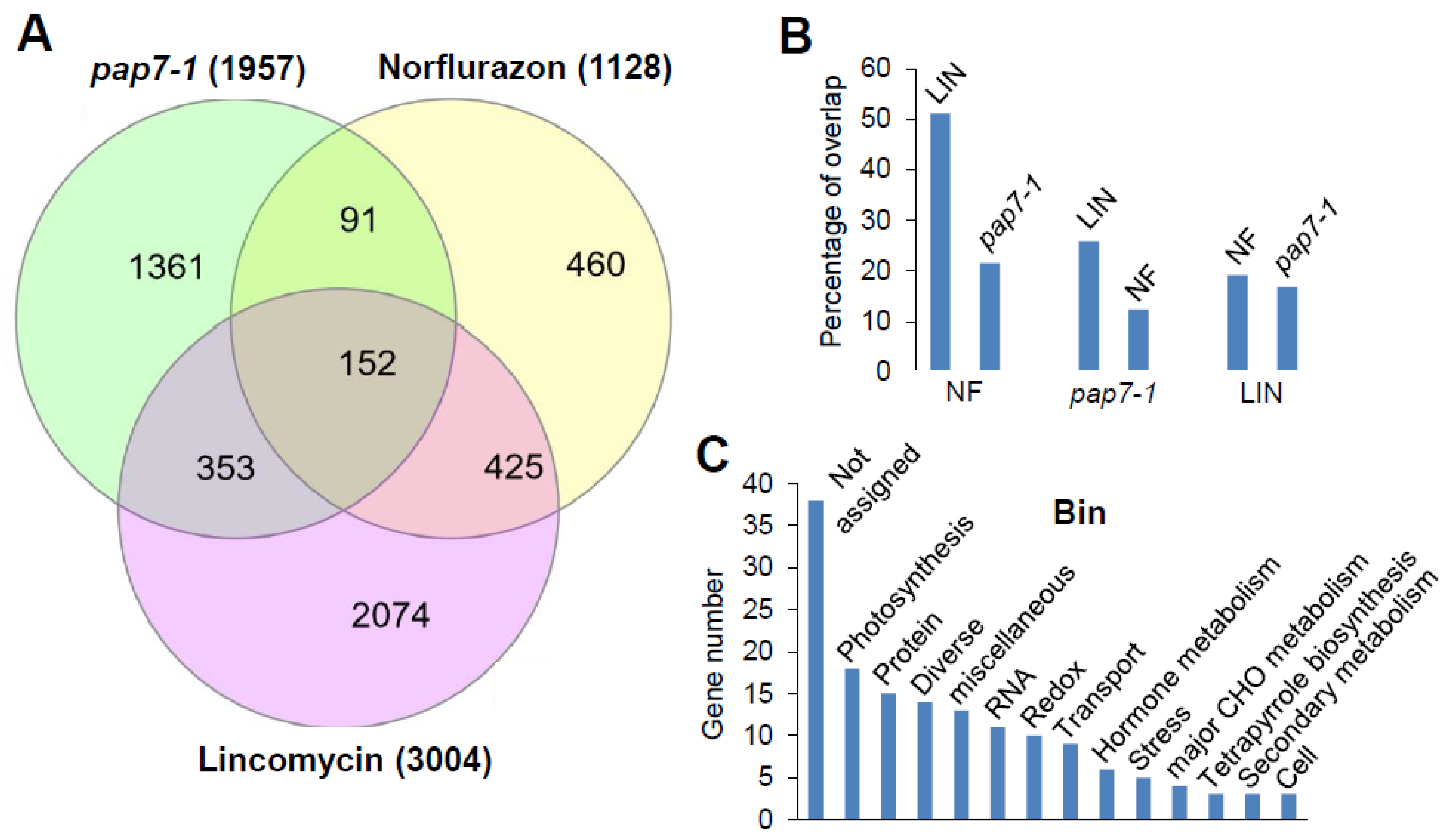
3.2. Identification of a Core-Module of Genes Controlled by Biogenic Signals
3.3. Functional Subsets within the Core Module
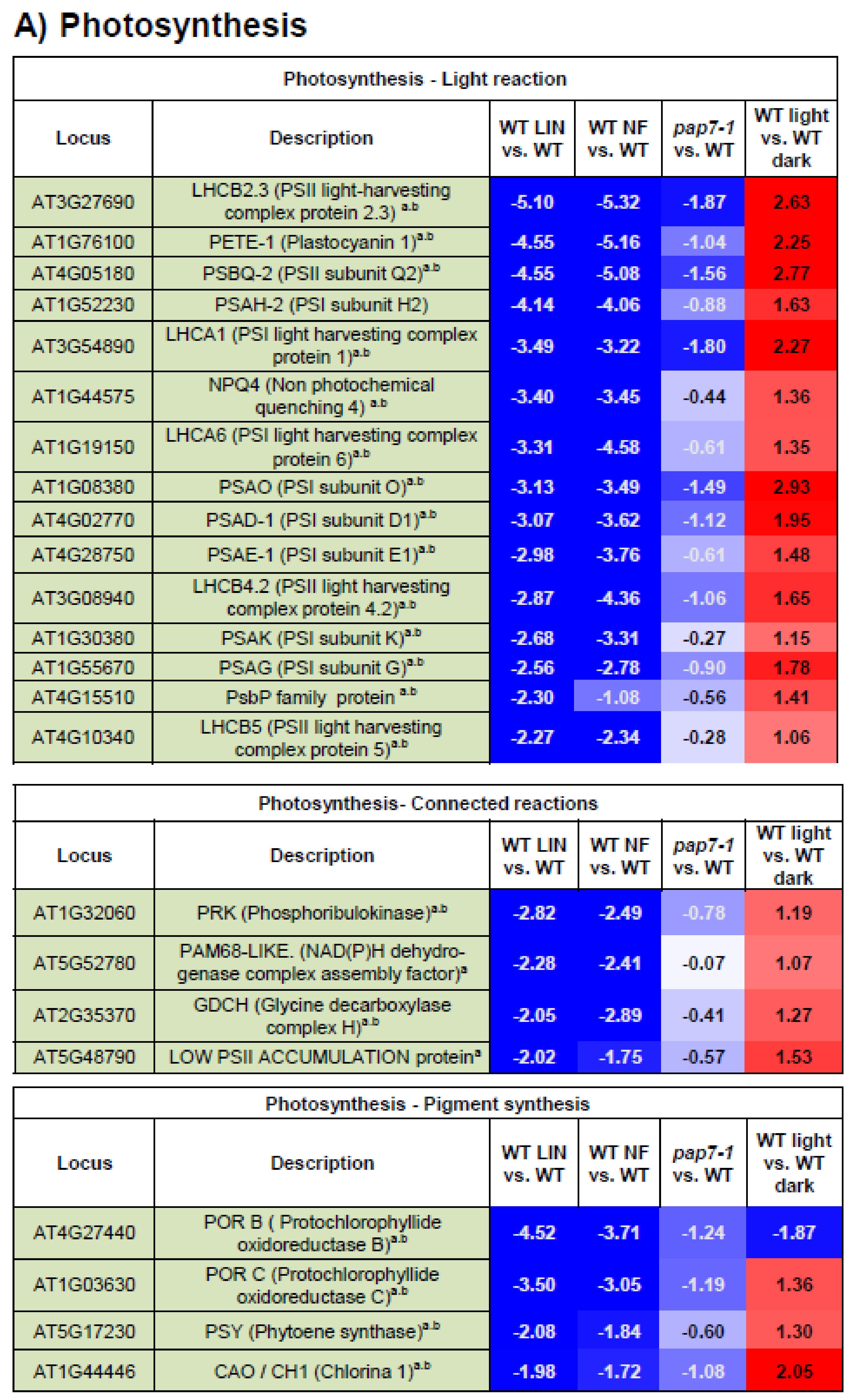
3.4. Photosynthesis
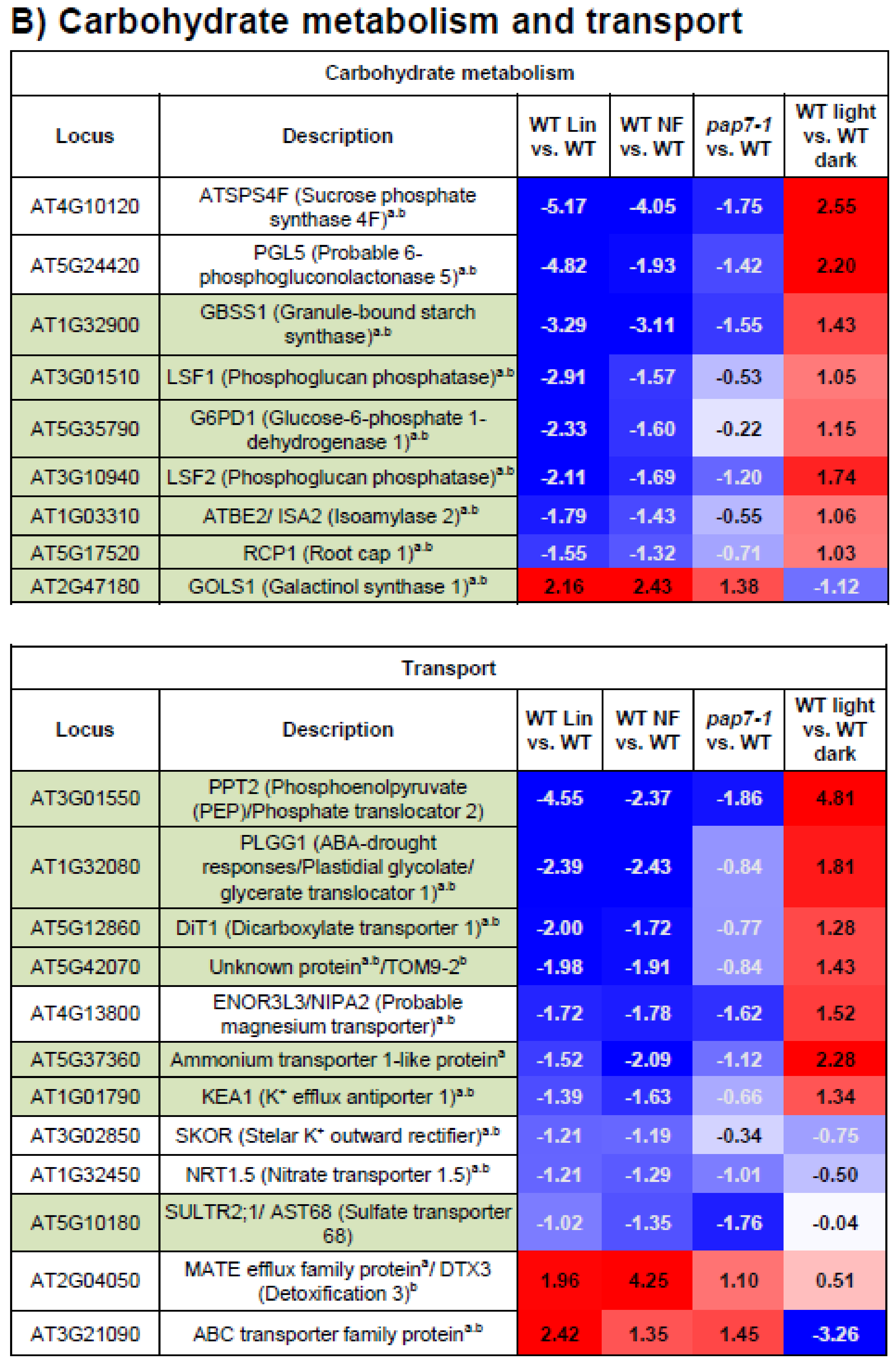
3.5. Carbohydrate Metabolism and Transport
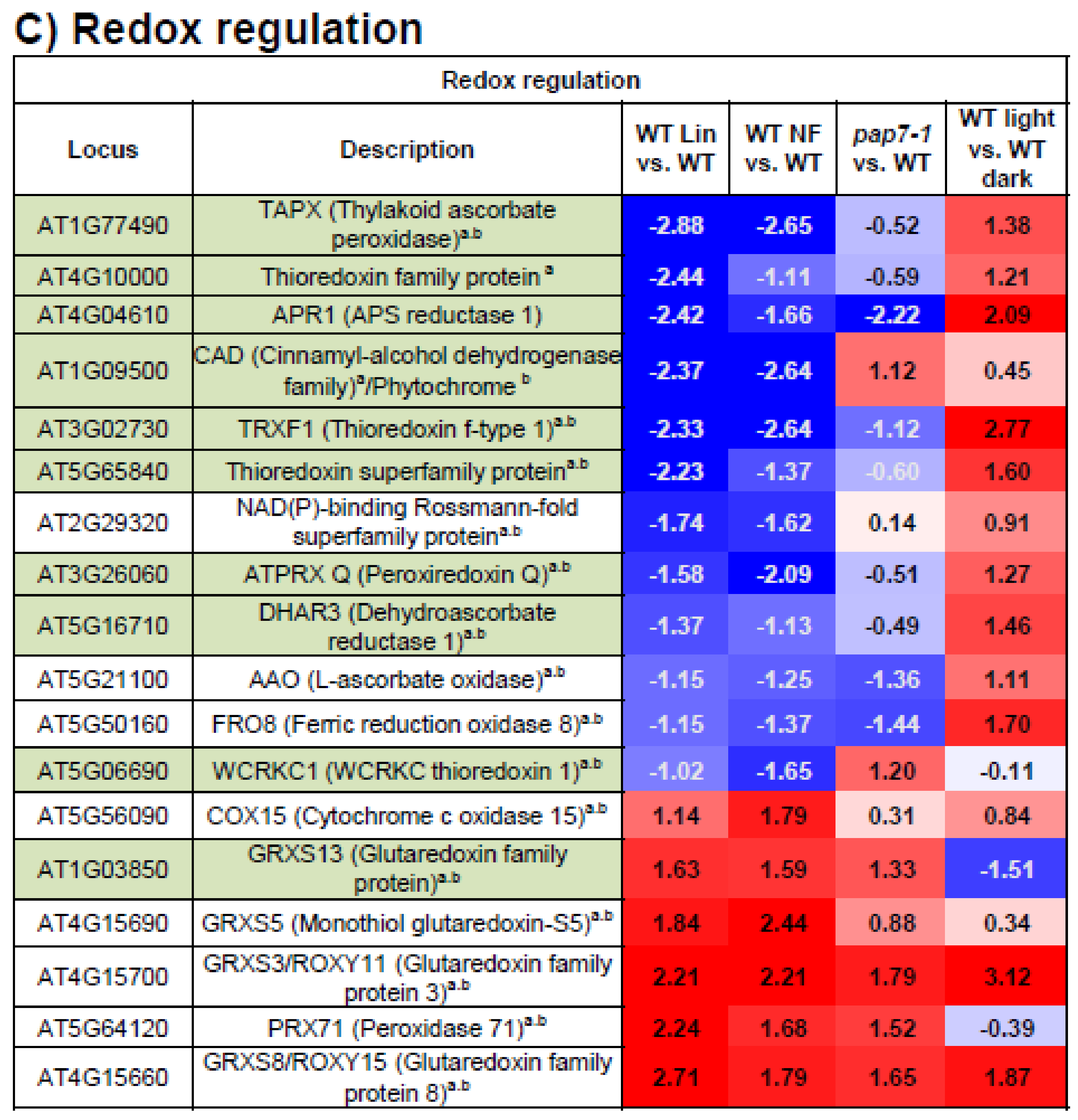
3.6. Redox Regulation
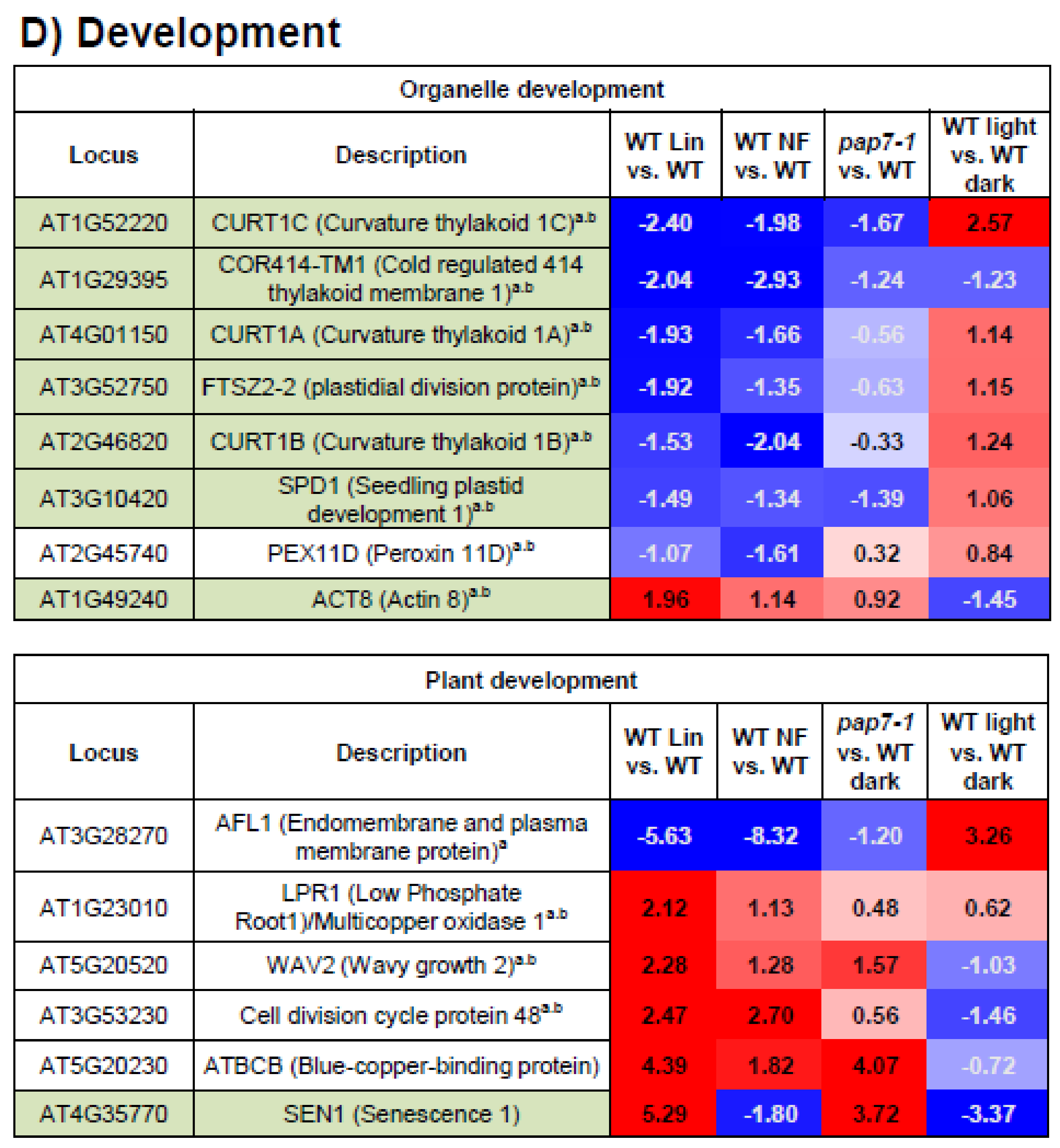
3.7. Development
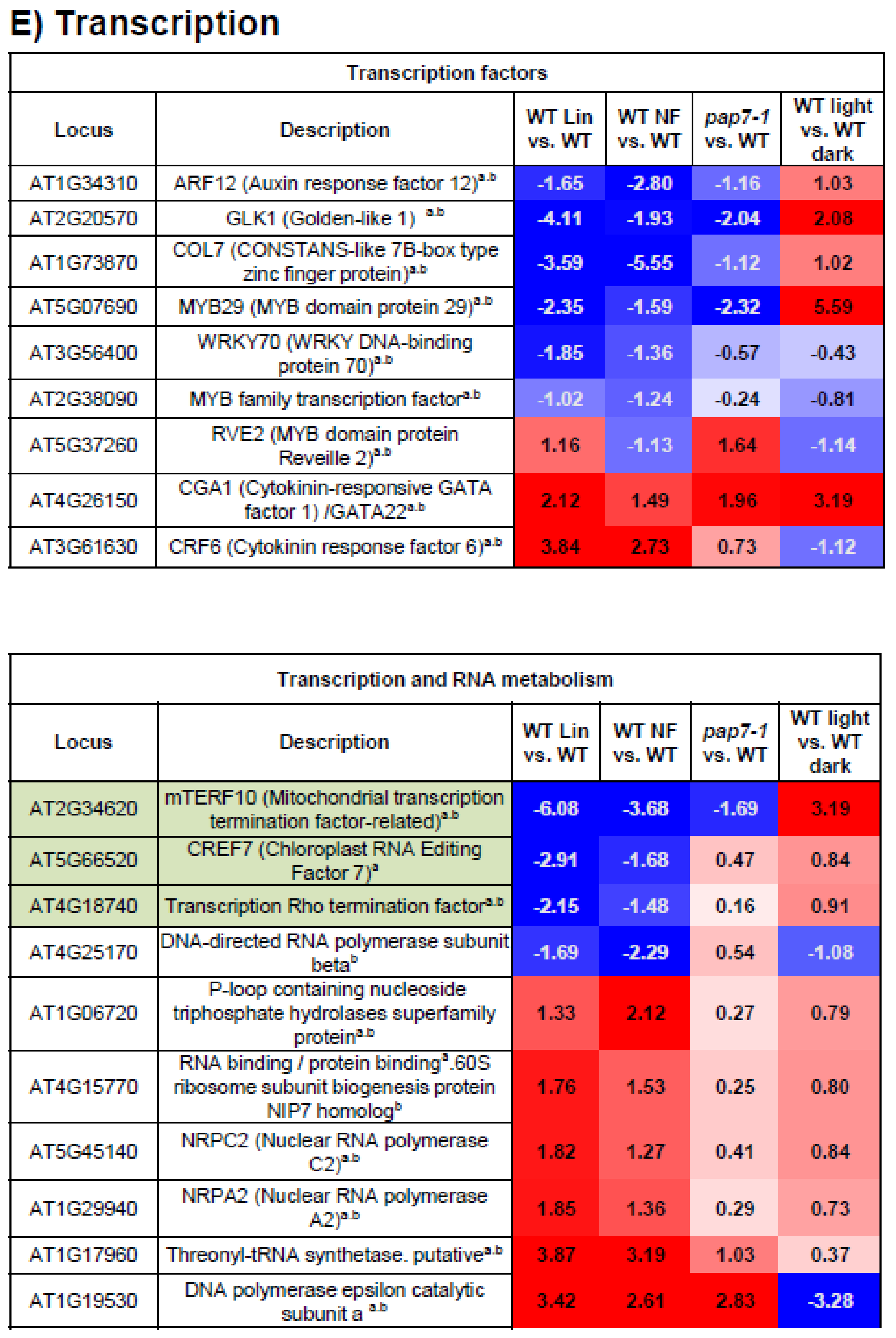
3.8. Transcription
3.9. Proteins and Stress
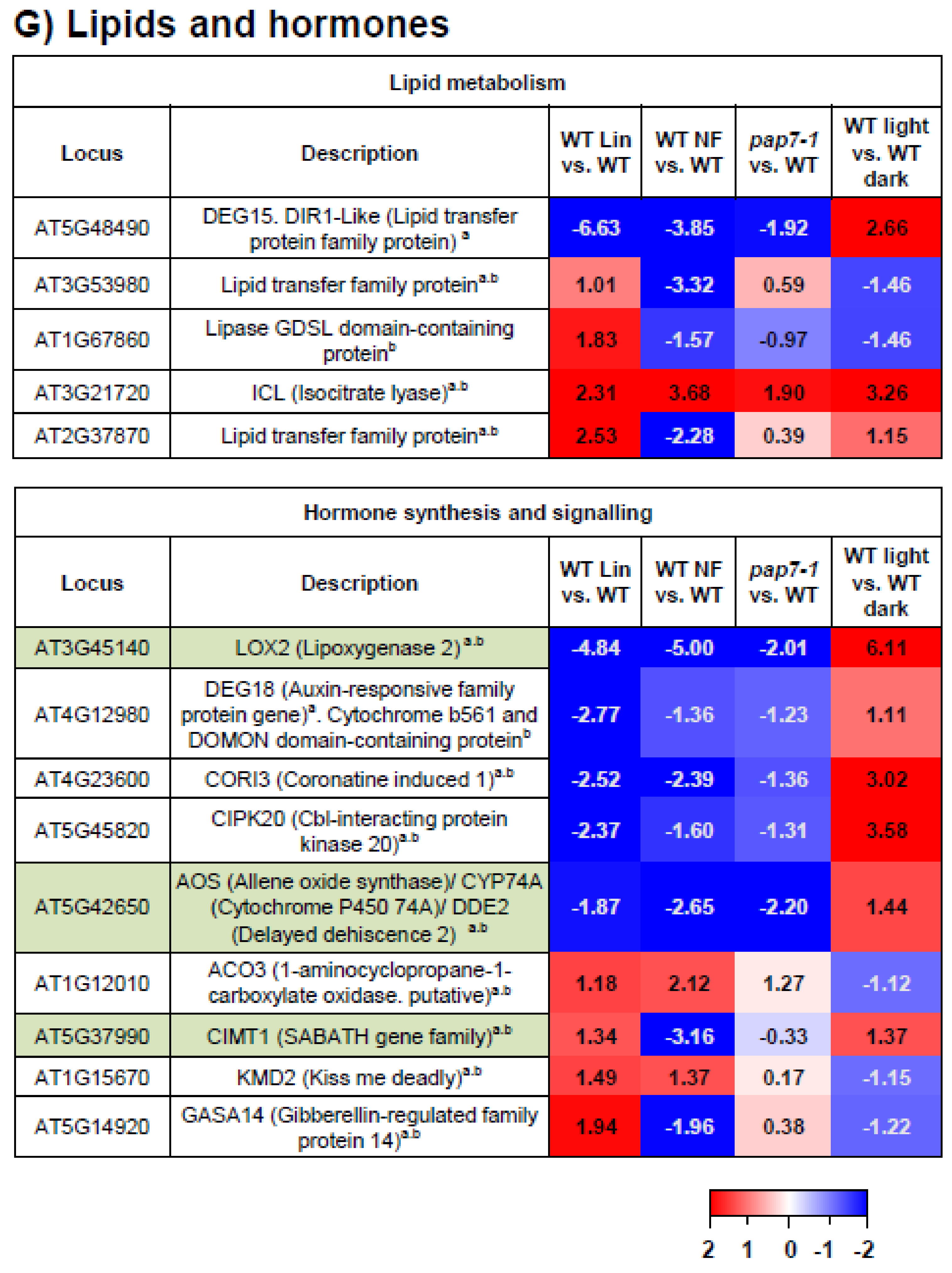
3.10. Lipids and Hormones
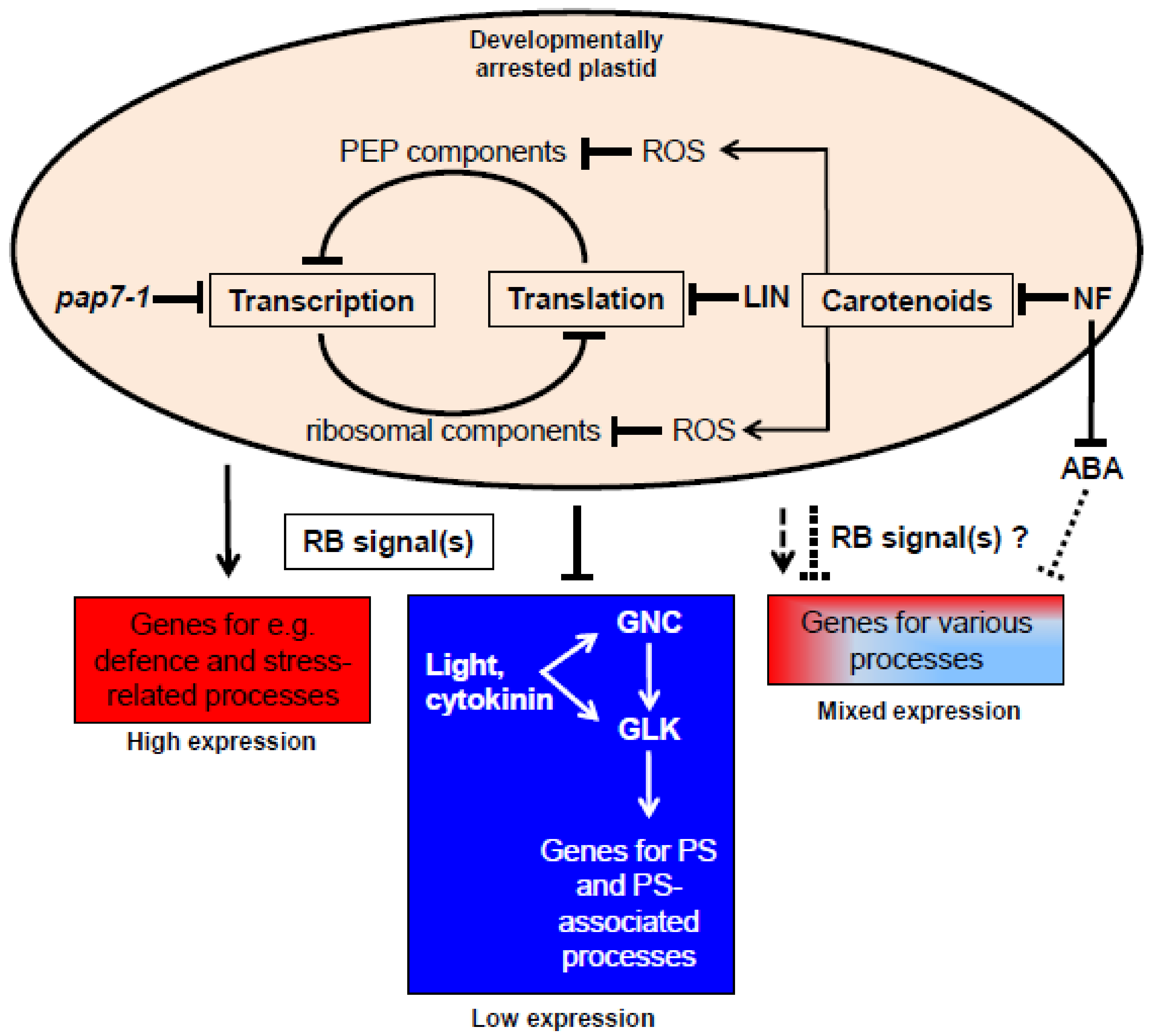
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787. [Google Scholar] [CrossRef]
- Liebers, M.; Grubler, B.; Chevalier, F.; Lerbs-Mache, S.; Merendino, L.; Blanvillain, R.; Pfannschmidt, T. Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front. Plant. Sci. 2017, 8, 23. [Google Scholar] [CrossRef]
- Solymosi, K.; Schoefs, B. Etioplast and etio-chloroplast formation under natural conditions: The dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 2010, 105, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, C.; Jin, X.; Barajas-Lopez, J.D.; Hewitt, T.C.; Tanz, S.K.; Dobrenel, T.; Schroder, W.P.; Hanson, J.; Pesquet, E.; Gronlund, A.; et al. Establishment of photosynthesis through chloroplast development is controlled by two distinct regulatory phases. Plant. Physiol. 2018, 176, 1199–1214. [Google Scholar] [CrossRef] [PubMed]
- Armarego-Marriott, T.; Kowalewska, L.; Burgos, A.; Fischer, A.; Thiele, W.; Erban, A.; Strand, D.; Kahlau, S.; Hertle, A.; Kopka, J.; et al. Highly resolved systems biology to dissect the etioplast-to-chloroplast transition in tobacco leaves. Plant. Physiol. 2019, 180, 654–681. [Google Scholar] [CrossRef] [PubMed]
- Armarego-Marriott, T.; Sandoval-Ibanez, O.; Kowalewska, L. Beyond the darkness: Recent lessons from etiolation and de-etiolation studies. J. Exp. Bot. 2020, 71, 1215–1225. [Google Scholar] [CrossRef]
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 2015, 1847, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Han, S.; Chen, M. Nucleus-to-plastid phytochrome signalling in controlling chloroplast biogenesis. Ann. Plant. Rev. 2020, 3, 251–280. [Google Scholar]
- Deng, X.W.; Matsui, M.; Wei, N.; Wagner, D.; Chu, A.M.; Feldmann, K.A.; Quail, P.H. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 1992, 71, 791–801. [Google Scholar] [CrossRef]
- Pfalz, J.; Pfannschmidt, T. Essential nucleoid proteins in early chloroplast development. Trends Plant. Sci. 2013, 18, 186–194. [Google Scholar] [CrossRef]
- Borner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 2015, 1847, 761–769. [Google Scholar] [CrossRef]
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant. Biol. 2016, 67, 25–53. [Google Scholar] [CrossRef]
- De Souza, A.; Wang, J.Z.; Dehesh, K. Retrograde Signals: Integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant. Biol. 2016, 68, 85–108. [Google Scholar] [CrossRef]
- Grimm, B.; Dehesh, K.; Zhang, L.; Leister, D. Intracellular communication. Mol. Plant. 2014, 7, 1071–1074. [Google Scholar] [CrossRef]
- Chi, W.; Sun, X.; Zhang, L. Intracellular signaling from plastid to nucleus. Annu. Rev. Plant. Biol. 2013, 64, 559–582. [Google Scholar] [CrossRef]
- Inaba, T.; Yazu, F.; Ito-Inaba, Y.; Kakizaki, T.; Nakayama, K. Retrograde signaling pathway from plastid to nucleus. Int. Rev. Cell Mol. Biol. 2011, 290, 167–204. [Google Scholar] [PubMed]
- Pfannschmidt, T.; Terry, M.J.; Van Aken, O.; Quiros, P.M. Retrograde signals from endosymbiotic organelles: A common control principle in eukaryotic cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190396. [Google Scholar] [CrossRef] [PubMed]
- Pogson, B.J.; Woo, N.S.; Forster, B.; Small, I.D. Plastid signalling to the nucleus and beyond. Trends Plant. Sci. 2008, 13, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Bradbeer, J.W.; Atkinson, Y.E.; Börner, T.; Hagemann, R. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid synthesized RNA. Nature 1979, 279, 816–817. [Google Scholar] [CrossRef]
- Oelmuller, R.; Levitan, I.; Bergfeld, R.; Rajasekhar, V.K.; Mohr, H. Expression of nuclear genes as affected by treatments acting on the plastids. Planta 1986, 168, 482–492. [Google Scholar] [CrossRef]
- Pfannschmidt, T. Plastidial retrograde signalling—A true “plastid factor” or just metabolite signatures? Trends Plant. Sci. 2010, 15, 427–435. [Google Scholar] [CrossRef]
- Hernandez-Verdeja, T.; Strand, A. Retrograde signals navigate the path to chloroplast development. Plant. Physiol. 2018, 176, 967–976. [Google Scholar] [CrossRef]
- Susek, R.E.; Ausubel, F.M.; Chory, J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 1993, 74, 787–799. [Google Scholar] [CrossRef]
- Sullivan, J.A.; Gray, J.C. Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant. Cell 1999, 11, 901–910. [Google Scholar] [PubMed]
- Grubler, B.; Merendino, L.; Twardziok, S.O.; Mininno, M.; Allorent, G.; Chevalier, F.; Liebers, M.; Blanvillain, R.; Mayer, K.; Lerbs-Mache, S.; et al. Light and plastid signals regulate different sets of genes in the albino mutant pap7-1. Plant. Physiol. 2017, 175, 1203–1219. [Google Scholar] [CrossRef]
- Page, M.T.; McCormac, A.C.; Smith, A.G.; Terry, M.J. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol. 2017, 213, 1168–1180. [Google Scholar] [CrossRef]
- Ruckle, M.E.; Burgoon, L.D.; Lawrence, L.A.; Sinkler, C.A.; Larkin, R.M. Plastids are major regulators of light signaling in Arabidopsis. Plant. Physiol. 2012, 159, 366–390. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant. Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Bruley, C.; Dupierris, V.; Salvi, D.; Rolland, N.; Ferro, M. AT_CHLORO: A chloroplast protein database dedicated to sub-plastidial localization. Front. Plant. Sci. 2012, 3, 205. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Nagel, A.; Thimm, O.; Redestig, H.; Blaesing, O.E.; Palacios-Rojas, N.; Selbig, J.; Hannemann, J.; Piques, M.C.; Steinhauser, D.; et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant. Physiol. 2005, 138, 1195–1204. [Google Scholar] [CrossRef]
- Ruckle, M.E.; DeMarco, S.M.; Larkin, R.M. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant. Cell 2007, 19, 3944–3960. [Google Scholar] [CrossRef]
- Burko, Y.; Seluzicki, A.; Zander, M.; Pedmale, U.V.; Ecker, J.R.; Chory, J. Chimeric Activators and Repressors Define HY5 Activity and Reveal a Light-Regulated Feedback Mechanism. Plant. Cell 2020, 32, 967–983. [Google Scholar] [CrossRef]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant. Cell 2001, 13, 2589–2607. [Google Scholar] [CrossRef] [PubMed]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Jang, J.H.; Shang, Y.; Kang, H.K.; Kim, S.Y.; Kim, B.H.; Nam, K.H. Arabidopsis galactinol synthases 1 (AtGOLS1) negatively regulates seed germination. Plant. Sci. 2018, 267, 94–101. [Google Scholar] [CrossRef]
- Dietz, K.J.; Pfannschmidt, T. Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant. Physiol. 2011, 155, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, N.; Holtmannspotter, M.; Mass, L.; O’Donoghue, M.; Busch, A.; Lauri, A.; Schubert, V.; Zachgo, S. Conserved redox-dependent DNA binding of ROXY glutaredoxins with TGA transcription factors. Plant. Direct. 2017, 1, e00030. [Google Scholar] [CrossRef] [PubMed]
- Ndamukong, I.; Abdallat, A.A.; Thurow, C.; Fode, B.; Zander, M.; Weigel, R.; Gatz, C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant. J. 2007, 50, 128–139. [Google Scholar] [CrossRef]
- Armbruster, U.; Labs, M.; Pribil, M.; Viola, S.; Xu, W.; Scharfenberg, M.; Hertle, A.P.; Rojahn, U.; Jensen, P.E.; Rappaport, F.; et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant. Cell 2013, 25, 2661–2678. [Google Scholar] [CrossRef]
- Pribil, M.; Labs, M.; Leister, D. Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 2014, 65, 1955–1972. [Google Scholar] [CrossRef]
- Ruppel, N.J.; Logsdon, C.A.; Whippo, C.W.; Inoue, K.; Hangarter, R.P. A mutation in Arabidopsis seedling plastid development1 affects plastid differentiation in embryo-derived tissues during seedling growth. Plant. Physiol. 2011, 155, 342–353. [Google Scholar] [CrossRef][Green Version]
- Lingard, M.J.; Gidda, S.K.; Bingham, S.; Rothstein, S.J.; Mullen, R.T.; Trelease, R.N. Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant. Cell 2008, 20, 1567–1585. [Google Scholar] [CrossRef]
- Sheahan, M.B.; Collings, D.A.; Rose, R.J.; McCurdy, A.D.W. ACTIN7 is required for perinuclear clustering of chloroplasts during arabidopsis protoplast culture. Plants 2020, 9, 225. [Google Scholar] [CrossRef]
- Fernandez-Calvino, L.; Guzman-Benito, I.; Del Toro, F.J.; Donaire, L.; Castro-Sanz, A.B.; Ruiz-Ferrer, V.; Llave, C. Activation of senescence-associated Dark-inducible (DIN) genes during infection contributes to enhanced susceptibility to plant viruses. Mol. Plant. Pathol. 2016, 17, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Lee, S.Y.; Chung, I.K.; Lee, C.H.; Nam, H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant. Mol. Biol. 1996, 30, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Kacprzak, S.M.; Mochizuki, N.; Naranjo, B.; Xu, D.; Leister, D.; Kleine, T.; Okamoto, H.; Terry, M.J. Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant. Physiol. 2019, 179, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant. Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef] [PubMed]
- Liebers, M.; Gillet, F.X.; Israel, A.; Pounot, K.; Chambon, L.; Chieb, M.; Chevalier, F.; Ruedas, R.; Favier, A.; Gans, P.; et al. Nucleo-plastidic PAP8/pTAC6 couples chloroplast formation with photomorphogenesis. EMBO J. 2020, 39, e104941. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, D.L.; Graham, N.S.; Klinder, A.; van Ommen Kloeke, A.E.E.; Marcotrigiano, A.R.; Wagstaff, C.; Verkerk, R.; Sonnante, G.; Aarts, M.G.M. Overexpression of the MYB29 transcription factor affects aliphatic glucosinolate synthesis in Brassica oleracea. Plant. Mol. Biol. 2019, 101, 65–79. [Google Scholar] [CrossRef]
- Zhang, X.; Ivanova, A.; Vandepoele, K.; Radomiljac, J.; Van de Velde, J.; Berkowitz, O.; Willems, P.; Xu, Y.; Ng, S.; Van Aken, O.; et al. The transcription factor MYB29 is a regulator of ALTERNATIVE OXIDASE1a. Plant. Physiol. 2017, 173, 1824–1843. [Google Scholar] [CrossRef]
- Jiang, Z.; Xu, G.; Jing, Y.; Tang, W.; Lin, R. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 2016, 7, 12377. [Google Scholar] [CrossRef]
- Lopez-Juez, E. Plastid biogenesis, between light and shadows. J. Exp. Bot. 2007, 58, 11–26. [Google Scholar] [CrossRef]
- Zubo, Y.O.; Blakley, I.C.; Franco-Zorrilla, J.M.; Yamburenko, M.V.; Solano, R.; Kieber, J.J.; Loraine, A.E.; Schaller, G.E. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant. Physiol. 2018, 178, 130–147. [Google Scholar] [CrossRef]
- Andreeva, A.A.; Vankova, R.; Bychkov, I.A.; Kudryakova, N.V.; Danilova, M.N.; Lacek, J.; Pojidaeva, E.S.; Kusnetsov, V.V. Cytokinin-regulated expression of arabidopsis thaliana PAP genes and its implication for the expression of chloroplast-encoded genes. Biomolecules 2020, 10, 1658. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Huang, J.; Chory, J. Unraveling the linkage between retrograde signaling and RNA metabolism in plants. Trends Plant. Sci. 2020, 25, 141–147. [Google Scholar] [CrossRef]
- Petrillo, E.; Godoy Herz, M.A.; Fuchs, A.; Reifer, D.; Fuller, J.; Yanovsky, M.J.; Simpson, C.; Brown, J.W.; Barta, A.; Kalyna, M.; et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science 2014, 344, 427–430. [Google Scholar] [CrossRef]
- Fang, X.; Zhao, G.; Zhang, S.; Li, Y.; Gu, H.; Li, Y.; Zhao, Q.; Qi, Y. Chloroplast-to-Nucleus Signaling Regulates MicroRNA Biogenesis in Arabidopsis. Dev. Cell 2019, 48, 371–382.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Huang, J.; Chory, J. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 10162–10167. [Google Scholar] [CrossRef] [PubMed]
- Llamas, E.; Pulido, P.; Rodriguez-Concepcion, M. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet. 2017, 13, e1007022. [Google Scholar] [CrossRef]
- Wu, G.Z.; Meyer, E.H.; Richter, A.S.; Schuster, M.; Ling, Q.; Schottler, M.A.; Walther, D.; Zoschke, R.; Grimm, B.; Jarvis, R.P.; et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 2019, 5, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Beaugelin, I.; Chevalier, A.; D’Alessandro, S.; Ksas, B.; Havaux, M. Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. Plant. J. 2020, 102, 1266–1280. [Google Scholar] [CrossRef]
- Tokumaru, M.; Adachi, F.; Toda, M.; Ito-Inaba, Y.; Yazu, F.; Hirosawa, Y.; Sakakibara, Y.; Suiko, M.; Kakizaki, T.; Inaba, T. Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastids signals. Plant. Physiol. 2017, 173, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Maynard, D.; Groger, H.; Dierks, T.; Dietz, K.J. The function of the oxylipin 12-oxophytodienoic acid in cell signaling, stress acclimation, and development. J. Exp. Bot. 2018, 69, 5341–5354. [Google Scholar] [CrossRef]
- Glasser, C.; Haberer, G.; Finkemeier, I.; Pfannschmidt, T.; Kleine, T.; Leister, D.; Dietz, K.J.; Hausler, R.E.; Grimm, B.; Mayer, K.F. Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol. Plant. 2014, 7, 1167–1190. [Google Scholar] [CrossRef] [PubMed]
- Leister, D.; Romani, I.; Mittermayr, L.; Paieri, F.; Fenino, E.; Kleine, T. Identification of target genes and transcription factors implicated in translation-dependent retrograde signaling in Arabidopsis. Mol. Plant. 2014, 7, 1228–1247. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leister, D.; Kleine, T. Definition of a core module for the nuclear retrograde response to altered organellar gene expression identifies GLK overexpressors as gun mutants. Physiol. Plant. 2016, 157, 297–309. [Google Scholar] [CrossRef]
- Terry, M.J.; Smith, A.G. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant. Sci. 2013, 4, 14. [Google Scholar] [CrossRef]
- Chen, M.; Galvao, R.M.; Li, M.; Burger, B.; Bugea, J.; Bolado, J.; Chory, J. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 2010, 141, 1230–1240. [Google Scholar] [CrossRef]
- Yang, E.J.; Yoo, C.Y.; Liu, J.; Wang, H.; Cao, J.; Li, F.W.; Pryer, K.M.; Sun, T.P.; Weigel, D.; Zhou, P.; et al. NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat. Commun. 2019, 10, 2630. [Google Scholar] [CrossRef]
- Krupinska, K.; Blanco, N.E.; Oetke, S.; Zottini, M. Genome communication in plants mediated by organelle-n-ucleus-located proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190397. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grübler, B.; Cozzi, C.; Pfannschmidt, T. A Core Module of Nuclear Genes Regulated by Biogenic Retrograde Signals from Plastids. Plants 2021, 10, 296. https://doi.org/10.3390/plants10020296
Grübler B, Cozzi C, Pfannschmidt T. A Core Module of Nuclear Genes Regulated by Biogenic Retrograde Signals from Plastids. Plants. 2021; 10(2):296. https://doi.org/10.3390/plants10020296
Chicago/Turabian StyleGrübler, Björn, Carolina Cozzi, and Thomas Pfannschmidt. 2021. "A Core Module of Nuclear Genes Regulated by Biogenic Retrograde Signals from Plastids" Plants 10, no. 2: 296. https://doi.org/10.3390/plants10020296
APA StyleGrübler, B., Cozzi, C., & Pfannschmidt, T. (2021). A Core Module of Nuclear Genes Regulated by Biogenic Retrograde Signals from Plastids. Plants, 10(2), 296. https://doi.org/10.3390/plants10020296




