Proteomic Profile of Glyphosate-Resistant Soybean under Combined Herbicide and Drought Stress Conditions
Abstract
1. Introduction
2. Materials & Methods
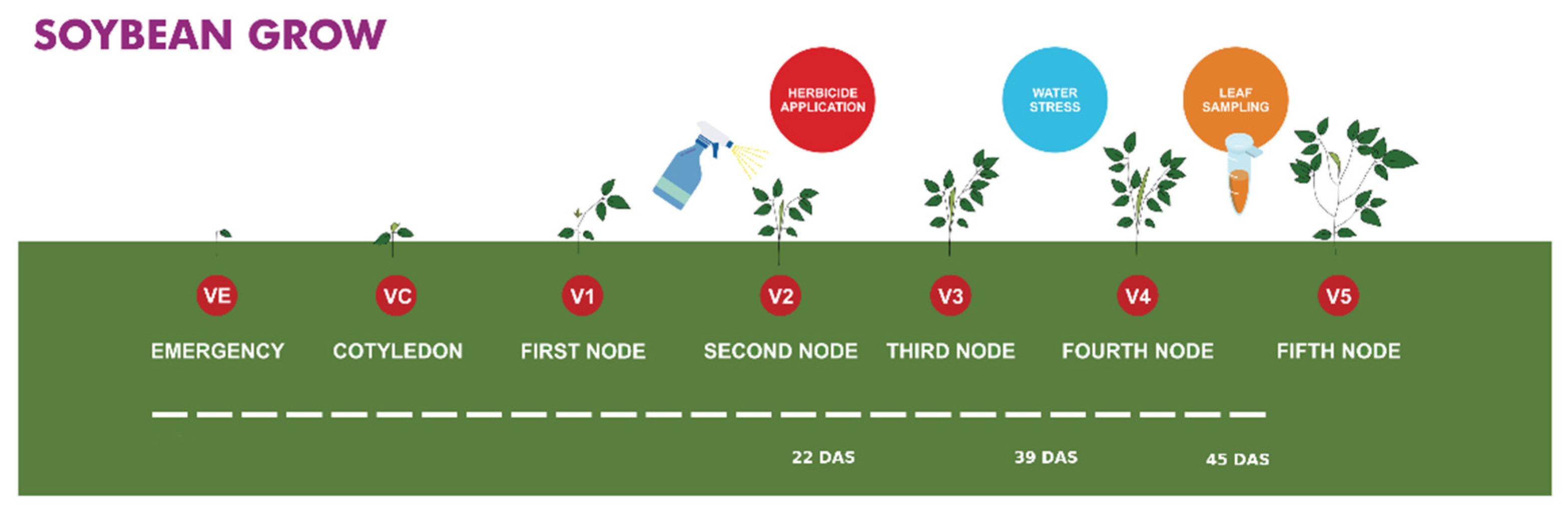
2.1. Plant Material and Growth Conditions
2.2. Sample Preparation for Proteomic Analysis
2.3. TMT Labelling and LC–MS/MS Analysis
2.4. Protein Identification and Quantification
2.5. Statistical Analysis
3. Results
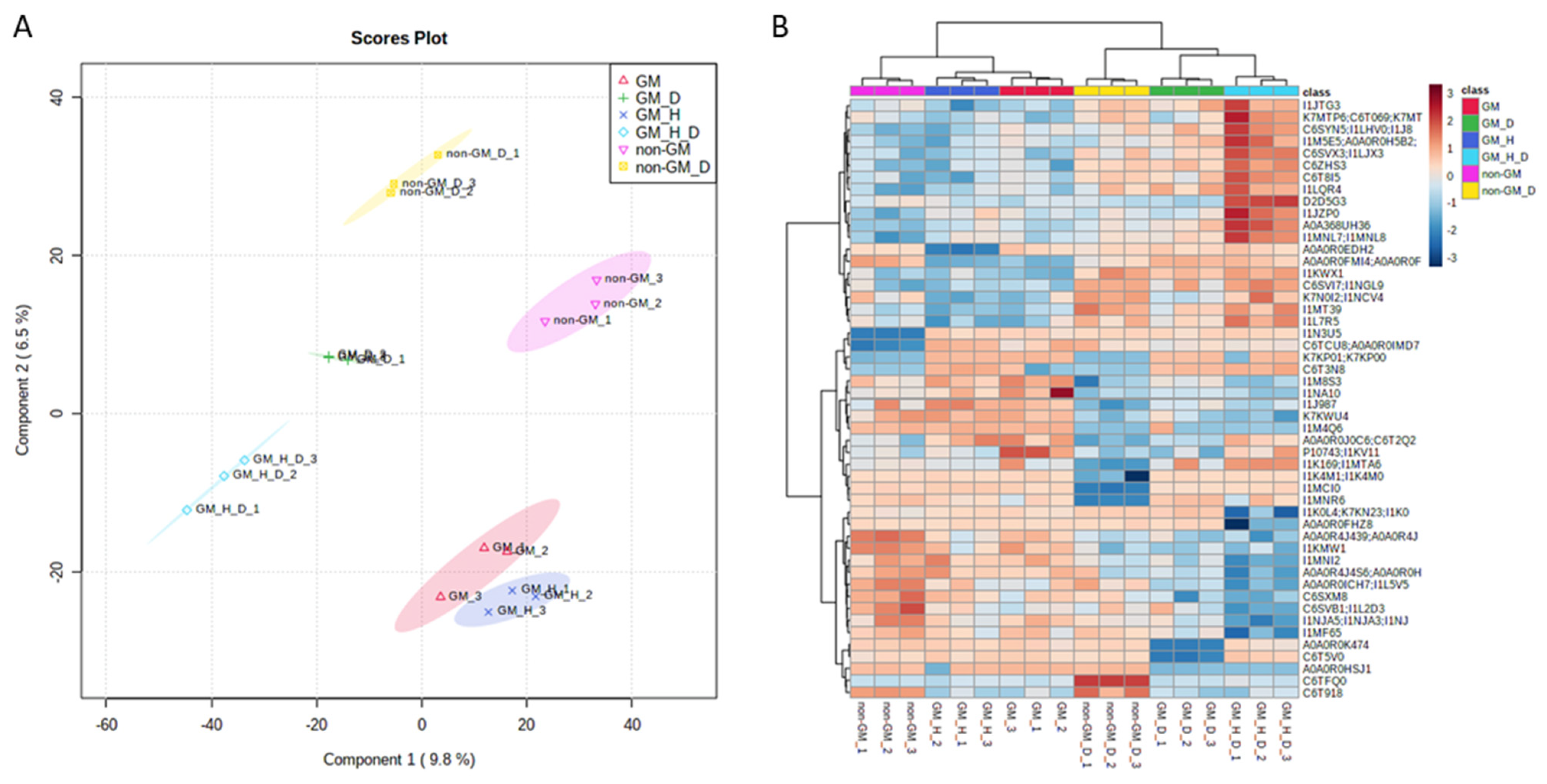
3.1. Exploratory Data Analysis
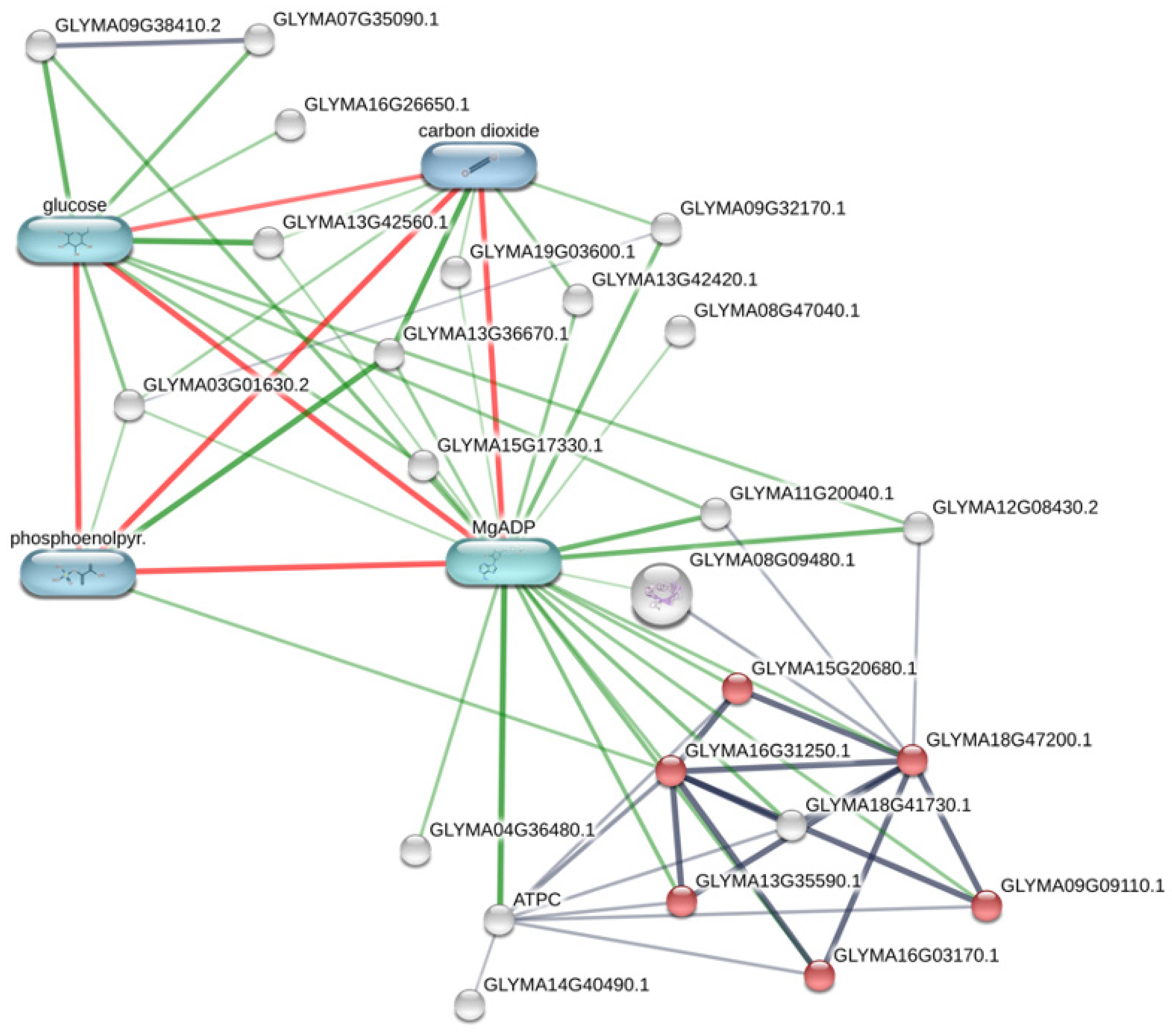
3.2. Metabolic Effects from Genetic Transformation
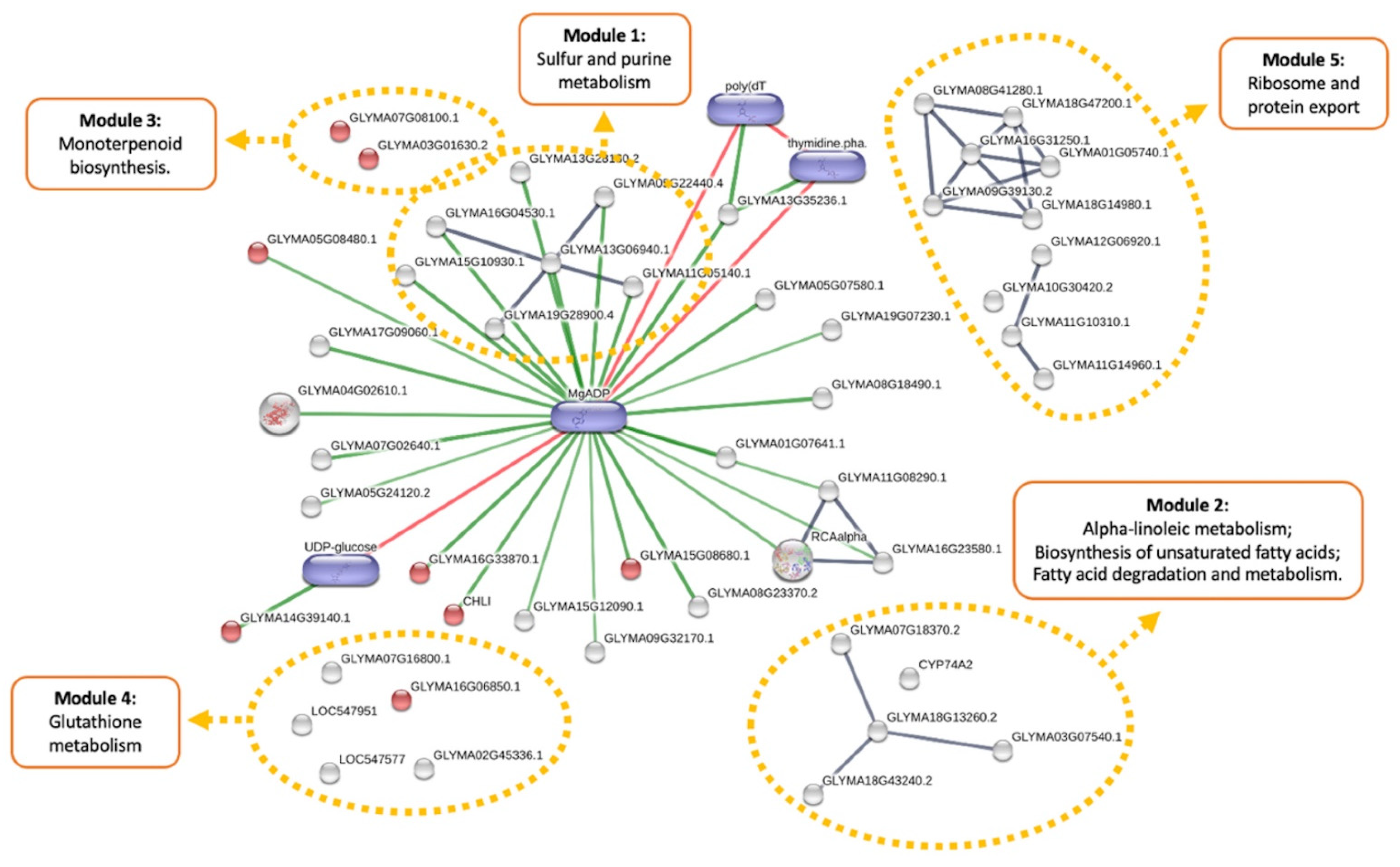
3.3. Metabolic Effects of Combined Herbicide and Drought Conditions
3.4. Cumulative Effects of Genetic Transformation and Abiotic Stress Conditions
4. Discussion
4.1. Metabolic Costs Inferred by Pleiotropic Effects in Genetically Modified Soybeans
4.2. Metabolic Costs from Combined Abiotic Stressors
4.3. Relevance of Abiotic Stress Testing for Risk Assessment of GM Plants
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GM | Genetically Modified |
| ATP | Adenosine Triphosphate |
| EFSA | European Food Safety Authority |
| PEP | Phosphoenolpyruvate |
| ER | Endoplasmic Reticulum |
| PEP | Phosphoenolpyruvate |
| MeJA | Methyl Ester of Jasmonic Acid |
| CA | Cinnamic Acid |
| JA | Jasmonic Acid |
| SA | Salicylic Acid hormones |
| ROS | Reactive Oxygen Species |
| GSTs | Glutathione S-transferases |
| GMO | Genetically Modified Organism |
| Co | cobalt |
| DAS | Days After Sowing |
| GBH | glyphosate-based herbicide |
| rCry | Recombinant Protein Cry |
| TMT | Tandem Mass Tag |
| LC-MS/MS | Liquid Chromatography-Tandem Mass Spectrometry |
| MS/MS | Tandem Mass Spectrometry |
| PLS-Tandem Mass Spectrometry | Partial Least Square-Discriminant Analysis |
References
- ISAAA. Global Status of Commercialized Biotech/Gm Approval Database. Executive Summary. 2020. Available online: http://www.isaaa.org/gmapprovaldatabase/ (accessed on 15 September 2021).
- Vats, S. Herbicides: History, Classification and Genetic Manipulation of Plants for Herbicide Resistance; Springer International Publishing: Cham, Switzerland, 2015; pp. 153–192. [Google Scholar] [CrossRef]
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. Former. Pestic. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Vila-Aiub, M.M.; Neve, P.; Powles, S. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 2009, 184, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2012, 69, 176–187. [Google Scholar] [CrossRef]
- Duhoux, A.; Délye, C. Reference Genes to Study Herbicide Stress Response in Lolium sp.: Up-Regulation of P450 Genes in Plants Resistant to Acetolactate-Synthase Inhibitors. PLoS ONE 2013, 8, e63576. [Google Scholar] [CrossRef]
- Reddy, K.N.; Rimando, A.M.; Duke, S.O. Aminomethylphosphonic acid, a metabolite of glyphosate, causes injury in glyphosate-treated, glyphosate-resistant soybean. J. Agric. Food Chem. 2004, 52, 5139–5143. [Google Scholar] [CrossRef] [PubMed]
- Zobiole, L.H.S.; Oliveira, R.S.; Visentainer, J.V.; Kremer, R.J.; Bellaloui, N.; Yamada, T. Glyphosate affects seed composition in glyphosate-resistant soybean. J. Agric. Food Chem. 2010, 58, 4517–4522. [Google Scholar] [CrossRef]
- Zobiole, L.H.S.; De Oliveira, R.S.; Huber, D.M.; Constantin, J.; De Castro, C.; De Oliveira, F.A.; De Oliveira, A. Glyphosate reduces shoot concentrations of mineral nutrients in glyphosate-resistant soybeans. Plant Soil 2009, 328, 57–69. [Google Scholar] [CrossRef]
- Ozturk, M.; Unal, B.T.; García-Caparrós, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2020, 172, 1321–1335. [Google Scholar] [CrossRef]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef]
- Ahmad, P.; Kumar, A.; Ashraf, M.; Akram, N.A. Salt-Induced Changes in Photosynthetic Activity and Oxidative Defense System of Three Cultivars of Mustard (Brassica juncea L.). Afr. J. Biotechnol. 2012, 11, 2694–2703. [Google Scholar]
- Vorasoot, N.; Songsri, P.; Akkasaeng, C.; Jogloy, S.; Patanothai, A. Effect of water stress on yield and agronomic characters of peanut (Arachis hypogaea L.). Songklanakarin J. Sci. Technol. 2003, 25, 283–288. [Google Scholar]
- Bauer-Panskus, A.; Miyazaki, J.; Kawall, K.; Then, C. Risk assessment of genetically engineered plants that can persist and propagate in the environment. Environ. Sci. Eur. 2020, 32, 1–15. [Google Scholar] [CrossRef]
- Benevenuto, R.F.; Agapito-Tenfen, S.; Vilperte, V.; Wikmark, O.-G.; van Rensburg, P.J.; Nodari, R. Molecular responses of genetically modified maize to abiotic stresses as determined through proteomic and metabolomic analyses. PLoS ONE 2017, 12, e0173069. [Google Scholar] [CrossRef]
- Mesnage, R.; Agapito-Tenfen, S.; Vilperte, V.; Renney, G.; Ward, M.; Séralini, G.-E.; Nodari, R.; Antoniou, M.N. An integrated multi-omics analysis of the NK603 Roundup-tolerant GM maize reveals metabolism disturbances caused by the transformation process. Sci. Rep. 2016, 6, 37855. [Google Scholar] [CrossRef]
- Zanatta, C.B.; Benevenuto, R.F.; Nodari, R.O.; Agapito-Tenfen, S.Z. Stacked genetically modified soybean harboring herbicide resistance and insecticide rCry1Ac shows strong defense and redox homeostasis disturbance after glyphosate-based herbicide application. Environ. Sci. Eur. 2020, 32, 1–17. [Google Scholar] [CrossRef]
- Barbosa, H.S.; Arruda, S.C.C.; Azevedo, R.A.; Arruda, M.A.Z. New insights on proteomics of transgenic soybean seeds: Evaluation of differential expressions of enzymes and proteins. Anal. Bioanal. Chem. 2011, 402, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Arruda, S.C.; Barbosa, H.S.; Azevedo, R.A.; Arruda, M.A. Comparative studies focusing on transgenic through cp4EPSPS gene and non-transgenic soybean plants: An analysis of protein species and enzymes. J. Proteom. 2013, 93, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Brandão, A.; Barbosa, H.; Arruda, M. Image analysis of two-dimensional gel electrophoresis for comparative proteomics of transgenic and non-transgenic soybean seeds. J. Proteom. 2010, 73, 1433–1440. [Google Scholar] [CrossRef]
- Goldsbrough, P.B.; Hatch, E.M.; Huang, B.; Kosinski, W.G.; Dyer, W.E.; Herrmann, K.M.; Weller, S.C. Gene amplification in glyphosate tolerant tobacco cells. Plant Sci. 1990, 72, 53–62. [Google Scholar] [CrossRef]
- Rogers, S.G.; Brand, L.A.; Holder, S.B.; Sharps, E.S.; Brackin, M.J. Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl. Environ. Microbiol. 1983, 46, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Shyr, Y.-Y.J.; Hepburn, A.G.; Widhohn, J.M. Glyphosate selected amplification of the 5-enolpyruvylshikimate-3-phosphate synthase gene in cultured carrot cells. Mol. Genet. Genom. 1992, 232, 377–382. [Google Scholar] [CrossRef]
- Jun, S.; Jianmin, C.; Dagang, T. A gene encodes 5-enol-pyruvylshikimate-3-phosphate Mutagenized by error-prone pcr conferred rice with high glyphosate-tolerance. Mol. Plant Breed. 2009, 6, 830–836. [Google Scholar]
- Yang, X.; Beres, Z.T.; Jin, L.; Parrish, J.T.; Zhao, W.; Mackey, D.; Snow, A.A. Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate resistance in Arabidopsis thaliana. PLoS ONE 2017, 12, e0175820. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.D.K.; Pedersen, P.; De Bruin, J.L.; Stuart, J.; Lux, J.; Franzenburg, D.; Grossnickle, D. Comparisons of genetically modified and non-genetically modified soybean cultivars and weed management systems. Crop Sci. 2010, 50, 2597–2604. [Google Scholar] [CrossRef]
- Zhou, H.; Berg, J.D.; Blank, S.E.; Chay, C.A.; Chen, G.; Eskelsen, S.R.; Fry, J.E.; Hoi, S.; Hu, T.; Isakson, P.J. Field efficacy assessment of transgenic roundup ready wheat. Crop Sci. 2003, 43, 1072–1075. [Google Scholar] [CrossRef]
- Wang, W.; Xia, H.; Yang, X.; Xu, T.; Si, H.J.; Cai, X.X.; Wang, F.; Su, J.; Snow, A.A.; Lu, B.R. A novel 5-enolpyruvoylshikimate-3-phosphate (Epsp) synthase transgene for glyphosate resistance stimulates growth and fecundity in weedy rice (O Ryza Sativa) without herbicide. New Phytol. 2014, 202, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Ashigh, J.; Tardif, F.J. Water and temperature stress impact fitness of acetohydroxyacid synthase–inhibiting herbicide-resistant populations of eastern black nightshade (Solanum ptychanthum). Weed Sci. 2011, 59, 341–348. [Google Scholar] [CrossRef]
- Khan, M.S.; Khan, M.A.; Ahmad, D. Assessing utilization and environmental risks of important genes in plant abiotic stress tolerance. Front. Plant Sci. 2016, 7, 792. [Google Scholar] [CrossRef]
- Choi, D.-H.; Ban, H.-Y.; Seo, B.-S.; Lee, K.-J.; Lee, B.-W. Phenology and seed yield performance of determinate soybean cultivars grown at elevated temperatures in a temperate region. PLoS ONE 2016, 11, e0165977. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Yang, C.; Du, S.; Yang, W. Photosynthetic performance of soybean plants to water deficit under high and low light intensity. S. Afr. J. Bot. 2016, 105, 279–287. [Google Scholar] [CrossRef]
- Júnior, C.S.M.; Da Silva, C.R.; Sanches, M.C.; Hamawaki, O.T.; De Sousa, L.B. Physiologic parameters of soybean of determinate and indeterminate growth habit subjected to levels of soil moisture. Plant Physiol. 2017, 52, 419–425. [Google Scholar] [CrossRef]
- Peirone, L.S.; Irujo, G.P.; Bolton, A.; Erreguerena, I.A.; Aguirrezábal, L.A.N. Assessing the efficiency of phenotyping early traits in a greenhouse automated platform for predicting drought tolerance of soybean in the field. Front. Plant Sci. 2018, 9, 587. [Google Scholar] [CrossRef]
- Jiang, L.-X.; Jin, L.-G.; Guo, Y.; Tao, B.; Qiu, L.-J. Glyphosate effects on the gene expression of the apical bud in soybean (Glycine max). Biochem. Biophys. Res. Commun. 2013, 437, 544–549. [Google Scholar] [CrossRef]
- Xu, C.; Xia, C.; Xia, Z.; Zhou, X.; Huang, J.; Huang, Z.; Liu, Y.; Jiang, Y.; Casteel, S.; Zhang, C. Physiological and transcriptomic responses of reproductive stage soybean to drought stress. Plant Cell Rep. 2018, 37, 1611–1624. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Deckers, P.; Swennen, R.; Panis, B. Preparation of protein extracts from recalcitrant plant tissues: An evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 2005, 5, 2497–2507. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 65–70. [Google Scholar]
- Darmency, H. Pleiotropic effects of herbicide-resistance genes on crop yield: A review. Pest Manag. Sci. 2013, 69, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Agapito-Tenfen, S.Z.; Guerra, M.P.; Wikmark, O.-G.; Nodari, R.O. Comparative proteomic analysis of genetically modified maize grown under different agroecosystems conditions in Brazil. Proteome Sci. 2013, 11, 46. [Google Scholar] [CrossRef]
- Christ, B.; Pluskal, T.; Aubry, S.; Weng, J.-K. Contribution of untargeted metabolomics for future assessment of biotech crops. Trends Plant Sci. 2018, 23, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Howard, D. A role for “Omics” technologies in food safety assessment. Food Control. 2010, 12, 1601–1610. [Google Scholar]
- Heinemann, J.A.; Kurenbach, B.; Quist, D. Molecular profiling—a tool for addressing emerging gaps in the comparative risk Assessment of gmos. Environ. Int. 2011, 37, 1285–1293. [Google Scholar] [CrossRef]
- Horiguchi, G.; Van Lijsebettens, M.; Candela, H.; Micol, J.L.; Tsukaya, H. Ribosomes and translation in plant developmental control. Plant Sci. 2012, 191–192, 24–34. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When defense Pathways collide. The response of arabidopsis to a combination of Drought and heat stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- Vásquez-Robinet, C.; Watkinson, J.I.; Sioson, A.; Ramakrishnan, N.; Heath, L.S.; Grene, R. Differential expression of heat shock protein genes in preconditioning for photosynthetic acclimation in water-stressed loblolly pine. Plant Physiol. Biochem. 2010, 48, 256–264. [Google Scholar] [CrossRef]
- Gupta, D.; Tuteja, N. Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal. Behav. 2011, 6, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2014, 72, 673–689. [Google Scholar] [CrossRef]
- Nio, S.; Cawthray, G.; Wade, L.; Colmer, T. Pattern of solutes accumulated during leaf osmotic adjustment as related to duration of water deficit for wheat at the reproductive stage. Plant Physiol. Biochem. 2011, 49, 1126–1137. [Google Scholar] [CrossRef]
- Biswal, U.C.; Raval, M.K. Chloroplast Biogenesis: From Proplastid to Gerontoplast; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Pandey, J.K.; Dash, S.K.; Biswal, B. Loss in photosynthesis during senescence is accompanied by an increase in the activity of β-galactosidase in leaves of Arabidopsis thaliana: Modulation of the enzyme activity by water stress. Protoplasma 2016, 254, 1651–1659. [Google Scholar] [CrossRef]
- Missihoun, T.; Missihoun, T.D.; Bartels, D. The role of Arabidopsis aldehyde dehydrogenase genes in response to high temperature and stress combinations. J. Exp. Bot. 2017, 68, 4295–4308. [Google Scholar] [CrossRef]
- Singhal, P.; Jan, A.T.; Azam, M.; Haq, Q.M.R. Plant abiotic stress: A prospective strategy of exploiting promoters as alternative to overcome the escalating burden. Front. Life Sci. 2015, 9, 52–63. [Google Scholar] [CrossRef]
- Zobiole, L.H.S.; Kremer, R.J.; de Oliveira, R.S., Jr.; Constantin, J. Glyphosate Effects on Photosynthesis, Nutrient Accumulation, and Nodulation in Glyphosate-Resistant Soybean. J. Plant Nutr. Soil Sci. 2012, 175, 319–330. [Google Scholar] [CrossRef]
- Picoli, G.J.; Carbonari, C.; Matos, A.; Rodrigues, L.; Velini, E. Influence of glyphosate on susceptible and resistant ryegrass populations to herbicide. Planta Daninha 2017, 35, e017163391. [Google Scholar] [CrossRef][Green Version]
- Arosio, P.; Levi, S. Ferritin, Iron Homeostasis, and Oxidative Damage. Free Radic. Biol. Med. 2002, 33, 457–463. [Google Scholar] [CrossRef]
- Kroh, G.E.; Pilon, M. Regulation of iron homeostasis and use in chloroplasts. Int. J. Mol. Sci. 2020, 21, 3395. [Google Scholar] [CrossRef]
- Crichton, R. Iron Metabolism: From Molecular Mechanisms to Clinical Consequences; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Tewari, R.K.; Hadacek, F.; Sassmann, S.; Lang, I. Iron deprivation-induced reactive oxygen species generation leads to non-autolytic PCD in Brassica napus leaves. Environ. Exp. Bot. 2013, 91, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Yadavalli, V.; Neelam, S.; Rao, A.S.; Reddy, A.R.; Subramanyam, R. Differential degradation of photosystem I subunits under iron deficiency in rice. J. Plant Physiol. 2012, 169, 753–759. [Google Scholar] [CrossRef]
- Balachandran, S.; Hurry, V.M.; Kelley, S.E.; Osmond, C.B.; Robinson, S.A.; Rohozinski, J.; Seaton, G.G.R.; Sims, D.A. Concepts of plant biotic stress. Some insights into the stress physiology of virus-Infected plants, from the Perspective of photosynthesis. Physiol. Plant. 1997, 100, 203–213. [Google Scholar] [CrossRef]
- Roitsch, T. Source-sink regulation by sugar and stress. Curr. Opin. Plant Biol. 1999, 2, 198–206. [Google Scholar] [CrossRef]
- Zeller, S.L.; Kalinina, O.; Brunner, S.; Keller, B.; Schmid, B. Transgene × Environment interactions in genetically modified wheat. PLoS ONE 2010, 5, e11405. [Google Scholar] [CrossRef] [PubMed]
- Trtikova, M.; Wikmark, O.G.; Zemp, N.; Widmer, A.; Hilbeck, A. Transgene expression and Bt protein content in transgenic Bt Maize (MON810) under optimal and stressful environmental conditions. PLoS ONE 2015, 10, e0123011. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, L.P.; Barbosa, A.P.; Silva, A.F.M.; Mendes, M.A.; Albrecht, A.J.P.; Ávila, M.R. RR soybean seed quality after application of glyphosate in different stages of crop development. Rev. Bras. Sement. 2012, 34, 373–381. [Google Scholar] [CrossRef][Green Version]
- Yu, W.; Zhang, R.; Li, R.; Guo, S. Isolation and characterization of glyphosate-regulated genes in soybean seedlings. Plant Sci. 2007, 172, 497–504. [Google Scholar] [CrossRef]
- Gomes, M.; Smedbol, E.; Chalifour, A.; Hénault-Ethier, L.; Labrecque, M.; Lepage, L.; Lucotte, M.; Juneau, P. Alteration of plant physiology by glyphosate and its by-product aminomethylphosphonic acid: An overview. J. Exp. Bot. 2014, 65, 4691–4703. [Google Scholar] [CrossRef] [PubMed]
- Vivancos, P.D.; Driscoll, S.P.; Bulman, C.A.; Ying, L.; Emami, K.; Treumann, A.; Mauve, C.; Noctor, G.; Foyer, C.H. Perturbations of amino acid metabolism associated with glyphosate-dependent inhibition of shikimic acid metabolism affect cellular redox homeostasis and alter the abundance of proteins involved in photosynthesis and photorespiration. Plant Physiol. 2011, 157, 256–268. [Google Scholar] [CrossRef]
- Miyazaki, J.; Bauer-Panskus, A.; Bøhn, T.; Reichenbecher, W.; Then, C. Insufficient risk assessment of herbicide-tolerant genetically engineered soybeans intended for import into the EU. Environ. Sci. Eur. 2019, 31, 1–21. [Google Scholar] [CrossRef]
- Séralini, G.-E.; Mesnage, R.; Clair, E.; Gress, S.; de Vendômois, J.S.; Cellier, D. Genetically modified crops safety assessments: Present limits and possible improvements. Environ. Sci. Eur. 2011, 23, 10. [Google Scholar] [CrossRef]
- Simó, C.; Ibáez, C.; Valdés, A.; Cifuentes, A.; García-Cañas, V. Metabolomics of genetically modified crops. Int. J. Mol. Sci. 2014, 15, 18941–18966. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benevenuto, R.F.; Zanatta, C.B.; Guerra, M.P.; Nodari, R.O.; Agapito-Tenfen, S.Z. Proteomic Profile of Glyphosate-Resistant Soybean under Combined Herbicide and Drought Stress Conditions. Plants 2021, 10, 2381. https://doi.org/10.3390/plants10112381
Benevenuto RF, Zanatta CB, Guerra MP, Nodari RO, Agapito-Tenfen SZ. Proteomic Profile of Glyphosate-Resistant Soybean under Combined Herbicide and Drought Stress Conditions. Plants. 2021; 10(11):2381. https://doi.org/10.3390/plants10112381
Chicago/Turabian StyleBenevenuto, Rafael Fonseca, Caroline Bedin Zanatta, Miguel Pedro Guerra, Rubens Onofre Nodari, and Sarah Z. Agapito-Tenfen. 2021. "Proteomic Profile of Glyphosate-Resistant Soybean under Combined Herbicide and Drought Stress Conditions" Plants 10, no. 11: 2381. https://doi.org/10.3390/plants10112381
APA StyleBenevenuto, R. F., Zanatta, C. B., Guerra, M. P., Nodari, R. O., & Agapito-Tenfen, S. Z. (2021). Proteomic Profile of Glyphosate-Resistant Soybean under Combined Herbicide and Drought Stress Conditions. Plants, 10(11), 2381. https://doi.org/10.3390/plants10112381







