Proteomics and Interspecies Interaction Analysis Revealed Abscisic Acid Signalling to Be the Primary Driver for Oil Palm’s Response against Red Palm Weevil Infestation
Abstract
1. Introduction
2. Results
2.1. Patterns of Protein Expressions
2.2. Literature Research on Differentially Expressed Proteins
2.3. Modelling and Docking Analysis
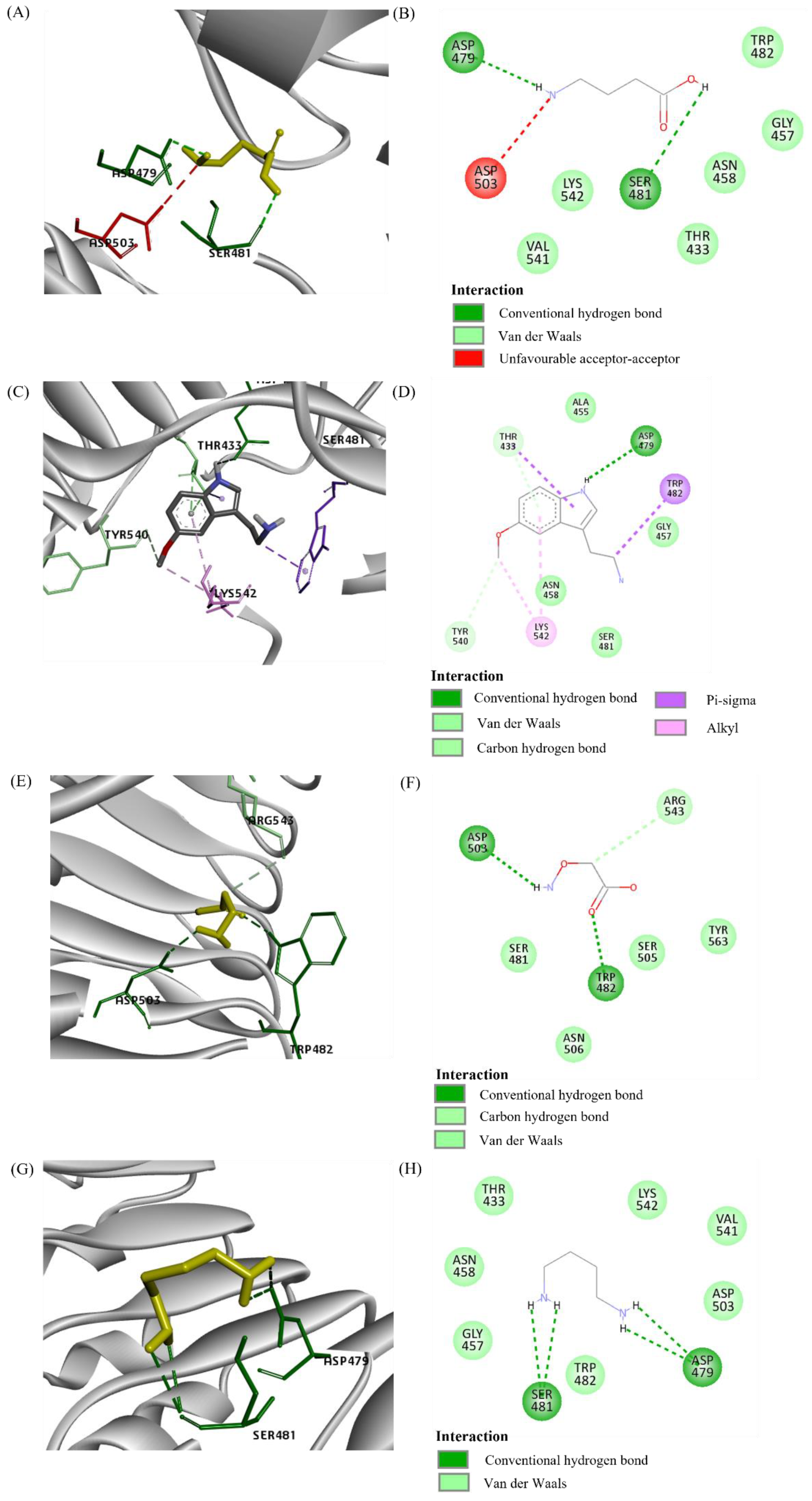
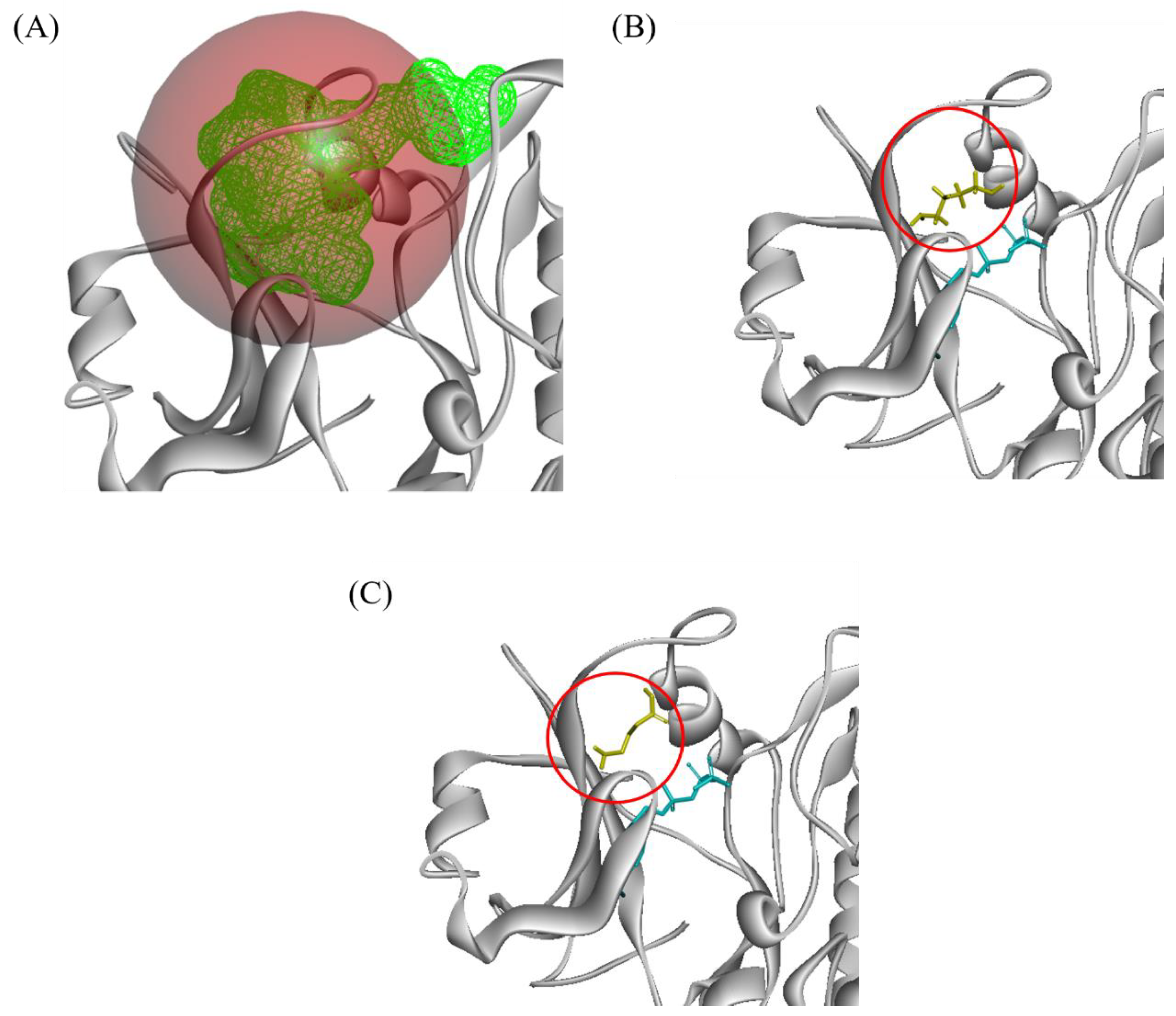
2.3.1. Overlapping Binding Region
2.3.2. Close Proximity Binding Region
3. Discussion
3.1. Photosynthesis-Related Proteins
3.2. Growth Related Proteins
3.3. Stress Response Related Proteins
3.4. Docking
3.5. Abscisic Acid Driving Systemic Response
4. Materials and Methods
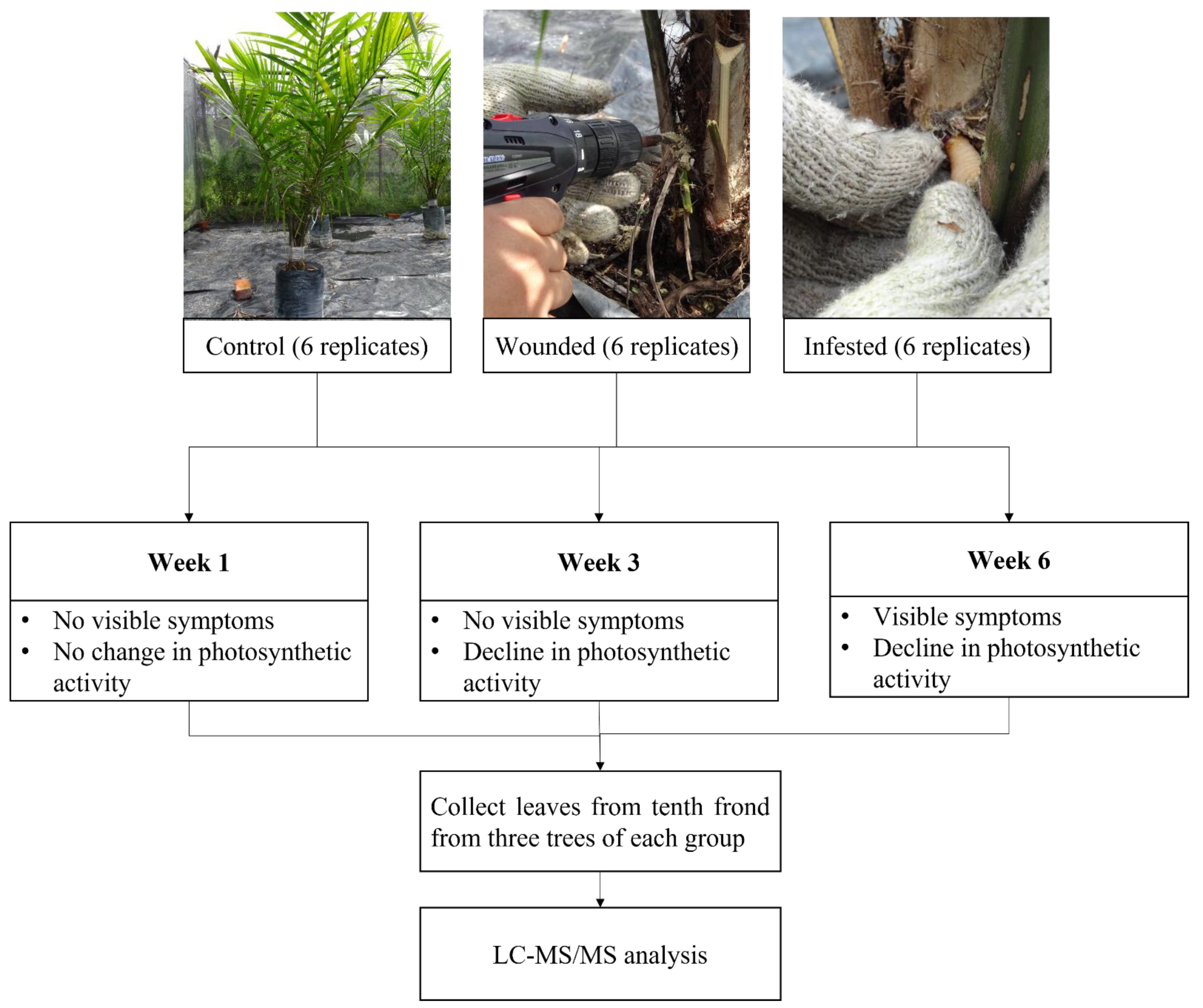
4.1. Artificial Infestation
4.2. Protein Extraction, SDS-PAGE and Peptide Digestion
4.3. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
4.4. Peptide Identification and Quantification
4.5. Statistical Analysis
4.6. Protein Enrichment Analysis and Literature Searches
4.7. Receptor Protein and Ligand Selection
4.8. Docking Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CABI. Red Palm Weevil Datasheet. Available online: http://www.cabi.org/isc/datasheet/47472 (accessed on 4 April 2017).
- Azmi, W.A.; Chik, Z.; Rahman, A.; Razak, A.R.; Tzzah, N.; Gham, A. A new invasive coconut pest in Malaysia: The red palm weevil (Curculionidae: Rhynchophorus ferrugineus). Plant. Kuala Lumpur 2013, 89, 97–110. [Google Scholar]
- Rochat, D.; Dembilio, O.; Jaques, J.A.; Suma, P.; La Pergola, A.; Hamidi, R.; Kontodimas, D.; Soroker, V. Rhynchophorus ferrugineus: Taxonomy, distribution, biology, and life cycle. In Handbook of Major Palm Pests: Biology and Management; Soroker, V., Colazza, S., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 69–101. ISBN 9781119057482. [Google Scholar]
- Jaques, J.A. Guidelines on visual inspection for early detection of red palm weevil in Canary Island palm (Phoenix canariensis). In Red Palm Weevil: Guidelines on Management Practices; Elkahy, M., Faleiro, J.R., Eds.; Food and Agriculture Organizations of United Nations: Rome, Italy, 2020; pp. 21–23. ISBN 9789251321898. [Google Scholar]
- Vidyasagar, P.S.P.V. Guidelines on visual inspection for early detection of red palm weevil in date palm (Phoenix dactylifera). In Red Palm Weevil: Guidelines on Management Practices; Elkahy, M., Faleiro, J.R., Eds.; Food and Agriculture Organizations of United Nations: Rome, Italy, 2020; pp. 11–19. ISBN 9789251321898. [Google Scholar]
- Harith-fadzilah, N.; Idris, M.H.; Ghani, A.; Zakaria, A.; Amit, S.; Zainal, Z.; Azmi, W.A.; Jalinas, J.; Hassan, M. Physical and physiological monitoring on red palm weevil-infested oil palms. Insects 2020, 11, 407. [Google Scholar] [CrossRef]
- Idris, A.B.; Mokhtaruddin, H.; Zazali, C.; Nurul Wahida, O.; Yaakop, S.; Hazmi, I.R. The potential of red palm weevil infesting and destroying oil palm industry in Malaysia. Planter 2014, 90, 329–335. [Google Scholar]
- Sticher, L.; Mauch-Mani, B.; Metraux, J. Systemic acquired resistance. Annu. Rev. Phytopathol. 1997, 35, 235–270. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Prasad, M. Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 2013, 38, 433–449. [Google Scholar] [CrossRef]
- Meena, R.K.; Jangra, S.; Wadhwa, Z. Role of plant volatiles in defense and communication. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 300–313. [Google Scholar] [CrossRef]
- Malik, N.A.A.; Kumar, I.S.; Nadarajah, K. Elicitor and receptor molecules: Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef]
- Mohr, P.G.; Cahill, D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 2003, 30, 461–469. [Google Scholar] [CrossRef]
- Kunz, B.A.; Dando, P.K.; Grice, D.M.; Mohr, P.G.; Schenk, P.M.; Cahill, D.M. UV-induced DNA damage promotes resistance to the biotrophic pathogen Hyaloperonospora parasitica in Arabidopsis. Plant Physiol. 2008, 148, 1021–1031. [Google Scholar] [CrossRef][Green Version]
- Von Dahl, C.C.; Baldwin, I.T. Deciphering the role of ethylene in plant-herbivore interactions. J. Plant Growth Regul. 2007, 26, 201–209. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J.G.M. How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Bos, J.I.B. Effector proteins that modulate plant-insect interactions. Curr. Opin. Plant Biol. 2011, 14, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.N.; Shafinaz, N.; Yaakop, S.; Othman, N.W. Distribution of serotonin (5-HT) and dopamine (DA) on digestive tract of red palm weevil larva, Rhynchophorus ferrugineus (Coleoptera:Dryophthoridae). Sains Malays. 2016, 21, 39–50. [Google Scholar]
- Pu, Y.C.; Xiang, H.J.; Liang, X.Y.; Wang, Y.; Hou, Y.M.; Fu, L.; Wang, R. External immune inhibitory efficiency of external secretions and their metabolic profiling in red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Front. Physiol. 2020, 10, 1624. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Nürnberger, T. A plant surface receptor for sensing insect herbivory. Proc. Natl. Acad. Sci. USA 2020, 117, 32839–32841. [Google Scholar] [CrossRef]
- Ranf, S. Pattern recognition receptors-versatile genetic tools for engineering broad-spectrum disease resistance in crops. Agronomy 2018, 8, 134. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, X.; Liang, S.; Fu, S.; Qi, Y.; Zhao, J.; Shao, J.; An, L.; Yu, F. Chloroplast translation initiation factors regulate leaf variegation and development. Plant Physiol. 2016, 172, 1117–1130. [Google Scholar] [CrossRef]
- Azarin, K.; Usatov, A.; Makarenko, M.; Kozel, N.; Kovalevich, A.; Dremuk, I.; Yemelyanova, A.; Logacheva, M.; Fedorenko, A.; Averina, N. A point mutation in the photosystem I P700 chlorophyll a apoprotein A1 gene confers variegation in Helianthus annuus L. Plant Mol. Biol. 2020, 103, 373–389. [Google Scholar] [CrossRef]
- Kim, C.; Keun, P.L.; Baruah, A.; Nater, M.; Göbel, C.; Feussner, I.; Apel, K. 1O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proc. Natl. Acad. Sci. USA 2009, 106, 9920–9924. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y. Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front. Plant Sci. 2016, 7, 168. [Google Scholar] [CrossRef]
- Sirhindi, G.; Mushtaq, R.; Gill, S.S.; Sharma, P.; Abd_Allah, E.F.; Ahmad, P. Jasmonic acid and methyl jasmonate modulate growth, photosynthetic activity and expression of photosystem II subunit genes in Brassica oleracea L. Sci. Rep. 2020, 10, 9322. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H.; Li, M.; Puthiyaveetil, S. Sublocalization of cytochrome b6f complexes in photosynthetic membranes. Trends Plant Sci. 2017, 22, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Thamil Arasan, S.K.; Park, J.I.; Ahmed, N.U.; Jung, H.J.; Hur, Y.; Kang, K.K.; Lim, Y.P.; Nou, I.S. Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Biochem. 2013, 67, 144–153. [Google Scholar] [CrossRef]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W.; et al. A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef]
- Huis, R.; Morreel, K.; Fliniaux, O.; Lucau-Danila, A.; Fénart, S.; Grec, S.; Neutelings, G.; Chabbert, B.; Mesnard, F.; Boerjan, W.; et al. Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol. 2012, 158, 1893–1915. [Google Scholar] [CrossRef]
- Villalobos, D.P.; Díaz-Moreno, S.M.; Said, E.S.S.; Cañas, R.A.; Osuna, D.; Van Kerckhoven, S.H.E.; Bautista, R.; Claros, M.G.; Cánovas, F.M.; Cantón, F.R. Reprogramming of gene expression during compression wood formation in pine: Coordinated modulation of S-adenosylmethionine, lignin and lignan related genes. BMC Plant Biol. 2012, 12, 100. [Google Scholar] [CrossRef]
- Jain, P.; Bhatla, S.C. Signaling role of phospholipid hydroperoxide glutathione peroxidase (PHGPX) accompanying sensing of NaCl stress in etiolated sunflower seedling cotyledons. Plant Signal. Behav. 2014, 9, e977746-1–e977746-7. [Google Scholar] [CrossRef]
- Gui, J.; Zheng, S.; Liu, C.; Shen, J.; Li, J.; Li, L. OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 2016, 38, 201–213. [Google Scholar] [CrossRef]
- Schaller, A.; Stintzi, A.; Rivas, S.; Serrano, I.; Chichkova, N.V.; Vartapetian, A.B.; Martínez, D.; Guiamét, J.J.; Sueldo, D.J.; van der Hoorn, R.A.L.; et al. From structure to function—A family portrait of plant subtilases. New Phytol. 2018, 218, 901–915. [Google Scholar] [CrossRef]
- Von Groll, U.; Berger, D.; Altmann, T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 2002, 14, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.; Torii, Y.; Hayashi, S.I.; Takimoto, K.; Matsui, K.; Nakamura, K.; Inzé, D.; Babiychuk, E.; Kushnir, S.; Asada, K. The NADPH:Quinone oxidoreductase P1-ζ-crystallin in Arabidopsis catalyzes the α,β-hydrogenation of 2-alkenals: Detoxication of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol. 2002, 43, 1445–1455. [Google Scholar] [CrossRef]
- Mano, J.; Belles-Boix, E.; Babiychuk, E.; Inzé, D.; Torii, Y.; Hiraoka, E.; Takimoto, K.; Slooten, L.; Asada, K.; Kushnir, S. Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase. Detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol. 2005, 139, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Wang, R.; Guan, Y.; Liu, Y.; Zhang, S. Sulfurtransferases 1 and 2 play essential roles in embryo and seed development in Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 7548–7557. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Cilia, M.; San Roman, A.; Thomas, C.; Maule, A.; Hearn, S.; Jackson, D. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 2009, 106, 3615–3620. [Google Scholar] [CrossRef]
- El-Shetehy, M.; Wang, C.; Shine, M.B.; Yu, K.; Kachroo, A.; Kachroo, P. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 2015, 10, e998544. [Google Scholar] [CrossRef]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 2018, 154, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Correa-Aragunde, N.; Cejudo, F.J.; Lamattina, L. Nitric oxide is required for the auxin-induced activation of NADPH-dependent thioredoxin reductase and protein denitrosylation during root growth responses in Arabidopsis. Ann. Bot. 2015, 116, 695–702. [Google Scholar] [CrossRef]
- Bashandy, T.; Meyer, Y.; Reichheld, J.P. Redox regulation of auxin signaling and plant development in Arabidopsis. Plant Signal. Behav. 2011, 6, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef]
- Liu, R.; Xu, Y.H.; Jiang, S.C.; Lu, K.; Lu, Y.F.; Feng, X.J.; Wu, Z.; Liang, S.; Yu, Y.T.; Wang, X.F.; et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 2013, 64, 5443–5456. [Google Scholar] [CrossRef] [PubMed]
- Barickman, T.C.; Kopsell, D.A.; Sams, C.E. Abscisic acid increases carotenoid and chlorophyll concentrations in leaves and fruit of two tomato genotypes. J. Am. Soc. Hortic. Sci. 2014, 139, 261–266. [Google Scholar] [CrossRef]
- Brausemann, A.; Gemmecker, S.; Koschmieder, J.; Ghisla, S.; Beyer, P.; Einsle, O. Structure of phytoene desaturase provides insights into herbicide binding and reaction mechanisms involved in carotene desaturation. Structure 2017, 25, 1222–1232.e3. [Google Scholar] [CrossRef] [PubMed]
- Koschmieder, J.; Fehling-Kaschek, M.; Schaub, P.; Ghisla, S.; Brausemann, A.; Timmer, J.; Beyer, P. Plant-type phytoene desaturase: Functional evaluation of structural implications. PLoS ONE 2017, 12, e0187628. [Google Scholar] [CrossRef] [PubMed]
- LeBrasseur, N.D.; MacIntosh, G.C.; Pérez-Amador, M.A.; Saitoh, M.; Green, P.J. Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. Plant J. 2002, 29, 393–403. [Google Scholar] [CrossRef]
- Sangaev, S.S.; Kochetov, A.V.; Ibragimova, S.S.; Levenko, B.A.; Shumny, V.K. Physiological role of extracellular ribonucleases of higher plants. Russ. J. Genet. Appl. Res. 2011, 1, 44–50. [Google Scholar] [CrossRef]
- Ifuku, K.; Yamamoto, Y.; Ono, T.A.; Ishihara, S.; Sato, F. PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol. 2005, 139, 1175–1184. [Google Scholar] [CrossRef]
- Hong, Y.; Wang, Z.; Liu, X.; Yao, J.; Kong, X.; Shi, H.; Zhu, J.K. Two chloroplast proteins negatively regulate plant drought resistance through separate pathways. Plant Physiol. 2020, 182, 1007–1021. [Google Scholar] [CrossRef]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 871, 1836. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Liu, H.Y.; Xu, H.M.; Li, M.; Kang, Z.S. Analysis of differential transcriptional profiling in wheat infected by Blumeria graminis f. sp. tritici using GeneChip. Mol. Biol. Rep. 2012, 39, 381–387. [Google Scholar] [CrossRef]
- DeRocher, A.E.; Helm, K.W.; Lauzon, L.M.; Vierling, E. Expression of a conserved family of cytoplasmic low molecular weight heat shock proteins during heat stress and recovery 1. Plant Physiol. 1991, 96, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Yu, G.X.; Meng, X.; Xu, J.; Stephenson, E.; Estrada, S.; Chilakamarri, S.; Zastrow-Hayes, G.; Thatcher, S. Developmental and transcriptional responses of maize to drought stress under field conditions. Plant Direct 2019, 3, e00129. [Google Scholar] [CrossRef] [PubMed]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.X.; Zhang, H.X.; Wei, A.M.; Gong, Z.H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Rehrig, E.M.; Appel, H.M.; Jones, A.D.; Schultz, J.C. Roles for jasmonate- and ethylene-induced transcription factors in the ability of Arabidopsis to respond differentially to damage caused by two insect herbivores. Front. Plant Sci. 2014, 5, 407. [Google Scholar] [CrossRef]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef]
- Scholz, S.S.; Reichelt, M.; Mekonnen, D.W.; Ludewig, F.; Mithöfer, A. Insect herbivory-elicited gaba accumulation in plants is a wound-induced, direct, systemic, and jasmonate-independent defense response. Front. Plant Sci. 2015, 6, 1128. [Google Scholar] [CrossRef]
- Steinite, I.; Gailite, A.; Ievinsh, G. Reactive oxygen and ethylene are involved in the regulation of regurgitant-induced responses in bean plants. J. Plant Physiol. 2004, 161, 191–196. [Google Scholar] [CrossRef]
- Rasool, K.G.; Khan, M.A.; Tufail, M.; Husain, M.; Mehmood, K.; Mukhtar, M.; Takeda, M.; Aldawood, A.S. Differential proteomic analysis of date palm leaves infested with the red palm weevil (Coleoptera: Curculionidae). Florida Entomol. 2018, 101, 290–298. [Google Scholar] [CrossRef]
- Sugiura, M. The chloroplast genome. Plant Mol. Biol. 1992, 19, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S. The light-harvesting chlorophyll ab-binding proteins. Biochim. Biophys. Acta Bioenerg. 1994, 1184, 1–19. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F. Systems biology approach in plant abiotic stresses. Plant Physiol. Biochem. 2017, 121, 58–73. [Google Scholar] [CrossRef]
- Wittmann, D.; Sinha, N.; Grimm, B. Thioredoxin-dependent control balances the metabolic activities of tetrapyrrole biosynthesis. Biol. Chem. 2021, 402, 379–397. [Google Scholar] [CrossRef]
- Reichheld, J.P.; Khafif, M.; Riondet, C.; Droux, M.; Bonnard, G.; Meyer, Y. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in arabidopsis development. Plant Cell 2007, 19, 1851–1865. [Google Scholar] [CrossRef] [PubMed]
- Ifuku, K.; Ishihara, S.; Sato, F. Molecular functions of oxygen-evolving complex family proteins in photosynthetic electron flow. J. Integr. Plant Biol. 2010, 52, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Bricker, T.M.; Roose, J.L.; Zhang, P.; Frankel, L.K. The PsbP family of proteins. Photosynth. Res. 2013, 116, 235–250. [Google Scholar] [CrossRef]
- Norhayati, Y.; Wahizatul, A.A.; Siti, N.J.S.; Nurul, W.M.R. Antioxidative responses of Cocos nucifera against infestation by the red palm weevil (RPW), Rhynchophorus ferrugineus, a new invasive coconut pest in Malaysia. Sains Malays. 2016, 45, 1035–1040. [Google Scholar]
- Li, W.J.; Feng, H.; Fan, J.H.; Zhang, R.Q.; Zhao, N.M.; Liu, J.Y. Molecular cloning and expression of a phospholipid hydroperoxide glutathione peroxidase homolog in Oryza sativa. Biochim. Biophys. Acta Gene Struct. Expr. 2000, 1493, 225–230. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Rakwal, R.; Jwa, N.S.; Agrawal, V.P. Effects of signaling molecules, protein phosphatase inhibitors and blast pathogen (Magnaporthe grisea) on the mRNA level of a rice (Oryza sativa L.) phospholipid hydroperoxide glutathione peroxidase (OsPHGPX) gene in seedling leaves. Gene 2002, 283, 227–236. [Google Scholar] [CrossRef]
- Jarsch, I.K.; Ott, T. Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant-Microbe Interact. 2011, 24, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Sun, Y.; Ma, Z.; Ke, M.; Cui, Y.; Chen, Z.; Chen, C.; Ji, C.; Tran, T.M.; Yang, L.; et al. Salicylic acid-mediated plasmodesmal closure via remorin-dependent lipid organization. Proc. Natl. Acad. Sci. USA 2019, 116, 21274–21284. [Google Scholar] [CrossRef]
- Weinmann, H.; Ottow, E. Recent development in novel anticancer therapies. In Comprehensive Medicinal Chemistry II; Taylor, J.B., Triggle, D.J., Eds.; Elsevier: Oxford, UK, 2007; pp. 221–251. ISBN 978-0-08-045044-5. [Google Scholar]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef]
- Tian, D.; Peiffer, M.; Shoemaker, E.; Tooker, J.; Haubruge, E.; Francis, F.; Luthe, D.S.; Felton, G.W. Salivary glucose oxidase from caterpillars mediates the induction of rapid and delayed-induced defenses in the tomato plant. PLoS ONE 2012, 7, e36168. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Assmann, S.M. The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5179–5189. [Google Scholar] [CrossRef] [PubMed]
- Frolov, A.; Bilova, T.; Paudel, G.; Berger, R.; Balcke, G.U.; Birkemeyer, C.; Wessjohann, L.A. Early responses of mature Arabidopsis thaliana plants to reduced water potential in the agar-based polyethylene glycol infusion drought model. J. Plant Physiol. 2017, 208, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, X.; Liu, X.; Yu, H.; Du, C.; Li, M.; He, D. Proteomic analysis of the compatible interaction of wheat and powdery mildew (Blumeria graminis f. sp. tritici). Plant Physiol. Biochem. 2017, 111, 234–243. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding affinity via docking: Fact and fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.L.; Rahman, A.; Baskin, T.I.; Kieber, J.J. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 2008, 20, 3065–3079. [Google Scholar] [CrossRef]
- Mariano, A.C.; Andrade, M.O.; Santos, A.A.; Carolino, S.M.B.; Oliveira, M.L.; Baracat-Pereira, M.C.; Brommonshenkel, S.H.; Fontes, E.P.B. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 2004, 318, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Fontes, E.P.B.; Santos, A.A.; Luz, D.F.; Waclawovsky, A.J.; Chory, J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004, 18, 2545–2556. [Google Scholar] [CrossRef]
- Kaufmann, C.; Motzkus, M.; Sauter, M. Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. J. Exp. Bot. 2017, 68, 1411–1423. [Google Scholar] [CrossRef]
- Rajamanickam, K.; Schönhof, M.D.; Hause, B.; Sauter, M. PSK signaling controls ABA homeostasis and signaling genes and maintains shoot growth under osmotic stress. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Dembilio, Ó.; Agut, B.; IbÁÑEz-Gual, M.V.; Flors, V.; Jaques, J.A. Could plant hormones provide a reliable tool for early detection of Rhynchophorus ferrugineus (coleoptera: Curculionidae) infested palms? J. Entomol. Res. Soc. 2019, 21, 1–9. [Google Scholar]
- Rasool, K.G.; Khan, M.A.; Aldawood, A.S.; Tufail, M.; Mukhtar, M.; Takeda, M. Identification of proteins modulated in the date palm stem infested with red palm weevil (Rhynchophorus ferrugineus oliv.) using two dimensional differential gel electrophoresis and mass spectrometry. Int. J. Mol. Sci. 2015, 16, 19326–19346. [Google Scholar] [CrossRef]
- Azzeme, A.M.; Abdullah, S.N.A.; Aziz, M.A.; Wahab, P.E.M. Oil palm leaves and roots differ in physiological response, antioxidant enzyme activities and expression of stress-responsive genes upon exposure to drought stress. Acta Physiol. Plant. 2016, 38, 52. [Google Scholar] [CrossRef]
- Giovino, A.; Bertolini, E.; Fileccia, V.; Al Hassan, M.; Labra, M.; Martinelli, F. Transcriptome analysis of Phoenix canariensis Chabaud in response to Rhynchophorus ferrugineus Olivier attacks. Front. Plant Sci. 2015, 6, 817. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, M.; Tomita, T.; Vogelstein, J.T.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Selected reaction monitoring approach for validating peptide biomarkers. Proc. Natl. Acad. Sci. USA 2017, 114, 13519–13524. [Google Scholar] [CrossRef]
- Hassan, H.; Lau, B.Y.C.; Ramli, U.S.; Palm, O. Extraction methods for analysis of oil palm leaf and root proteins by two- dimensional gel electrophoresis. J. Oil Palm Res. 2014, 26, 54–61. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havliš, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2007, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Hamezah, H.S.; Durani, L.W.; Yanagisawa, D.; Ibrahim, N.F.; Aizat, W.M.; Bellier, J.P.; Makpol, S.; Ngah, W.Z.W.; Damanhuri, H.A.; Tooyama, I. Proteome profiling in the hippocampus, medial prefrontal cortex, and striatum of aging rat. Exp. Gerontol. 2018, 111, 53–64. [Google Scholar] [CrossRef]
- Cox, J. First Steps with MaxQuant. Available online: http://www.coxdocs.org/doku.php?id=maxquant:manual:beginner (accessed on 14 December 2019).
- Draga, M. Label-Free Interaction Data. Available online: http://www.coxdocs.org/doku.php?id=perseus:user:use_cases:interactions (accessed on 14 December 2019).
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Lam, S.D.; Das, S.; Sillitoe, I.; Orengo, C. An overview of comparative modelling and resources dedicated to large-scale modelling of genome sequences. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 628–640. [Google Scholar] [CrossRef]
- Söding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.Y.; Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008, 426, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; John, B.; Mirkovic, N.; Fiser, A.; Ilyin, V.A.; Pieper, U.; Stuart, A.C.; Marti-Renom, M.A.; Madhusudhan, M.S.; Yerkovich, B.; et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003, 31, 3375–3380. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Glob. Food Secur. Gov. 2015, 1263, 1–11. [Google Scholar] [CrossRef]
- Dallakian, S. PyRx Ligand Docking Tutorial. Available online: https://pyrx.sourceforge.io/blog/121-pyrx-ligand-docking-tutorial (accessed on 14 December 2019).
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
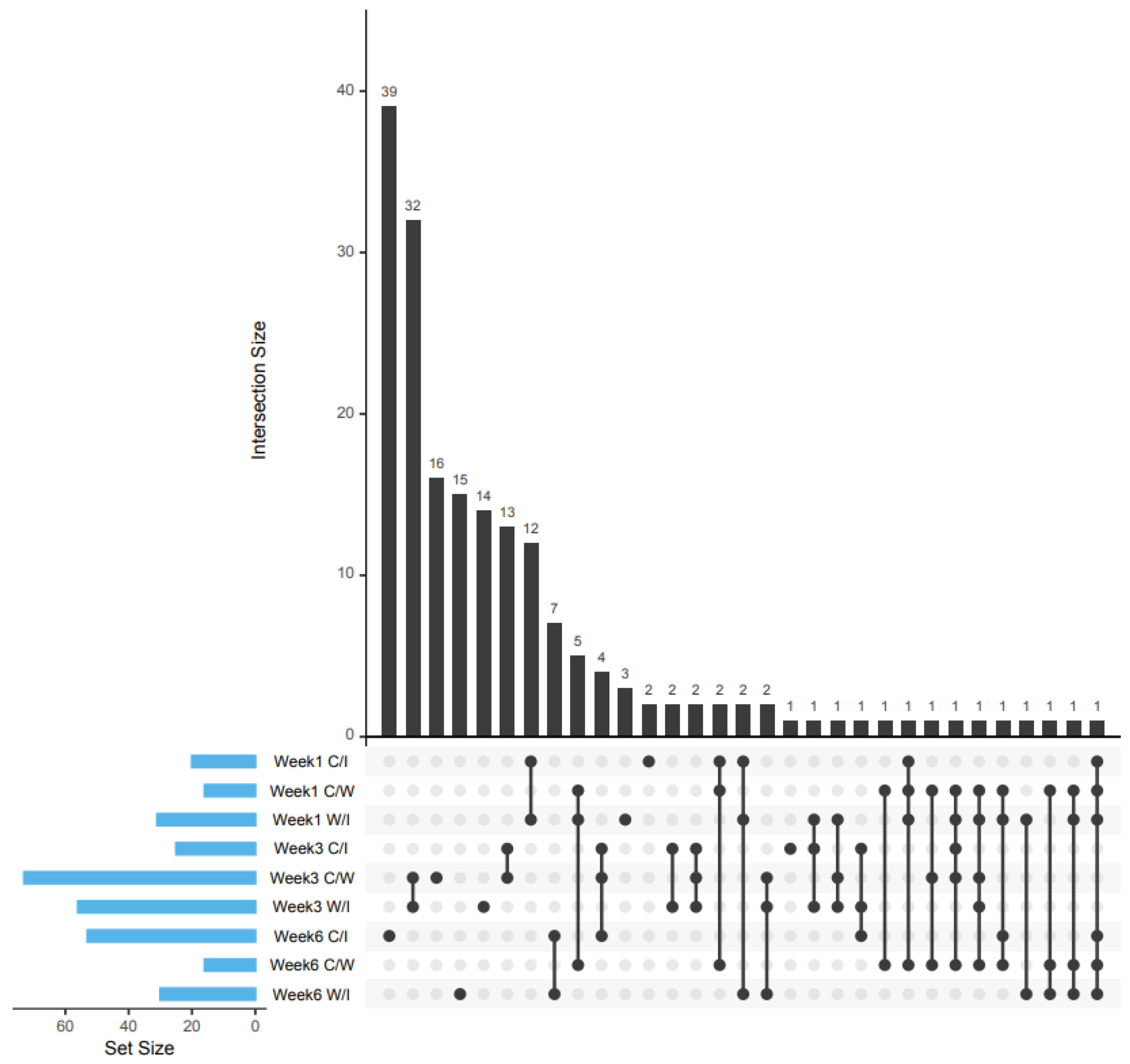




| Week | Comparison Group | ||
|---|---|---|---|
| Control/Infested | Wounded/Infested | Control/Wounded | |
| 1 |
|
|
|
| 3 |
|
|
|
| 6 |
|
|
|
| NCBI Accession ID | Description | Abbreviation | Week | Function | Hormone Influence | Log2 FC W/I | Reference |
|---|---|---|---|---|---|---|---|
| XP_010905021.1 | Translation initiation factor IF3-2, chloroplastic isoform X1 | IF3-2 | 1 | Photosynthesis | - | −1.622 | [23] |
| YP_006073104.1 | Photosystem I P700 apoprotein A1 (chloroplast) | PsaA | 1 | Photosynthesis | ABA(+) | −3.323 | [24,25] |
| YP_006073130.1 | Photosystem II CP47 chlorophyll apoprotein (chloroplast) | PsbB | 1 | Photosynthesis | MeJA(–); ABA(+) | −2.005 | [25,26,27] |
| YP_006073134.1 | Cytochrome b6 (chloroplast) | PetB | 1 | Photosynthesis | - | −2.962 | [28] |
| XP_010912515.1 | Dirigent protein 19 | DIR19 | 1 | Stress response | ABA(+,–); JA(+); MeJA(+) | −2.106 | [29,30,31,32,33] |
| XP_010918555.1 | Probable phospholipid hydroperoxide glutathione peroxidase | PHGPX | 1 | Stress response | JA(+); SA(+); ABA(+) | −2.365 | [34] |
| XP_010905109.1 | Remorin | REM | 1 | Stress response | ABA(+); SA(+) | −1.652 | [35] |
| XP_010906967.1 | Subtilisin-like protease SBT1.2 | SBT1.2 | 1 | Stress response | - | −2.776 | [36,37,38,39] |
| XP_029118427.1 | NADP(+) dependent 2-alkenal reductase | DBR | 1 | Stress response | - | −1.572 | [39] |
| XP_010923778.1 | Thiosulfate/3-mercaptopyruvate sulfurtransferase 2 | 3-MST | 3 | Growth | - | −1.515 | [40] |
| XP_010930644.1 | Thioredoxin M-type, chloroplastic | TRXM | 3 | Growth; photosynthesis | - | −2.021 | [41,42,43] |
| XP_010908796.1 | NADPH-dependent thioredoxin reductase | NTRB | 3 | Growth | - | 1.787 | [44,45] |
| XP_010936352.2 | Chlorophyll a-b binding protein 5, chloroplastic | CAB5 | 3 | Photosynthesis; Stress response | ABA(+) | −1.808 | [46,47,48,49,50] |
| XP_010916973.1 | 15-cis-phytoene desaturase, chloroplastic/chromoplastic | PDS | 3 | Photosynthesis; Stress response | SA(+) | −1.635 | [48,50] |
| XP_010925305.2 | Extracellular ribonuclease LE | RNase LE | 3 | Stress response | - | 2.389 | [51,52] |
| XP_010906401.1 | Psbp domain-containing protein 6, chloroplastic | PPD6 | 6 | Growth | ABA(–) | 1.512 | [53,54] |
| YP_006073134.1 | Cytochrome b6 (chloroplast) | PetB | 6 | Photosynthesis | ABA(+) | −1.546 | [28] |
| XP_010912634.1 | Dirigent protein 2 | DIR2 | 6 | Stress response | ABA(+,–); JA(+), MeJA(+) | −2.105 | [29,30,32,33] |
| XP_010935284.1 | Dirigent protein 7 | DIR7 | 6 | Stress response | ABA(+,–); JA(+), MeJA(+) | 1.924 | [29,30,32,33] |
| XP_010910894.1 | Glutathione S-transferase F11 | GSTF11 | 6 | Stress response | SA(+) | 2.431 | [55,56,57] |
| XP_010912721.1 | 22.7 kDa class IV heat shock protein | HSP22 | 6 | Stress response | ABA[+]; MeJA(+) | −1.844 | [58,59,60] |
| XP_010925290.1 | 18.1 kDa class I heat shock protein | HSP18 | 6 | Stress response | ABA[+]; MeJA(+) | −1.687 | [58,59,60] |
| XP_010925996.1 | 16.9 kDa class I heat shock protein 2 | HSP16.9 | 6 | Stress response | ABA[+]; MeJA(+) | −1.660 | [58,59,60] |
| XP_019708948.1 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | PGM-I | 6 | Stress response | ABA(+) | 2.581 | [51] |
| Compound | Abbreviation | Reference |
|---|---|---|
| Putrescine | PUT | [61] |
| 5-Methoxytryptamine | 5-MT | [62] |
| γ-aminobutyric acid | GABA | [63] |
| Aminooxyacetic acid | AAO | [64] |
| Identity | Accession ID | Template Uniprot ID | Binding Affinity (kcal/mol) | ||||
|---|---|---|---|---|---|---|---|
| T | GABA | 5-MT | AAO | PUT | |||
| probable LRR receptor-like serine/threonine-protein kinase At5g45780 isoform X2 | XP_010925612.1 | 3UIM | −6.9 | n/a | −6.1 | −3.8 | n/a |
| protein NSP-INTERACTING KINASE 1 | XP_010929457.1 | 3UIM | −6.9 | n/a | n/a | n/a | n/a |
| LRR receptor kinase SERK2 isoform X1 | XP_010937435.1 | 3UIM | −6.9 | n/a | n/a | n/a | n/a |
| LRR receptor kinase SERK2 isoform X1 | XP_010937436.1 | 3UIM | −6.9 | n/a | n/a | n/a | n/a |
| LRR receptor kinase SERK2 | XP_010939661.1 | 3UIM | −6.9 | −4.8 | −5.8 | n/a | n/a |
| protein NSP-INTERACTING KINASE 1 | XP_010942232.1 | 3UIM | −6.9 | −4.4 | n//a | n/a | −4.2 |
| probable LRR receptor-like serine/threonine-protein kinase At5g45780 isoform X1 | XP_029121312.1 | 3UIM | −6.9 | n/a | −6.1 | n/a | n/a |
| probable LRR receptor-like serine/threonine-protein kinase At3g47570 | XP_010907375.1 | 4MN8 | −5.2 | n/a | n/a | −4.3 | n/a |
| probable LRR receptor-like serine/threonine-protein kinase At3g47570 | XP_010908730.1 | 4MN8 | −5.2 | n/a | −5.3 | n/a | n/a |
| probable leucine-rich repeat receptor-like protein kinase At5g63930 | XP_010933136.2 | 4MN8 | −5.2 | n/a | n/a | n/a | n/a |
| probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 precursor | NP_001290509.1 | 4Z63 | −6.8 | n/a | n/a | −4.4 | n/a |
| phytosulfokine receptor 2 | XP_010929346.1 | 4Z63 | −6.8 | −4.1 | −5.8 | −3.7 | −3.6 |
| receptor-like protein kinase | XP_010930679.2 | 4Z63 | −6.8 | n/a | n/a | n/a | −3.9 |
| LRR receptor-like serine/threonine-protein kinase GHR1 | XP_010906523.1 | 5UV4 | −8.2 | n/a | n/a | n/a | n/a |
| probable leucine-rich repeat receptor-like protein kinase At5g63930 | XP_010910517.1 | 5UV4 | −8.2 | n/a | −6.1 | n/a | n/a |
| probable inactive receptor kinase At4g23740 | XP_010915720.1 | 5UV4 | −8.2 | −3.8 | −5.6 | n/a | n/a |
| probable inactive receptor kinase At4g23740 | XP_010915721.1 | 5UV4 | −8.2 | −3.8 | −5.5 | n/a | n/a |
| probable inactive receptor kinase At4g23740 | XP_010925786.1 | 5UV4 | −8.2 | n/a | −5.6 | n/a | −3.6 |
| probable leucine-rich repeat receptor-like protein kinase At1g68400 | XP_010933300.1 | 5UV4 | −8.2 | −4.2 | −5.9 | n/a | n/a |
| probable LRR receptor-like serine/threonine-protein kinase At1g53440 | XP_010934669.1 | 5UV4 | −8.2 | n/a | n/a | n/a | n/a |
| putative kinase-like protein TMKL1 | XP_010940648.1 | 5UV4 | −8.2 | n/a | n/a | n/a | −3.7 |
| LRR receptor-like serine/threonine-protein kinase FEI 1 isoform X1 | XP_010942956.1 | 5UV4 | −8.2 | n/a | −6 | −4 | n/a |
| probable inactive receptor kinase At4g23740 | XP_019707070.1 | 5UV4 | −8.2 | n/a | −5.9 | n/a | −4.1 |
| probable inactive receptor kinase At4g23740 | XP_029121337.1 | 5UV4 | −8.2 | n/a | n/a | n/a | −3.7 |
| probable inactive receptor kinase At5g58300 isoform X2 | XP_010931391.1 | 6BRJ | −8.9 | n/a | −5.9 | n/a | n/a |
| probable inactive receptor kinase At5g58300 isoform X2 | XP_010931392.1 | 6BRJ | −8.9 | n/a | −6 | n/a | n/a |
| probable inactive receptor kinase At5g58300 isoform X2 | XP_010931393.1 | 6BRJ | −8.9 | n/a | −6 | n/a | n/a |
| LOW QUALITY PROTEIN: receptor protein kinase TMK1 | XP_010910643.2 | 6BSD | −9.5 | n/a | n/a | n/a | n/a |
| probable inactive receptor kinase At1g48480 | XP_010910915.1 | 6BSD | −9.5 | n/a | n/a | n/a | n/a |
| probable inactive receptor kinase At2g26730 | XP_010916177.1 | 6BSD | −9.5 | n/a | −5.6 | n/a | n/a |
| receptor-like protein 51 | XP_010924732.1 | 6TME | −5.1 | −4.1 | n/a | −4.1 | −3.5 |
| probable LRR receptor-like serine/threonine-protein kinase At2g16250 | XP_010936262.1 | 6TME | −5.1 | n/a | n/a | n/a | n/a |
| Accession ID | Identity | Abbreviation | Week | Log2 Ratio W/I |
|---|---|---|---|---|
| YP_006073104.1 | Photosystem I P700 apoprotein A1 (chloroplast) | PsaA | 1 | 3.323 |
| YP_006073130.1 | Photosystem II CP47 chlorophyll apoprotein (chloroplast) | PsbB | 1 | 2.005 |
| XP_010905109.1 | Remorin | REM | 1 | 1.652 |
| XP_010912515.1 | Dirigent protein 19 | DIR19 | 1 | 2.106 |
| XP_010918555.1 | Probable phospholipid hydroperoxide glutathione peroxidase | PHGPX | 1 | 2.365 |
| XP_010936352.2 | Chlorophyll ab binding protein 5 | CAB5 | 3 | 1.808 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harith-Fadzilah, N.; Lam, S.D.; Haris-Hussain, M.; Ghani, I.A.; Zainal, Z.; Jalinas, J.; Hassan, M. Proteomics and Interspecies Interaction Analysis Revealed Abscisic Acid Signalling to Be the Primary Driver for Oil Palm’s Response against Red Palm Weevil Infestation. Plants 2021, 10, 2574. https://doi.org/10.3390/plants10122574
Harith-Fadzilah N, Lam SD, Haris-Hussain M, Ghani IA, Zainal Z, Jalinas J, Hassan M. Proteomics and Interspecies Interaction Analysis Revealed Abscisic Acid Signalling to Be the Primary Driver for Oil Palm’s Response against Red Palm Weevil Infestation. Plants. 2021; 10(12):2574. https://doi.org/10.3390/plants10122574
Chicago/Turabian StyleHarith-Fadzilah, Nazmi, Su Datt Lam, Mohammad Haris-Hussain, Idris Abd Ghani, Zamri Zainal, Johari Jalinas, and Maizom Hassan. 2021. "Proteomics and Interspecies Interaction Analysis Revealed Abscisic Acid Signalling to Be the Primary Driver for Oil Palm’s Response against Red Palm Weevil Infestation" Plants 10, no. 12: 2574. https://doi.org/10.3390/plants10122574
APA StyleHarith-Fadzilah, N., Lam, S. D., Haris-Hussain, M., Ghani, I. A., Zainal, Z., Jalinas, J., & Hassan, M. (2021). Proteomics and Interspecies Interaction Analysis Revealed Abscisic Acid Signalling to Be the Primary Driver for Oil Palm’s Response against Red Palm Weevil Infestation. Plants, 10(12), 2574. https://doi.org/10.3390/plants10122574






