A Roadmap to Modulated Anthocyanin Compositions in Carrots
Abstract
1. Introduction
2. Genetic Transformation Tools Suggested for Modulating Anthocyanin Compositions in Carrots
3. Genes Responsible for Anthocyanin Biosynthesis in Black Carrots
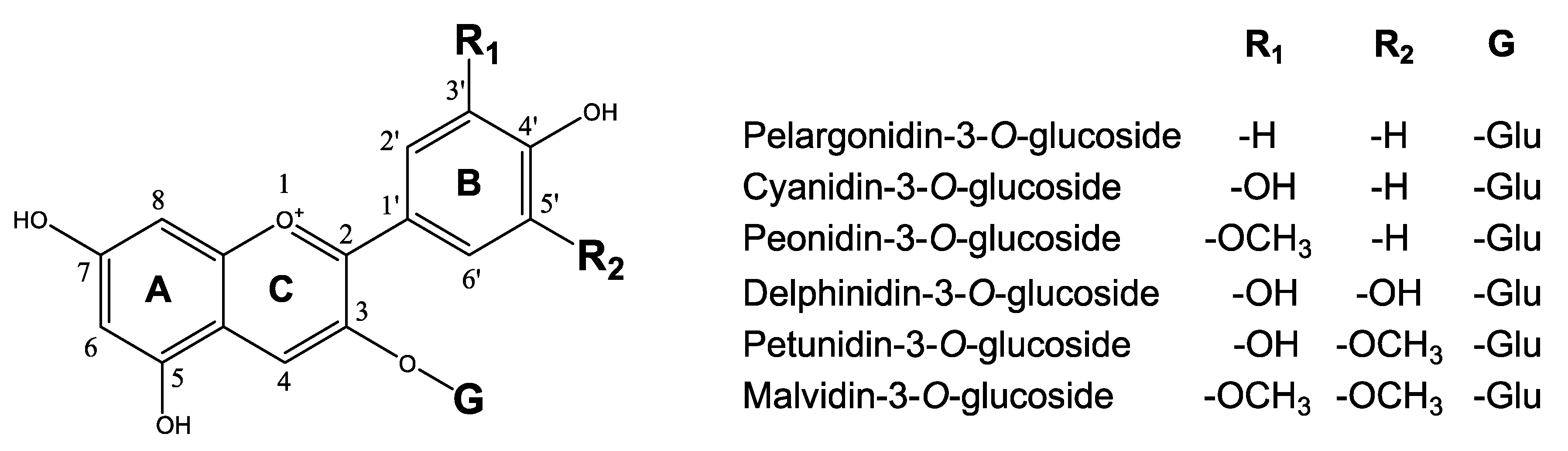
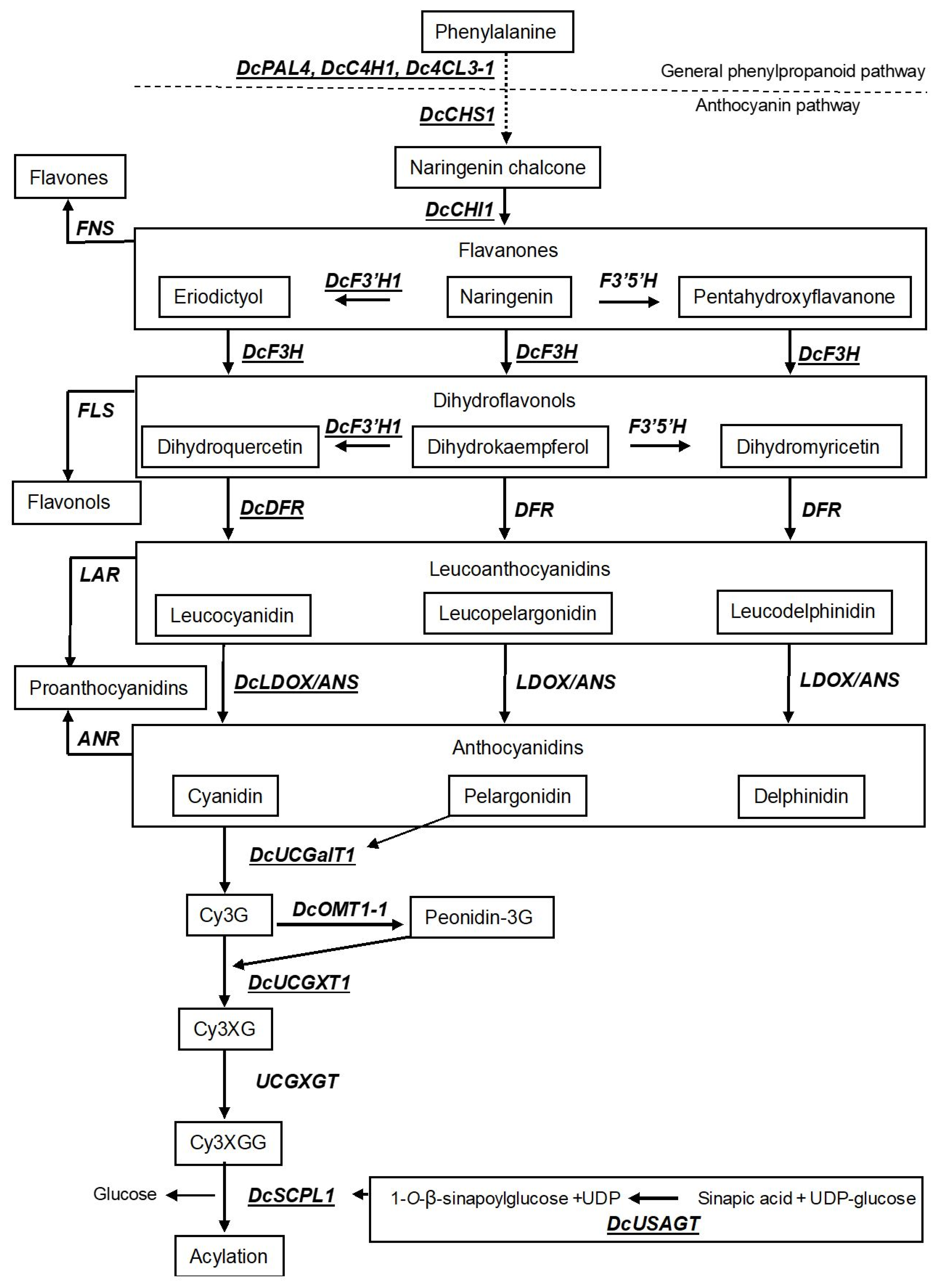
3.1. Structural Genes
3.2. Transcriptional Regulatory Activating Genes
4. Modulating Anthocyanin Composition in Black Carrots
4.1. Changing the Anthocyanin Color in Black Carrots
4.2. Increasing the Level of Acylation Anthocyanins in Black Carrots
4.3. Increasing the Total Amount of Anthocyanins in Black Carrots
5. Induction of the Anthocyanin Pathway in Orange Carrot
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Potera, C. The artificial food dye blues. Environ. Health Perspect. 2010, 118, A428. [Google Scholar] [CrossRef]
- Akhtar, S.; Rauf, A.; Imran, M.; Qamar, M.; Riaz, M.; Mubarak, M.S. Black carrot (Daucus carota L.), dietary and health promoting perspectives of its polyphenols: A review. Trends Food Sci. Technol. 2017, 66, 36–47. [Google Scholar] [CrossRef]
- Stevens, L.J.; Kuczek, T.; Burgess, J.R.; Stochelski, M.A.; Arnold, L.E.; Galland, L. Mechanisms of behavioral, atopic, and other reactions to artificial food colors in children. Nutr. Rev. 2013, 71, 268–281. [Google Scholar] [CrossRef] [PubMed]
- McCann, D.; Barrett, A.; Cooper, A.; Crumpler, D.; Dalen, L.; Grimshaw, K.; Kitchin, E.; Lok, K.; Porteous, L.; Prince, E.; et al. Food additives and hyperactive behaviour in 3-year-old and 8/9-year-old children in the community: A randomised, double-blinded, placebo-controlled trial. Lancet 2007, 3, 1560–1567. [Google Scholar] [CrossRef]
- Wrolstad, R.E.; Culver, C.A. Alternatives to those artificial FD&C food colorants. Annu. Rev. Food Sci. Technol. 2012, 3, 59–77. [Google Scholar] [CrossRef]
- Sigurdson, G.T.; Peipei, T.; Giusti, M.M. Natural Colorants: Food colorants from natural source. Annu. Rev. Food Sci. Technol. 2017, 8, 261–280. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Mu, L.; Yan, G.L.; Liang, N.N.; Pan, Q.H.; Wang, J.; Reeves, M.J.; Duan, C.Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Zafra-Stone, S.; Yasmin, T.; Bagchi, M.; Chatterjee, A.; Vinson, J.A.; Bagchi, D. Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol. Nutr. Food Res. 2007, 51, 675–683. [Google Scholar] [CrossRef]
- Rodriguez-Amaya, D.B. Natural food pigments and colorants. Curr. Opin. Food Sci. 2016, 7, 20–26. [Google Scholar] [CrossRef]
- Kirca, A.; Özkan, M.; Cemeroǧlu, B. Stability of black carrot anthocyanins in various fruit juices and nectars. Food Chem. 2006, 97, 598–605. [Google Scholar] [CrossRef]
- Patras, A.; Brunton, N.P.; O’Donnell, C.; Tiwari, B.K. Effect of thermal processing on anthocyanin stability in foods; mechanisms and kinetics of degradation. Trends Food Sci. Technol. 2010, 21, 3–11. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Trouillas, P.; Sancho-García, J.C.; De Freitas, V.; Gierschner, J.; Otyepka, M.; Dangles, O. Stabilizing and Modulating Color by Copigmentation: Insights from Theory and Experiment. Chem. Rev. 2016, 116, 4937–4982. [Google Scholar] [CrossRef]
- Malien-Aubert, C.; Dangles, O.; Amiot, M.J. Color stability of commercial anthocyanin-based extracts in relation to the phenolic composition. Protective effects by intra- and intermolecular copigmentation. J. Agric. Food Chem. 2001, 49, 170–176. [Google Scholar] [CrossRef]
- Gras, C.C.; Bause, K.; Leptihn, S.; Carle, R.; Schweiggert, R.M. Effect of chlorogenic acid on spectral properties and stability of acylated and non-acylated cyanidin-3-O-glycosides. Food Chem. 2018, 240, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Giusti, M.M.; Wrolstad, R.E. Acylated anthocyanins from edible sources and their application in food systems. Biochem. Eng. J. 2003, 14, 217–225. [Google Scholar] [CrossRef]
- Kırca, A.; Özkan, M.; Cemeroğlu, B. Effects of temperature, solid content and pH on the stability of black carrot anthocyanins. Food Chem. 2007, 101, 212–218. [Google Scholar] [CrossRef]
- Barba-Espín, G.; Glied, S.; Crocoll, C.; Dzhanfezova, T.; Joernsgaard, B.; Okkels, F.; Lütken, H.; Müller, R. Foliar-applied ethephon enhances the content of anthocyanin of black carrot roots (Daucus carota ssp. sativus var. atrorubens Alef). BMC Plant Biol. 2017, 17, 70. [Google Scholar] [CrossRef]
- Kammerer, D.; Carle, R.; Schieber, A. Quantification of anthocyanins in black carrot extracts (Daucus carota ssp. sativus var. atrorubens Alef.) and evaluation of their color properties. Eur. Food Res. Technol. 2004, 219, 479–486. [Google Scholar] [CrossRef]
- Montilla, E.C.; Arzaba, M.R.; Hillebrand, S.; Winterhalter, P. Anthocyanin Composition of Black Carrot (Daucus carota ssp. sativus var. atrorubens Alef.) Cultivars Antonina, Beta Sweet, Deep Purple, and Purple Haze. J. Agric. Food Chem. 2011, 59, 3385–3390. [Google Scholar] [CrossRef] [PubMed]
- Algarra, M.; Fernandes, A.; Mateus, N.; de Freitas, V.; Esteves da Silva, J.C.G.; Casado, J. Anthocyanin profile and antioxidant capacity of black carrots (Daucus carota L. ssp. sativus var. atrorubens Alef.) from Cuevas Bajas, Spain. J. Food Compos. Anal. 2014, 33, 71–76. [Google Scholar] [CrossRef]
- Koley, T.K.; Srivastava, S.; Tripathi, Y.B.; Banerjee, K.; Oulkar, D.; Goon, A.; Tripathi, A.; Singh, B. High-Resolution LCMS profiling of phenolic compounds of indian black carrot and evaluation of its effect on antioxidant defense and glucose metabolism in animal model. Agric. Res. 2019, 8, 481–489. [Google Scholar] [CrossRef]
- Vaerbak, S. (Chr. Hansen Natural Colors A/S, Hoersholm, Denmark). Personal communication, 2019.
- Xu, Z.S.; Tan, H.W.; Wang, F.; Hou, X.L.; Xiong, A.S. CarrotDB: A genomic and transcriptomic database for carrot. Database J. Biol. Databases Curation 2014, 2014, bau096. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Curaba, J.; Pottorff, M.; Ferruzzi, M.G.; Simon, P.; Cavagnaro, P.F. Carrot Anthocyanins Genetics and Genomics: Status and Perspectives to Improve Its Application for the Food Colorant Industry. Genes 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Draper, J. Transformation of carrot tissues derived from proembryogenic suspension cells: A useful model system for gene expression studies in plants. Plant Mol. Biol. 1987, 8, 265–274. [Google Scholar] [CrossRef]
- Baranski, R.; Lukasiewicz, A. Genetic Engineering of carrot. In The Carrot Genome. Compendium of Plant Genomes; Simon, P., Iorizzo, M., Grzebelus, D., Baranski, R., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Pawlicki, N.; Sangwan, R.S.; Sangwan-Norreel, B.S. Factors influencing the Agrobacterium tumefaciens-mediated transformation of carrot (Daucus carota L.). Plant Cell Tissue Organ Cult. 1992, 31, 129–139. [Google Scholar] [CrossRef]
- Hardegger, M.; Sturm, A. Transformation and regeneration of carrot (Daucus carota L.). Mol. Breed. 1998, 4, 119–127. [Google Scholar] [CrossRef]
- Monreal-Escalante, E.; Govea-Alonso, D.O.; Hernández, M.; Cervantes, J.; Salazar-Gonzalez, J.A.; Romero-Maldonado, A.; Rosas, G.; Garate, T.; Fragoso, G.; Sciutto, E. Towards the development of an oral vaccine against porcine cysticercosis: Expression of the protective HP6/TSOL18 antigen in transgenic carrots cells. Planta 2016, 243, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Holme, I.B.; Dionisio, G.; Kodama, M.; Dzhanfezova, T.; Joernsgaard, B.; Brinch-Pedersen, H. Cyanidin based anthocyanin biosynthesis in orange carrot is restored by expression of AmRosea1 and AmDelila, MYB and bHLH transcription factors. Plant Mol. Biol. 2020, 103, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Qi, Y.; Baranski, R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018, 37, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Annon, A.; Rathore, K.; Crosby, K. Overexpression of a tobacco osmotin gene in carrot (Daucus carota L.) enhances drought tolerance. In Vitro Cell. Dev. Biol. Plant 2014, 50, 299–306. [Google Scholar] [CrossRef]
- Tokuji, Y.; Fukuda, H.A. Rapid Method for Transformation of Carrot (Daucus carota L.) by Using Direct Somatic Embryogenesis. Biosci. Biotechnol. Biochem. 1999, 63, 519–523. [Google Scholar] [CrossRef]
- Arango, J.; Salazar, B.; Welsch, R.; Sarmiento, F.; Beyer, P.; Al-Babili, S. Putative storage root specific promoters from cassava and yam: Cloning and evaluation in transgenic carrots as a model system. Plant Cell Rep. 2010, 29, 651–659. [Google Scholar] [CrossRef]
- Dirk, R.; Sidorov, V.; Tulmans, C.A. A new protoplast culture system in Daucus carota L. and its applications for mutant selection and transformation. Theor. Appl. Genet. 1996, 93, 809–815. [Google Scholar] [CrossRef]
- Droge, W.; Broer, I.; Pühler, A. Transgenic plants containing the phosphinothricin-N-acetyltransferase gene metabolize the herbicide L-phosphinothricin (glufosinate) differently from untransformed plants. Planta 1992, 187, 142–151. [Google Scholar] [CrossRef]
- Matzke, M.; Matzke, A.J.; Kooter, J.M. RNA: Guiding gene silencing. Science 2001, 10, 1080–1083. [Google Scholar] [CrossRef] [PubMed]
- Voytas, D.F.; Gao, C. Precision Genome Engineering and Agriculture: Opportunities and Regulatory Challenges. PLoS Biol. 2014, 12, e1001877. [Google Scholar] [CrossRef] [PubMed]
- Voytas, D.F. Plant genome engineering with sequence specific nucleases. Annu. Rev. Plant Biol. 2013, 64, 327–350. [Google Scholar] [CrossRef] [PubMed]
- Epinat, J.C.; Arnould, S.; Chames, P.; Rochaix, P.; Desfontaines, D.; Puzin, C.; Patin, A.; Zanghellini, A.; Pâques, F.; Lacroix, E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003, 31, 2952–2962. [Google Scholar] [CrossRef]
- Bibikova, M.; Beumer, K.; Trautman, J.K.; Carroll, D. Enhancing gene targeting with designer zinc finger nucleases. Science 2003, 300, 764. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Voytas, D.F. TAL effectors: Customizable proteins for DNA targeting. Science 2011, 333, 1843–1846. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.; Kim, S.; Choe, S.; Kim, J. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef]
- Nicolia, A.; Andersson, M.; Hofvander, P.; Festa, G.; Cardi, T. Tomato protoplasts as cell target for ribonucleoprotein (RNP)-mediated multiplexed genome editing. Plant Cell Tissue Organ Cult. 2021, 144, 463–467. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Menz, J.; Modrzejewski, D.; Sprink, T. DNA-free genome editing: Past, present and future. Front. Plant Sci. 2019, 9, 1957. [Google Scholar] [CrossRef]
- Eriksson, D.; Brinch-Pedersen, H.; Chawade, A.; Holme, I.B.; Hvoslef-Eide, T.A.; Ritala, A.; Teeri, T.H.; Thorstensen, T. Scandinavian perspectives on plant gene technology: Applications, policies and progress. Physiol. Plant. 2018, 162, 219–238. [Google Scholar] [CrossRef]
- Holme, I.B.; Gregersen, P.L.; Brinch-Pedersen, H. Induced Genetic Variation in Crop Plants by Random or Targeted Mutagenesis: Convergence and Differences. Front. Plant Sci. 2019, 10, 1468. [Google Scholar] [CrossRef] [PubMed]
- Rommens, C.M. All-native DNA transformation: A new approach to plant genetic engineering. Trends Plant Sci. 2004, 9, 457–464. [Google Scholar] [CrossRef]
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Cisgenic plants are similar to traditionally bred plants: International regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep. 2006, 7, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Lusk, J.L.; Sullivan, P. Consumer acceptance of genetically modified foods. Food Technol. Mag. 2002, 56, 32–37. [Google Scholar]
- Lusk, J.L.; Rozan, A. Consumer acceptance of ingenic foods. Biotechnol. J. 2006, 1, 1433–1434. [Google Scholar] [CrossRef] [PubMed]
- Gaskell, G.; Allansdottir, A.; Allum, N.; Castro, P.; Esmer, Y.; Fischler, C.; Jackson, J.; Kronberger, N.; Hampel, J.; Mejlgaard, N.; et al. The 2010 Eurobarometer on the life sciences. Nat. Biotechnol. 2011, 29, 113–114. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.D. Challenging regulations: Managing risks in crop biotechnology. Food Energy Secur. 2015, 4, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Holme, I.B.; Wendt, T.; Holm, P.B. Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 2013, 11, 395–407. [Google Scholar] [CrossRef]
- Afolabi, A.S.; Worland, B.; Snape, J.W.; Vain, P. A large-scale study of rice plants transformed with different T-DNAs provides new insights into locus composition and T-DNA linkage configurations. Theor. Appl. Genet. 2004, 109, 815–826. [Google Scholar] [CrossRef]
- Matthews, P.R.; Wang, M.B.; Waterhouse, P.M.; Thomton, S.; Fieg, S.J.; Gubler, F.; Jacobsen, J.V. Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNAs’ on a standard Agrobacterium transformation vector. Mol. Breed. 2001, 7, 195–202. [Google Scholar] [CrossRef]
- Holme, I.B.; Dionisio, G.; Brinch-Pedersen, H.; Wendt, T.; Madsen, C.K.; Vincze, E.; Holm, P.B. Cisgenic barley with improved phytase activity. Plant Biotechnol. J. 2012, 10, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Bannoud, F.; Ellison, S.; Paolinelli, M.; Horejsi, T.; Senalik, D.; Fanzone, M.; Iorizzo, M.; Simon, P.W.; Cavagnaro, P.F. Dissecting the genetic control of root and leaf tissue-specific anthocyanin pigmentation in carrot (Daucus carota L.). Theor. Appl. Genet. 2019, 132, 2485–2507. [Google Scholar] [CrossRef]
- Iorizzo, M.; Cavagnaro, P.F.; Bostan, H.; Zhao, Y.; Zhang, J.; Simon, P.W. A Cluster of MYB Transcription Factors Regulates Anthocyanin Biosynthesis in Carrot (Daucus carota L.) Root and Petiole. Front. Plant Sci. 2019, 9, 1927. [Google Scholar] [CrossRef]
- Curaba, J.; Bostan, H.; Cavagnaro, P.F.; Douglas, S.; Fentie, M.M.; Yunyang, Z.; Simon, P.W.; Iorizzo, M. Identification of an SCPL Gene Controlling Anthocyanin Acylation in Carrot (Daucus carota L.) Root. Front. Plant Sci. 2020, 10, 1770. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, M.; Willis, D.K.; Cavagnaro, P.F.; Iorizzo, M.; Abak, K.; Simon, P.W. Expression and mapping of anthocyanin biosynthesis genes in carrot. Theor. Appl. Genet. 2013, 126, 1689–1702. [Google Scholar] [CrossRef]
- Meng, G.; Clausen, S.K.; Rasmussen, S.K. Transcriptome Analysis Reveals Candidate Genes Related to Anthocyanin Biosynthesis in Different Carrot Genotypes and Tissues. Plants 2020, 9, 344. [Google Scholar] [CrossRef]
- Xu, Z.S.; Huang, Y.; Wang, F.; Song, X.; Wang, G.; Xiong, A.S.S. Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol. 2014, 14, 262. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, Q.; Feng, K.; Xiong, A. Changing Carrot Color: Insertions in DcMYB7 Alter the Regulation of Anthocyanin Biosynthesis and Modification. Plant Phys. 2019, 118, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yang, Q.; Feng, K.; Yu, X.; Xiong, A. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 2020, 18, 1585–1597. [Google Scholar] [CrossRef]
- Kodama, M.; Brinch-Pedersen, H.; Sharma, S.; Holme, I.B.; Joernsgaard, B.; Dzhanfezova, T.; Amby, D.B.; Vieira, F.G.; Liu, S.; Gilbert, M.T.P. Identification of transcription factor genes involved in anthocyanin biosynthesis in carrot (Daucus carota L.) using RNA-Seq. BMC Genom. 2018, 19, 811. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Silva, S.; dos Santos, A.L.; Chalfun-Júnior, A.; Zhao, J.; Peres, L.E.P.; Benedito, V.A. Understanding the genetic regulation of anthocyanin biosynthesis in plants—Tools for breeding purple varieties of fruits and vegetables. Phytochemistry 2018, 153, 11–27. [Google Scholar] [CrossRef]
- Xu, Z.S.; Ma, J.; Wang, F.; Ma, H.Y.; Wang, Q.X.; Xiong, A.S. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Sci. Rep. 2016, 6, 27356. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Xu, Z.S.; Xiong, A.S. Identification and Characterization of DcUSAGT1, a UDP-Glucose: Sinapic Acid Glucosyltransferase from Purple Carrot Taproots. PLoS ONE 2016, 11, e0154938. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R. The TRANSPARENT TESTA GLABRA1 Locus, Which Regulates Trichome Differentiation and Anthocyanin Biosynthesis in Arabidopsis, Encodes a WD40 Repeat Protein. Plant Cell 1999, 11, 1337–1350. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Cavagnaro, P.F.; Iorizzo, M.; Yildiz, M.; Senalik, D.; Parsons, J.; Ellison, S.; Simon, P.W. A gene-derived SNP-based high resolution linkage map of carrot including the location of QTL conditioning root and leaf anthocyanin pigmentation. BMC Genom. 2014, 15, 1118. [Google Scholar] [CrossRef]
- Xu, Z.; Feng, K.; Xiong, A. CRISPR/Cas9-Mediated Multiply Targeted Mutagenesis in Orange and Purple Carrot Plants. Mol. Biotechnol. 2019, 61, 191–199. [Google Scholar] [CrossRef]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Meyer, P.; Heidmann, I.; Forkmann, G.; Saedler, H. A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef]
- Helariutta, Y.; Elomaa, P.; Kotilainen, M.; Seppanen, P.; Teeri, T.H. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 1993, 22, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Katsumoto, Y.; Fukuchi-Mizutani, M.; Fukui, Y.; Brugliera, F.; Holton, T.A.; Karan, M.; Nakamura, N.; Yonekura-Sakakibara, K.; Togami, J.; Pigeaire, A.; et al. Engineering of the Rose Flavonoid Biosynthetic Pathway Successfully Generated Blue-Hued Flowers Accumulating Delphinidin. Plant Cell Physiol. 2007, 48, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Brugliera, F.; Tao, G.Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/Blue Chrysanthemums—Metabolic Engineering of the Anthocyanin Biosynthetic Pathway Results in Novel Petal Colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A. Transgenic Plants Exhibiting Altered Flower Color and Methods for Producing Same. International Publication Number WO 96/36716, 27 June 2000. [Google Scholar]
- Chandler, S.; Tanaka, Y. Genetic Modification in Floriculture. CRC Crit. Rev. Plant Sci. 2007, 26, 169–197. [Google Scholar] [CrossRef]
- Du, H.; Wu, J.; Ji, K.X.; Zeng, Q.Y.; Bhuiya, M.W.; Su, S.; Shu, Q.Y.; Ren, H.X.; Liu, Z.A.; Wang, L.S. Methylation mediated by an anthocyanin, O-methyltransferase, is involved in purple flower coloration in Paeonia. J. Exp. Bot. 2015, 66, 6563–6577. [Google Scholar] [CrossRef]
- Netzel, M.; Netzel, G.; Kammerer, D.R.; Schieber, A.; Carle, R.; Simons, L.; Bitsch, I.; Bitsch, R.; Konczak, I. Cancer cell antiproliferation activity and metabolism of black carrot anthocyanins. Innov. Food Sci. Emerg. Technol. 2007, 8, 365–372. [Google Scholar] [CrossRef]
- Bontpart, T.; Cheynier, V.; Ageorges, A.; Terrier, N. BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 2015, 208, 695–707. [Google Scholar] [CrossRef]
- Simon, P.W. Inheritance and Expression of Purple and Yellow Storage Root Color in Carrot. J. Hered. 1996, 87, 63–66. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 926–980. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, J.; Cho, K.; Cheon-Ill, C.; Sung, M.; Choung, M.; Roh, K. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep. 2008, 27, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zou, Q.; Yang, G.; Jiang, S.; Fang, H.; Wang, H.; Wang, Y.; Zhang, J.; Zhang, Z.; Wang, N.; et al. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 2020, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Li, C.; Ma, X.; Luo, K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017, 36, 1263–1276. [Google Scholar] [CrossRef]
- Sharma, S. Anthocyanin Biosynthesis in Carrot (Daucus carota L.). Ph.D. Thesis, Aarhus University, Aarhus, Denmark, 14 January 2019. [Google Scholar]
- Thill, J.; Miosic, S.; Ahmed, R.; Schlangen, K.; Muster, G.; Stich, K.; Halbwirth, H. ‘Le Rouge et le Noir’: A decline in flavone formation correlates with the rare color of black dahlia (Dahlia variabilis hort.) flowers. BMC Plant Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.F.; Ma, J.; Zhang, X.Y.; Xu, Z.S.; Xiong, A.S. AgFNS overexpression increase apigenin and decrease anthocyanins in petioles of transgenic celery. Plant Sci. 2017, 263, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Lewis, D.H.; Bloor, S.J.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of Flavonol Synthase and Dihydroflavonol-4-Reductase Expression Associated Tightly to White vs. Red Color Flower Formation in Plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef]
- Leja, M.; Kamińska, I.; Kramer, M.; Maksylewics-Kaul, A.; Kammerer, D.; Carle, R.; Baranski, R. The Content of Phenolic Compounds and Radical Scavenging Activity Varies with Carrot Origin and Root Color. Plant Foods Hum. Nutr. 2013, 68, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Bangà, O. Origin of the Euripean cultivated carrot. Euphytica 1957, 6, 54–63. [Google Scholar] [CrossRef]
- Bangà, O. The development of the original European carrot material. Euphytica 1957, 6, 64–76. [Google Scholar] [CrossRef]
- Baranski, R.; Maksylewicz-Kaul, A.; Nothnagel, T.; Cavagnaro, P.E.; Simon, P.W.; Grzebelus, D. Genetic diversity of carrot (Daucus carota L.) cultivars revealed by analysis of SSR loci. Genet. Resour. Crop Evol. 2012, 59, 163–170. [Google Scholar] [CrossRef]
| Trait | Desired | Approach no. | CRISPR/Cas DNA-Constructs or RNP Knock-Out Approaches | Transgenic Approaches | Intra-/Cisgenic Approaches * |
|---|---|---|---|---|---|
| Changing colors | Pelargonidin | 1a | KO of DcF3′H1 | - | - |
| 1b | KO of DcF3′H1 | Simultaneous insertion of a DFR gene encoding an enzyme with substrate preference for dihydro-kaempferol from any species. | - | ||
| 1c | KO of DcF3′H1 | - | Simultaneous insertion of a DcDFR gene upregulated in cultivars synthesizing pelargonidin. | ||
| Delphinidin | 2a | KO of DcF3′H1 | Insertion of a F3′5′H gene from any species. | - | |
| 2b | KO of DcF3′H1 | - | Insertion of a DcF3′5′H gene present and upregulated in cultivars synthesizing delphinidin. | ||
| 2c | KO of DcF3′H1 | Simultaneous insertion of a F3′5′H gene and a DFR gene encoding an enzyme with substrate preference for dihydro-myricetin from any species. | - | ||
| 2d | KO of DcF3′H1 | - | Simultaneous insertion of a DcF3′5′H and a DcDFR-gene upregulated in cultivars synthesizing delphinidin. | ||
| Peonidin | 3a | - | Isolation and insertion of an OMT-gene from any species. | - | |
| 3b | - | - | Isolation and insertion of the DcOMT1-1 gene. | ||
| Acylation | Increased Cy3XSGG acylation | 4a | - | - | Overexpression of the DcUSAGT gene. |
| Increased acylation in cultivars homozygous for allele DcSCPL1-2 | 4b | - | - | Insertion of the DcSCPL1-1 allele. | |
| Increased total content of anthocyanins | Induction of anthocyanin in the non-purple tissue of the taproot | 5a | KO of potential repressors in non-purple taproot tissue: DcMYB13 DcMYB14 DcMYB15 DcMYB1R1-1 DcMYB1R1-2 | - | - |
| 5b | - | - | Overexpression of the DcR2R3-MYB responsible for anthocyanin production in a given carrot cultivar. | ||
| 5c | KO of potential competing enzymes: DcFNS-like1 DcFNS-like2 | - | - | ||
| 5d | KO of potential competing enzymes: FLS1 FLS2 | - | - | ||
| 5e | KO of potential competing enzymes: FLS1 FLS2 | - | Simultaneous overexpression of the DcDRF1 gene for cyanidin production. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bæksted Holme, I.; Dionisio, G.; Brinch-Pedersen, H. A Roadmap to Modulated Anthocyanin Compositions in Carrots. Plants 2021, 10, 472. https://doi.org/10.3390/plants10030472
Bæksted Holme I, Dionisio G, Brinch-Pedersen H. A Roadmap to Modulated Anthocyanin Compositions in Carrots. Plants. 2021; 10(3):472. https://doi.org/10.3390/plants10030472
Chicago/Turabian StyleBæksted Holme, Inger, Giuseppe Dionisio, and Henrik Brinch-Pedersen. 2021. "A Roadmap to Modulated Anthocyanin Compositions in Carrots" Plants 10, no. 3: 472. https://doi.org/10.3390/plants10030472
APA StyleBæksted Holme, I., Dionisio, G., & Brinch-Pedersen, H. (2021). A Roadmap to Modulated Anthocyanin Compositions in Carrots. Plants, 10(3), 472. https://doi.org/10.3390/plants10030472







