Genome-Wide Analysis of MDHAR Gene Family in Four Cotton Species Provides Insights into Fiber Development via Regulating AsA Redox Homeostasis
Abstract
1. Introduction
2. Results
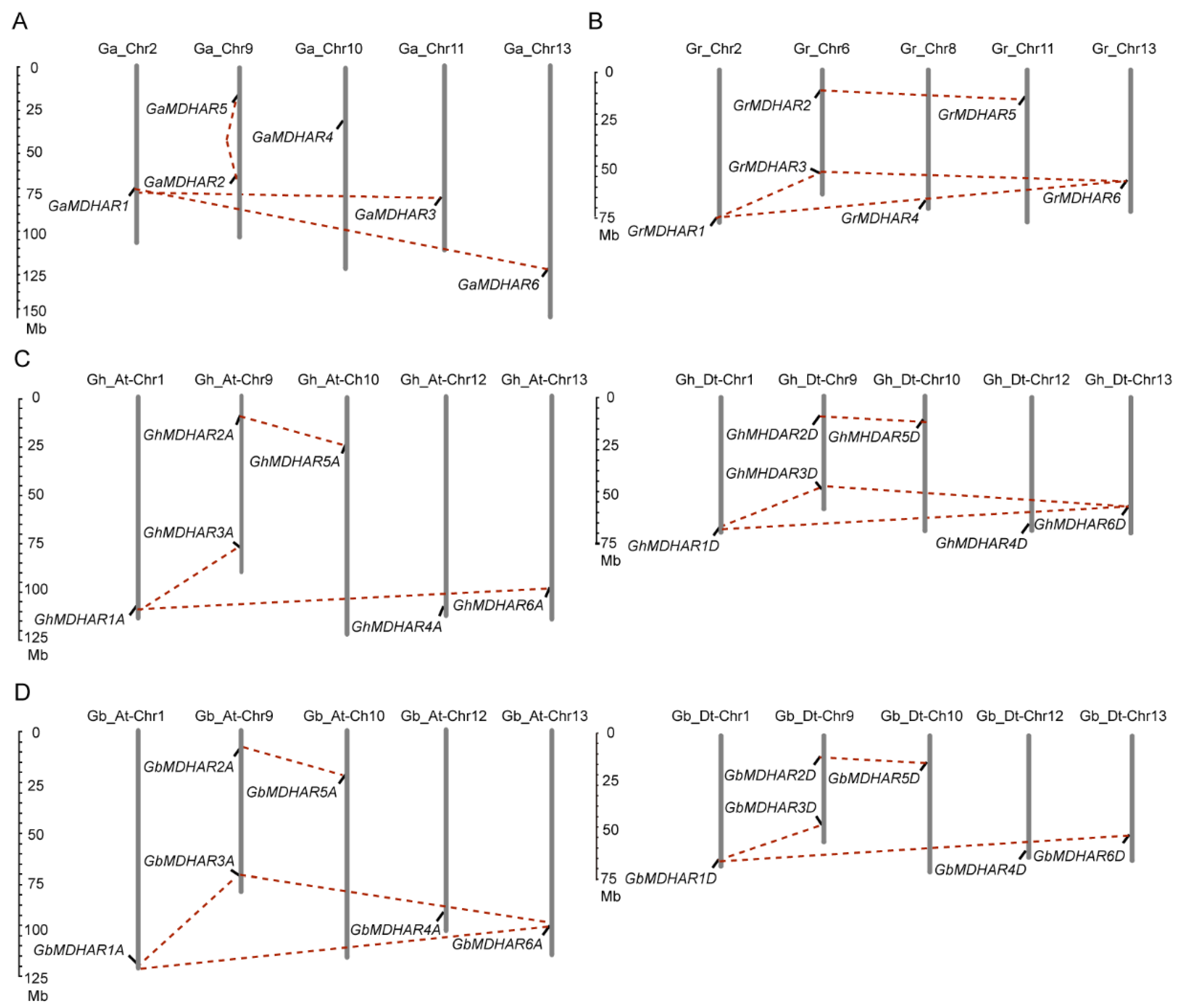
2.1. Identification and Chromosome Location of MDHAR Genes of Four Gossypium Species
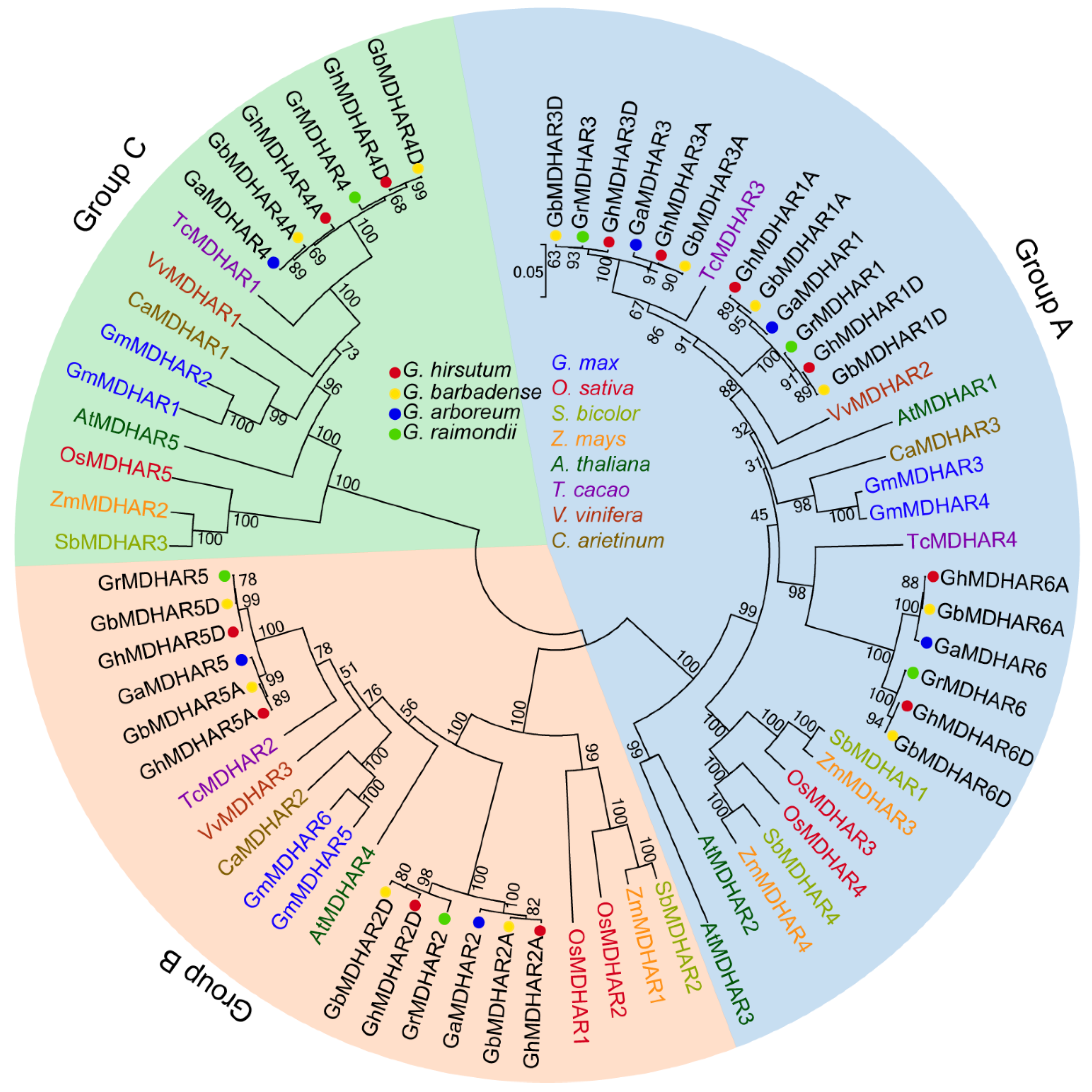
2.2. Phylogenetic and Evolutionary Analyses of MDHAR Members
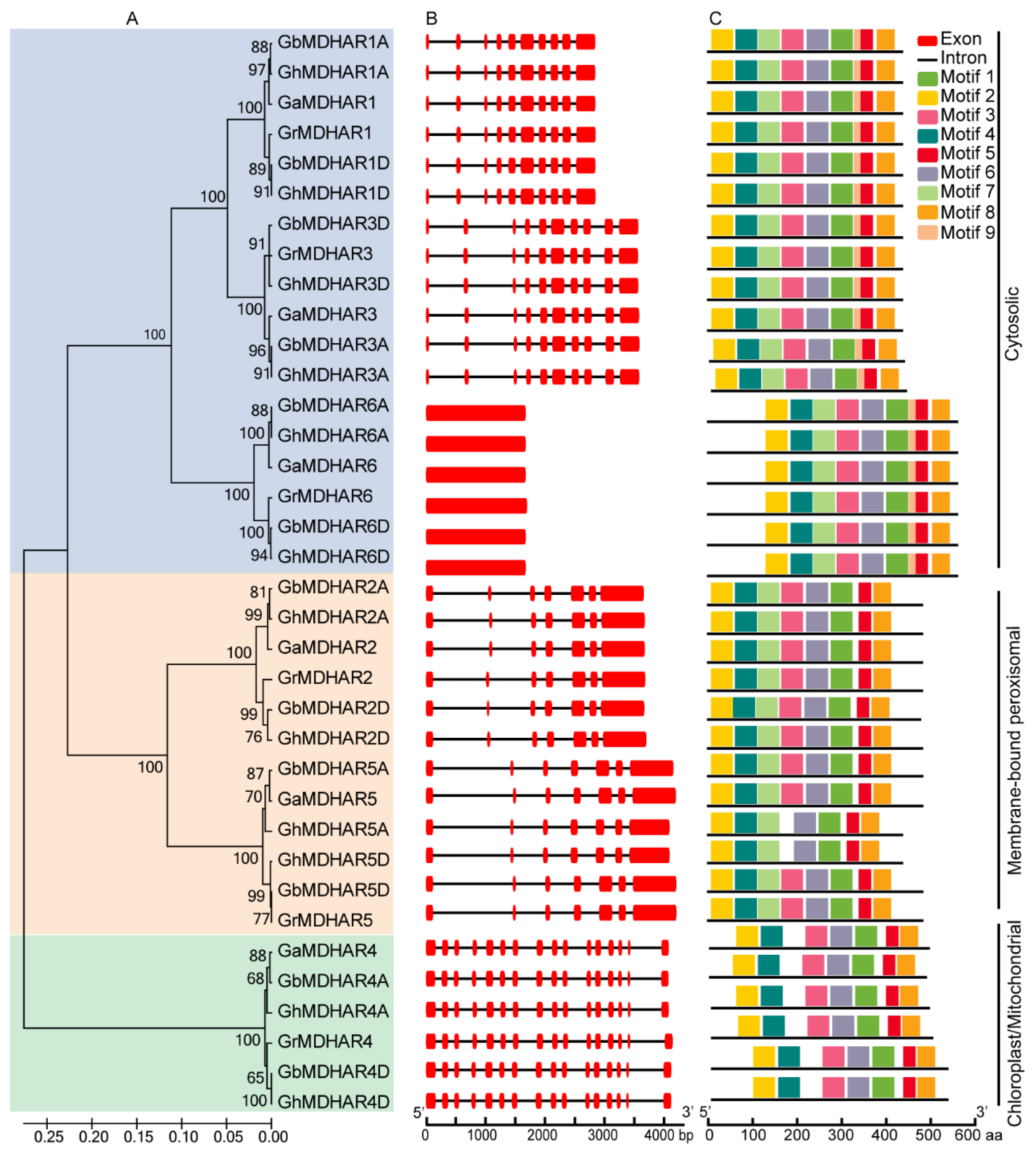
2.3. Analyses of Gene Structure, Conserved Motif, and Promoter Cis-Acting Element of MDHAR Genes
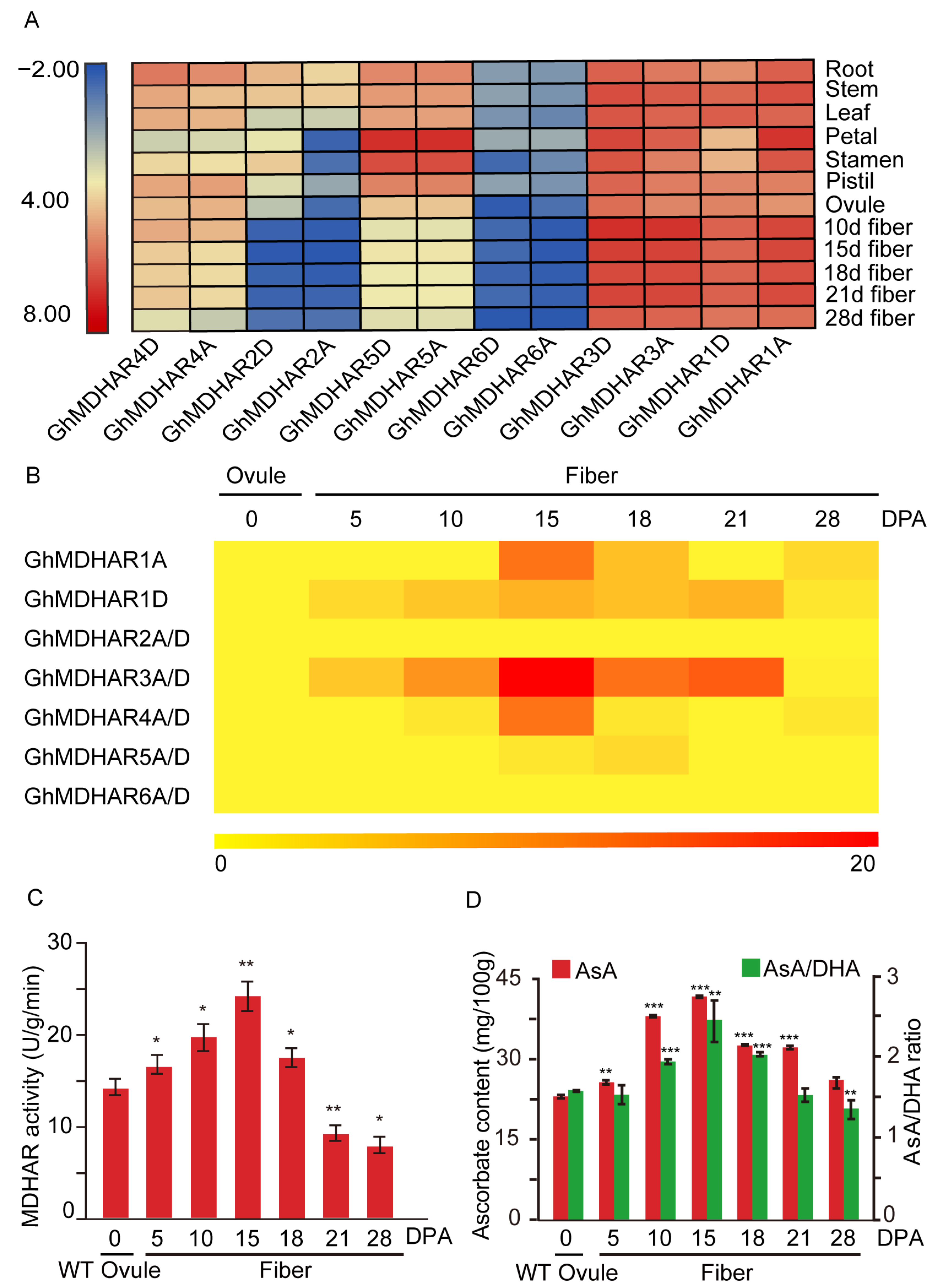
2.4. Tissue-Specific and Fiber-Developmental Expression Analysis of G. hirsutum GhMDHARs
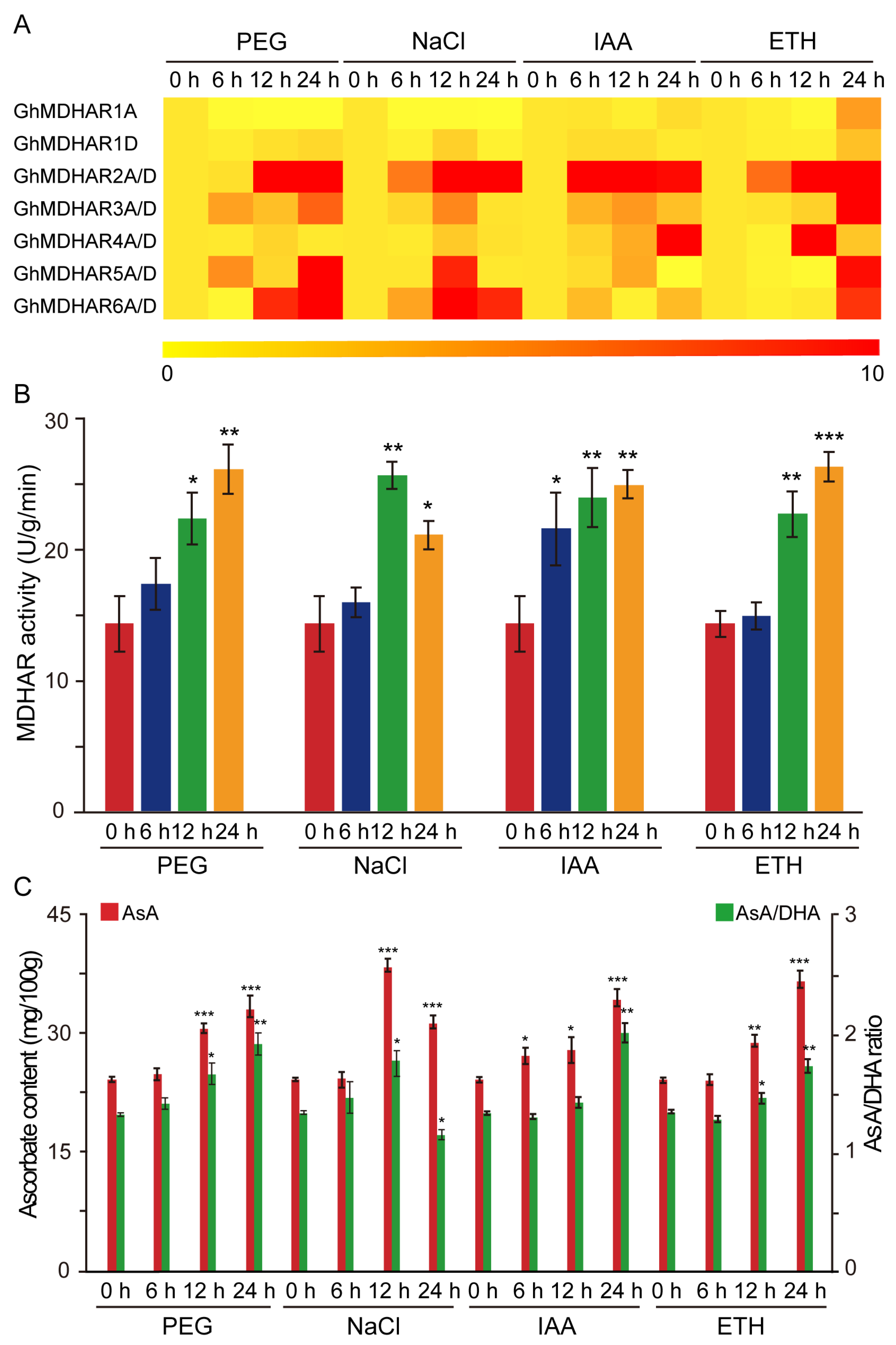
2.5. Expression Analysis of GhMDHARs under Abiotic and Phytohormone Treatments
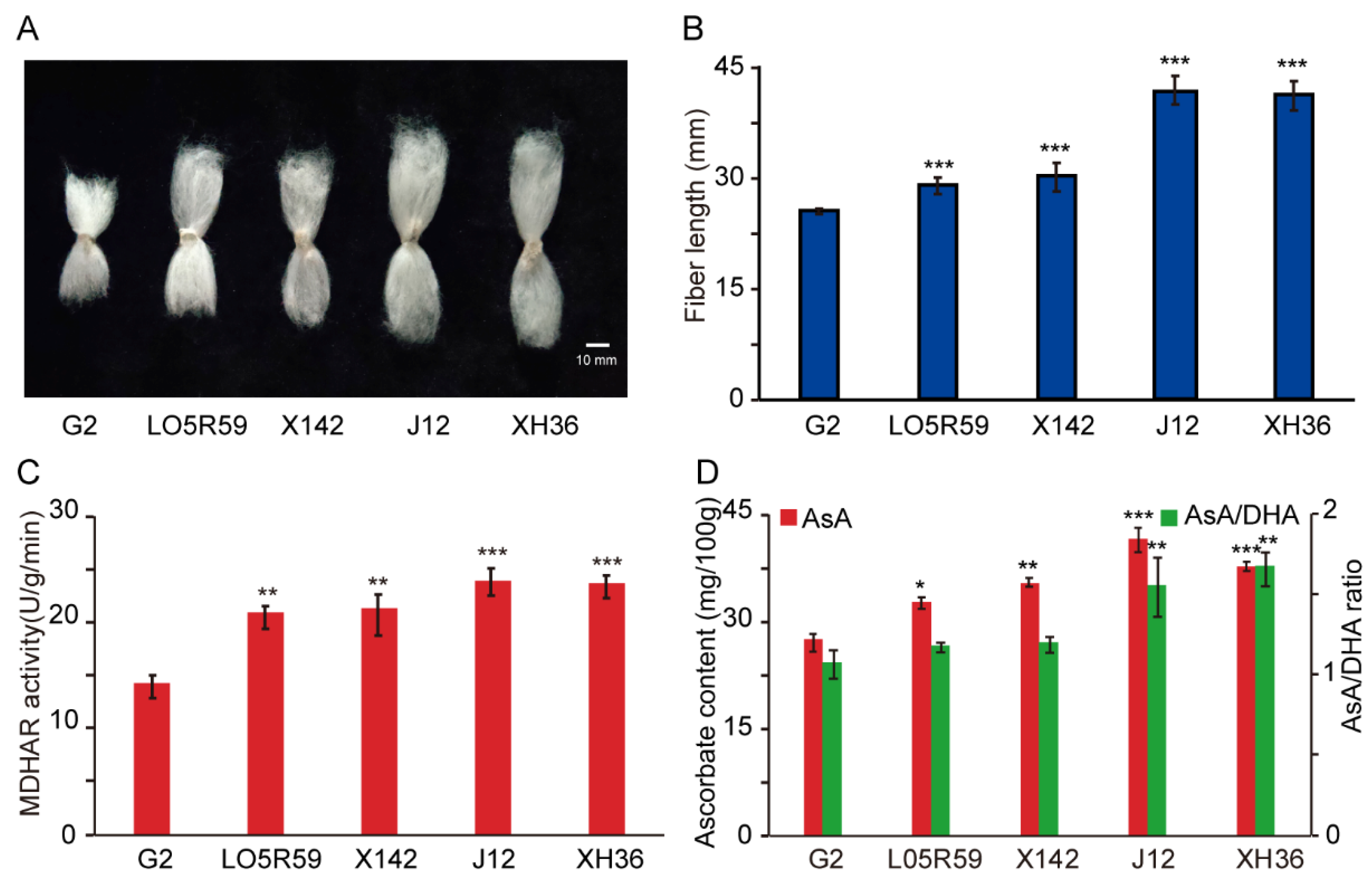
2.6. Analysis of GhMDHAR Involving in Fiber Elongation Development
2.7. Analysis of AsA Antioxidative Enzymes under H2O2 Stimulation
3. Discussion
4. Materials and Methods
4.1. Sequence Acquirement and Chromosomal Distribution of MDHAR Genes in Cotton
4.2. Evolutionary Analysis and Promoter Cis-Element Identification
4.3. Cotton Planting and Material Treatment
4.4. RNA Extraction and qRT-PCR Analysis
4.5. Enzyme Assays
4.6. Measurement of Asc, AsA and DHA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACO | 1-aminocyclopropane-1-carboxylic acid oxidase |
| ANOVA | Analysis of Variance |
| AO | Ascorbate oxidase |
| APX | Ascorbate peroxidase |
| AsA | Ascorbic acid |
| Asc | Ascorbate |
| CAT | Catalase |
| DHA | Dehydroascorbic acid |
| DPA | Day post anthesis |
| DTT | Dithiothreitol |
| ETH | Ethephon |
| FAD | Flavin adenine dinucleotide |
| FPKM | Fragments per kilobase of exon per million reads mapped |
| GSDS | Gene Structure Display Server |
| GSH | Glutathione |
| HRGP | Hydroxyproline rich glycoprotein |
| IAA | Indole-3-acetic acid |
| Ka | Nonsynonymous substitution rate |
| kDa | KiloDalton |
| Ks | Synonymous substitution rate |
| MDHAR | Monodehydroasorbate reductase |
| MEGA | Molecular evolutionary genetics analysis |
| MEME | Multiple EM for Motif Elicitation |
| MIPS | Myo-inositol-1-phosphate synthase |
| MW | Molecular weight |
| NAD | Nicotinamide adenine dinucleotide |
| NADH | Nicotinamide adenine dinucleotide |
| NADPH | Dinucleotide phosphate |
| NCBI | National Center for Biotechnology Information |
| N—J | Neighbor-joining |
| ORF | Open reading frame |
| PEG | Polyethylene glycol |
| pI | Isoelectric point |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| ROS | Reactive oxygen species |
| SOD | Superoxide dismutase |
| TCA | Trichloroacetic acid |
| TNT | 2,4,6-trinitrotoluene |
| UBQ | Ubiquitin |
References
- Pastori, G.M.; Kiddle, G.; Antoniw, J.; Bernard, S.; Veljovic-Jovanovic, S.; Verrier, P.J.; Noctor, G.; Foyer, C.H. Leaf vitamin C contentsmodulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 2003, 15, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Qin, Y.M.; Pang, Y.; Song, W.Q.; Mei, W.Q.; Zhu, Y.X. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 2007, 175, 462–471. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Pignocchi, C.; Foyer, C.H. Apoplastic ascorbate metabolism and its role in the regulation of cell signaling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef]
- Raven, E.L. Peroxidase-catalyzed oxidation of ascorbate. Structural, spectroscopic and mechanistic correlations in ascorbate peroxidase. Subcell. Biochem. 2000, 35, 317–349. [Google Scholar]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Conklin, P.L.; Pallanca, J.E.; Last, R.L.; Smirnoff, N. L-ascorbic acid metabolism in the ascorbate-deficient Arabidopsis mutant vtc1. Plant Physiol. 1997, 115, 1277–1285. [Google Scholar] [CrossRef]
- Conklin, P.L.; Saracco, S.A.; Norris, S.R.; Last, R.L. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 2000, 154, 847–856. [Google Scholar] [PubMed]
- Smirnoff, N.; Wheeler, G.L. Ascorbic acid in plants: Biosynthesis and function. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 291–314. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.L.; Barth, C. Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ. 2004, 27, 959–970. [Google Scholar] [CrossRef]
- Wolucka, B.A.; Goossens, A.; Inzé, D. Methyl jasmonate stimulates the de novo biosynthesis of vitamin C in plant cell suspensions. J. Exp. Bot. 2005, 56, 2527–2538. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Xia, Y.; Kai, G.; Chen, W.; Tang, K. Metabolic engineering of plant L–ascorbic acid biosynthesis: Recent trends and applications. Crit. Rev. Biotechnol. 2007, 27, 173–182. [Google Scholar] [CrossRef]
- Li, R.; Xin, S.; Tao, C.C.; Jin, X.; Li, H.B. Cotton Ascorbate Oxidase Promotes Cell Growth in Cultured Tobacco Bright Yellow-2 Cells through Generation of Apoplast Oxidation. Int. J. Mol. Sci. 2017, 18, 1346. [Google Scholar] [CrossRef]
- Foyer, C.H.; Halliwell, B. The presence of glutathione and glutathione reductase in chloroplasts: A proposed role in ascorbic acid metabolism. Planta 1976, 133, 21–25. [Google Scholar] [CrossRef]
- Potters, G.; Horemans, N.; Bellone, S.; Caubergs, R.J.; Trost, P.; Guisez, Y.; Asard, H. Dehydroascorbate influences the plant cell cycle through a glutathione independent reduction mechanism. Plant Physiol. 2004, 134, 1479–1487. [Google Scholar] [CrossRef]
- Green, M.A.; Fry, S.C. Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-l-threonate. Nature 2005, 433, 83–87. [Google Scholar] [CrossRef]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef]
- Gallie, D.R. L-ascorbic Acid: A multifunctional molecule supporting plant growth and development. Scientifica 2013, 2013, 795964. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.; Page, D.; Gouble, B.; Garchery, C.; Zamir, D.; Causse, M. Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ. 2008, 31, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, Q.Y.; Sun, Y.L.; Wang, L.Y.; Yang, X.H.; Meng, Q.W. Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen-mediated oxidative stresses. Physiol. Plant. 2010, 139, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.L.; Lin, T.H.; Chen, C.C.; Cheng, T.X.; Chang, H.Y.; Lee, T.M. Monodehydroascorbate Reductase Plays a Role in the Tolerance of Chlamydomonas Reinhardtii to Photooxidative Stress. Plant Cell Physiol. 2019, 60, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- Hossain, M.; Nakano, Y.; Asada, K. Monodehydroascorbate reductase in spinach chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen peroxide. Plant Cell Physiol. 1984, 25, 385–395. [Google Scholar]
- Jimenez, A.; Hernandez, J.; del Rio, L.; Sevilla, F. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 1997, 114, 275. [Google Scholar] [CrossRef]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant Cell Environ. 2003, 26, 845–856. [Google Scholar] [CrossRef]
- Eastmond, P.J. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 2007, 19, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Dalton, D.A.; Baird, L.M.; Langeberg, L.; Taugher, C.Y.; Anyan, W.R.; Vance, C.P.; Sarath, G. Subcellular Localization of Oxygen Defense Enzymes in Soybean (Glycine max [L.] Merr.) Root Nodules. Plant Physiol. 1993, 102, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Obara, K.; Sumi, K.; Fukuda, H. The use of multiple transcription starts causes the dual targeting of Arabidopsis putative monodehydroascorbate reductase to both mitochondria and chloroplasts. Plant Cell Physiol. 2002, 43, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.; Zilinskas, B. Molecular cloning and characterization of a cDNA encoding pea monodehydroascorbate reductase. J. Biol. Chem. 1994, 269, 31129–31133. [Google Scholar] [CrossRef]
- Grantz, A.A.; Brummell, D.A.; Bennett, A.B. Ascorbate free radical reductase mRNA levels are induced by wounding. Plant Physiol. 1995, 108, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Tao, S.; Endo, Y.; Inaba, T.; Hossain, M.A.; Miyake, C.; Matsuo, M.; Aoki, H.; Asada, K.; Saito, K. Purification and cDNA cloning of chloroplastic monodehydroascorbate reductase from spinach. Biosci. Biotechnol. Biochem. 2005, 69, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Asada, K. Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. J. Biol. Chem. 1985, 260, 12920–12926. [Google Scholar] [CrossRef]
- Borracino, G.; Dipierro, S.; Arrigoni, O. Purification and properties of ascorbate free radical reductase from potato tubers. Planta 1986, 167, 521–526. [Google Scholar] [CrossRef]
- Dalton, D.A.; Langeberg, L.; Robbins, M. Purification and characterization of monodehydroascorbate reductase from soybean root nodules. Arch. Biochem. Biophys. 1992, 292, 281–286. [Google Scholar] [CrossRef]
- De Leonardis, S.; De Lorenzo, G.; Borraccino, G.; Dipierro, S. A specific ascorbate free radical reductase isozyme participates in the regeneration of ascorbate for scavenging toxic oxygen species in potato tuber mitochondria. Plant Physiol. 1995, 109, 847–851. [Google Scholar] [CrossRef][Green Version]
- Lunde, C.; Baumann, U.; Shirley, N.J.; Drew, D.P.; Fincher, G.B. Gene structure and expression pattern analysis of three monodehydroascorbate reductase (Mdhar) genes in Physcomitrella patens: Implications for the evolution of the MDHAR family in plants. Plant Mol. Biol. 2006, 60, 259–275. [Google Scholar] [CrossRef]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005, 43, 900–914. [Google Scholar] [CrossRef]
- Negi, B.; Salvi, P.; Bhatt, D.; Majee, M.; Arora, S. Molecular cloning, in-silico characterization and functional validation of monodehydroascorbate reductase gene in Eleusine coracana. PLoS ONE 2017, 12, e0187793. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Liu, W.; Zhang, Q.; Wang, X.; Wang, X.; Duan, X.; Li, F.; Huang, L.; Kang, Z. TaMDHAR4, a monodehydroascorbate reductase gene participates in the interactions between wheat and Puccinia striiformis f. sp. tritici. Plant Physiol. Biochem. 2014, 76, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Johnston, E.J.; Rylott, E.L.; Beynon, E.; Lorenz, A.; Chechik, V.; Bruce, N.C. Monodehydroascorbate reductase mediates TNT toxicity in plants. Science 2015, 349, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Sakihama, Y.; Mano, J.; Sano, S.; Asada, K.; Yamasaki, H. Reduction of phenoxyl radicals mediated by monodehydroascorbate reductase. Biochem. Biophys. Res. Commun. 2000, 279, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Kim, Y.S.; Kim, Y.H.; Park, A.K.; Kim, H.W.; Lee, J.H.; Yoon, H.S. Potential Application of the Oryza sativa Monodehydroascorbate Reductase Gene (OsMDHAR) to Improve the Stress Tolerance and Fermentative Capacity of Saccharomyces cerevisiae. PLoS ONE 2016, 11, e0158841. [Google Scholar]
- Truffault, V.; Riqueau, G.; Garchery, C.; Gautier, H.; Stevens, R.G. Is monodehydroascorbate reductase activity in leaf tissue critical for the maintenance of yield in tomato? J. Plant Physiol. 2018, 222, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Rus, E.; Amaya, I.; Sánchez-Sevilla, J.F.; Botella, M.A.; Valpuesta, V. Regulation of L-ascorbic acid content in strawberry fruits. J. Exp. Bot. 2011, 62, 4191–4201. [Google Scholar] [CrossRef]
- Tao, C.; Jin, X.; Zhu, L.; Xie, Q.; Wang, X.; Li, H. Genome-wide investigation and expression profiling of APX gene family in Gossypium hirsutum provide new insights in redox homeostasis maintenance during different fiber development stages. Mol. Genet. Genom. 2018, 293, 685–697. [Google Scholar] [CrossRef]
- Yang, Z.; Qanmber, G.; Wang, Z.; Yang, Z.; Li, F. Gossypium Genomics: Trends, Scope, and Utilization for Cotton Improvement. Trends Plant. Sci. 2020, 25, 488–500. [Google Scholar] [CrossRef]
- Song, W.; Wang, F.; Chen, L.; Ma, R.; Zuo, X.; Cao, A.; Xie, S.; Chen, X.; Jin, X.; Li, H. GhVTC1, the Key Gene for Ascorbate Biosynthesis in Gossypium hirsutum, Involves in Cell Elongation Under Control of Ethylene. Cells 2019, 8, 1039. [Google Scholar] [CrossRef]
- Ma, R.; Song, W.; Wang, F.; Cao, A.; Xie, S.; Chen, X.; Jin, X.; Li, H. A Cotton (Gossypium hirsutum) Myo-Inositol-1-Phosphate Synthase (GhMIPS1D) Gene Promotes Root Cell Elongation in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 1224. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, K.; George, S.; Venkataraman, G.; Parida, A. A salt-inducible chloroplastic monodehydroascorbate reductase from halophyte Avicennia marina confers salt stress tolerance on transgenic plants. Biochimie 2010, 92, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Qin, Y.; Song, W.; Li, J.; Zhu, Y. Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics 2009, 36, 141–150. [Google Scholar] [CrossRef]
- Chen, Z.; Young, T.E.; Ling, J.; Chang, S.C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc. Natl. Acad. Sci. USA 2003, 100, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Bulley, S.; Laing, W. The regulation of ascorbate biosynthesis. Curr. Opin. Plant Biol. 2016, 33, 15–22. [Google Scholar] [CrossRef]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Río, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef]
- Barba-Espin, G.; Diaz-Vivancos, P.; Clemente-Moreno, M.J.; Albacete, A.; Faize, L.; Faize, M.; Pérez-Alfocea, F.; Hernández, J.A. Interaction between hydrogen peroxide and plant hormones during germination and the early growth of pea seedlings. Plant Cell Environ. 2010, 33, 981–994. [Google Scholar] [CrossRef]
- Gest, N.; Garchery, C.; Gautier, H.; Jiménez, A.; Stevens, R. Light-dependent regulation of ascorbate in tomato by a monodehydroascorbate reductase localized in peroxisomes and the cytosol. Plant Biotechnol. J. 2013, 11, 344–354. [Google Scholar] [CrossRef]
- Sultana, S.; Khew, C.Y.; Morshed, M.M.; Namasivayam, P.; Napis, S.; Ho, C.L. Overexpression of monodehydroascorbate reductase from a mangrove plant (AeMDHAR) confers salt tolerance on rice. J. Plant Physiol. 2012, 169, 311–318. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Du, X.; Tu, L.; Tang, W.; Wang, P.; Wang, M.; Liu, Z.; Zhang, X. Fibre elongation requires normal redox homeostasis modulated by cytosolic ascorbate peroxidase in cotton (Gossypium hirsutum). J. Exp. Bot. 2016, 67, 3289–3301. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.Y.; Wang, H.; Pang, Y.; Xu, C.; Jiao, Y.; Qin, Y.M.; Western, T.L.; Yu, S.X.; Zhu, Y.X. Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol. Cell Proteomics 2010, 9, 2019–2033. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Eberhard, S.; Pattathil, S.; Warder, C.; Glushka, J.; Yuan, C.; Hao, Z.; Zhu, X.; Avci, U.; Miller, J.S.; et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 2013, 25, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Kavi Kishor, P.B.; Hima Kumari, P.; Sunita, M.S.; Sreenivasulu, N. Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front. Plant Sci. 2015, 6, 544. [Google Scholar] [CrossRef]
- Marzol, E.; Borassi, C.; Bringas, M.; Sede, A.; Rodríguez Garcia, D.R.; Capece, L.; Estevez, J.M. Filling the Gaps to Solve the Extensin Puzzle. Mol. Plant 2018, 11, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Carol, R.J.; Dolan, L. The role of reactive oxygen species in cell growth: Lessons from root hairs. J. Exp. Bot. 2006, 57, 1829–1834. [Google Scholar] [CrossRef]
- Pitts, R.J.; Cernac, A.; Estelle, M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998, 16, 553–560. [Google Scholar] [CrossRef]
- Achard, P.; Vriezen, W.H.; Van Der Straeten, D.; Harberd, N.P. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 2003, 15, 2816–2825. [Google Scholar] [CrossRef]
- Grauwe, L.D.; Vandenbussche, F.; Tietz, O.; Palme, K.; Straeten, D.V.D. Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 2005, 46, 827–836. [Google Scholar] [CrossRef]
- Shi, Y.H.; Zhu, S.W.; Mao, X.Z.; Feng, J.X.; Qin, Y.M.; Zhang, L.; Cheng, J.; Wei, L.P.; Wang, Z.Y.; Zhu, Y.X. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006, 18, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.M.; Hu, C.Y.; Pang, Y.; Kastaniotis, A.J.; Hiltunen, J.K.; Zhu, Y.X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 2007, 19, 3692–3704. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Chen, G.L.; Bartel, B. Ethylene directs auxin to control root cell expansion. Plant J. 2010, 64, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, X.; Song, S.; Zeng, Q.; Hou, L.; Li, D.; Zhao, J.; Wei, Y.; Li, X.; Luo, M.; et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 2011, 29, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xu, P.; Li, B.S.; Li, P.P.; Wen, X.; An, F.Y.; Gong, Y.; Xin, Y.; Zhu, Z.Q.; Wang, Y.C.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci USA 2017, 114, 13834–13839. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, Y.; Fu, Y.; Guo, H. Ethylene-induced microtubule reorientation is essential for fast inhibition of root elongation in Arabidopsis. J. Integr. Plant Biol. 2018, 60, 864–877. [Google Scholar] [CrossRef]
- Davey, M.W.; Van Montagu, M.; Inze, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.J.J.; Strain, J.J.; Favell, D.; Fletcher, J. Plant l–ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Conklin, P.L. Recent advances in the role and biosynthesis of ascorbic acid in plants. Plant Cell Environ. 2001, 24, 383–394. [Google Scholar] [CrossRef]
- Arrigoni, O.; De Tullio, M.C. Ascorbic acid: Much more than an antioxidant. Biochim. Biophys. Acta. 2002, 1569, 1–9. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L.M.; Hulse-Kemp, A.M.; Schmutz, J. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Schmutz, J. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Tong, X.; Cao, A.; Wang, F.; Chen, X.; Xie, S.; Shen, H.; Jin, X.; Li, H. Calcium-Dependent Protein Kinase Genes in Glycyrrhiza Uralensis Appear to be Involved in Promoting the Biosynthesis of Glycyrrhizic Acid and Flavonoids under Salt Stress. Molecules 2019, 24, 1837. [Google Scholar] [CrossRef]
- Zhu, L.; Jin, X.; Xie, Q.; Yao, Q.; Wang, X.; Li, H. Calcium-Dependent Protein Kinase Family Genes Involved in Ethylene-Induced Natural Rubber Production in Different Hevea brasiliensis Cultivars. Int. J. Mol. Sci. 2018, 19, 947. [Google Scholar] [CrossRef]
- Yadav, C.B.; Bonthala, V.S.; Muthamilarasan, M.; Pandey, G.; Khan, Y.; Prasad, M. Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res. 2015, 22, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Chen, L.; Wang, F.; Song, W.; Cao, A.; Xie, S.; Chen, X.; Jin, X.; Li, H. Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence. Int. J. Mol. Sci. 2019, 20, 6167. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vanacker, H.; Guichard, M.; Bohrer, A.S.; Issakidis-Bourguet, E. Redox Regulation of Monodehydroascorbate Reductase by Thioredoxin y in Plastids Revealed in the Context of Water Stress. Antioxidants 2018, 7, 183. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, F.; Zheng, B.; Wang, F.; Cao, A.; Xie, S.; Chen, X.; Schick, J.A.; Jin, X.; Li, H. Genome-Wide Analysis of MDHAR Gene Family in Four Cotton Species Provides Insights into Fiber Development via Regulating AsA Redox Homeostasis. Plants 2021, 10, 227. https://doi.org/10.3390/plants10020227
Zhou F, Zheng B, Wang F, Cao A, Xie S, Chen X, Schick JA, Jin X, Li H. Genome-Wide Analysis of MDHAR Gene Family in Four Cotton Species Provides Insights into Fiber Development via Regulating AsA Redox Homeostasis. Plants. 2021; 10(2):227. https://doi.org/10.3390/plants10020227
Chicago/Turabian StyleZhou, Fangfang, Bowen Zheng, Fei Wang, Aiping Cao, Shuangquan Xie, Xifeng Chen, Joel A. Schick, Xiang Jin, and Hongbin Li. 2021. "Genome-Wide Analysis of MDHAR Gene Family in Four Cotton Species Provides Insights into Fiber Development via Regulating AsA Redox Homeostasis" Plants 10, no. 2: 227. https://doi.org/10.3390/plants10020227
APA StyleZhou, F., Zheng, B., Wang, F., Cao, A., Xie, S., Chen, X., Schick, J. A., Jin, X., & Li, H. (2021). Genome-Wide Analysis of MDHAR Gene Family in Four Cotton Species Provides Insights into Fiber Development via Regulating AsA Redox Homeostasis. Plants, 10(2), 227. https://doi.org/10.3390/plants10020227




