Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development
Abstract
:1. Introduction
2. Results
2.1. Identification of PdNAC Gene Family
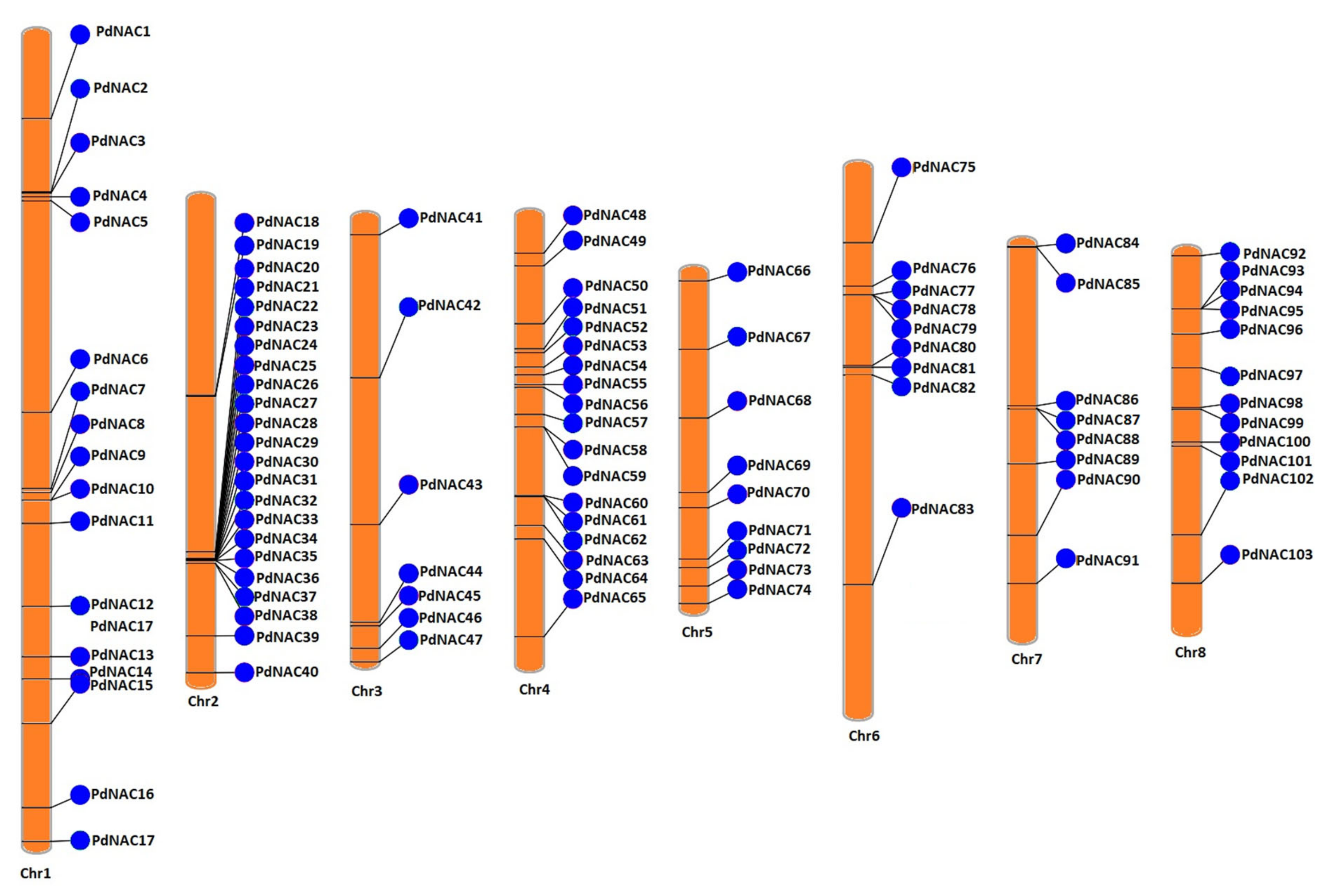
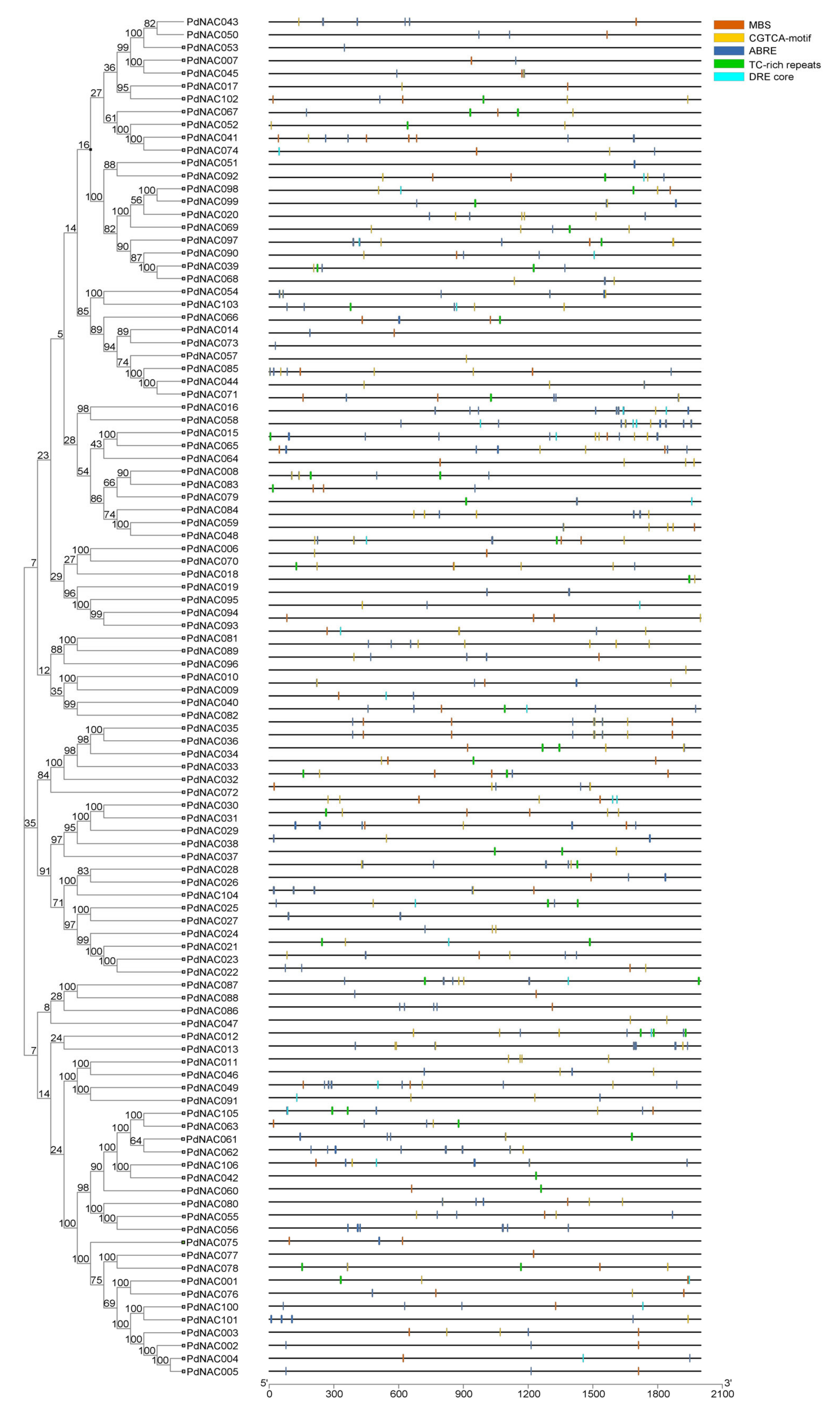
2.2. Chromosomal Mapping and Cis-Acting Regulatory Analysis of NAC Genes
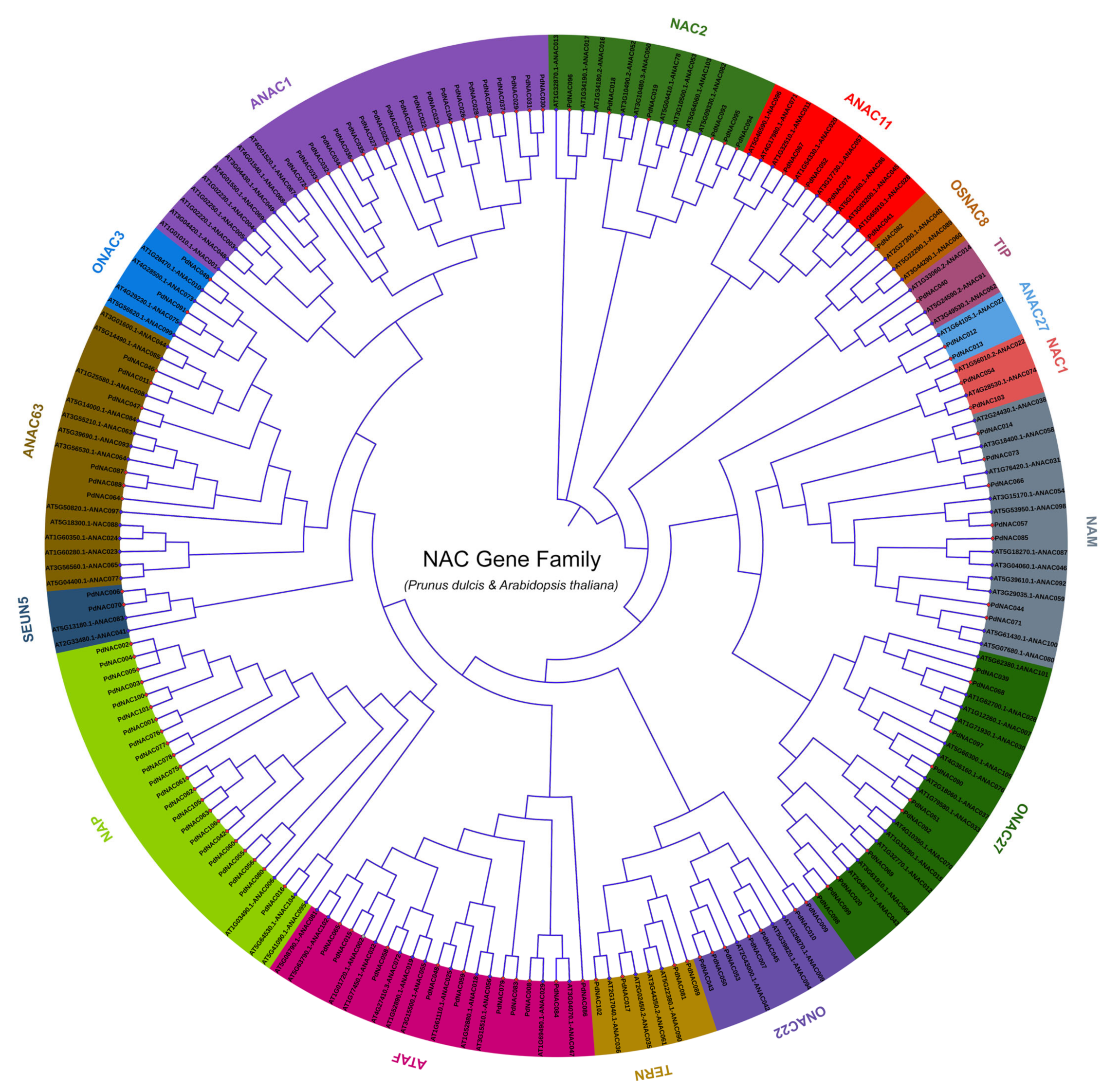
2.3. Phylogenetic Analysis of PdNAC and AtNAC
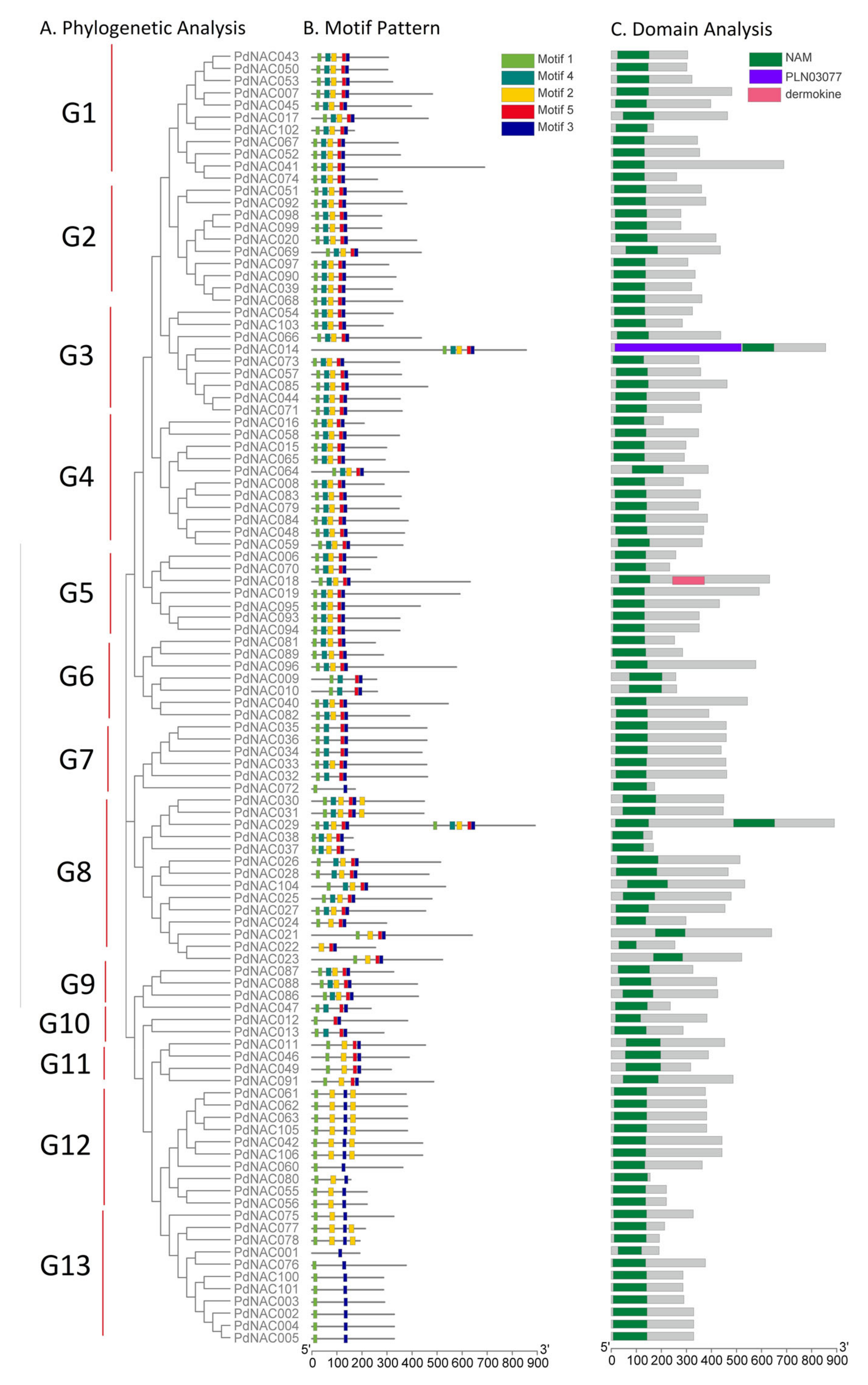
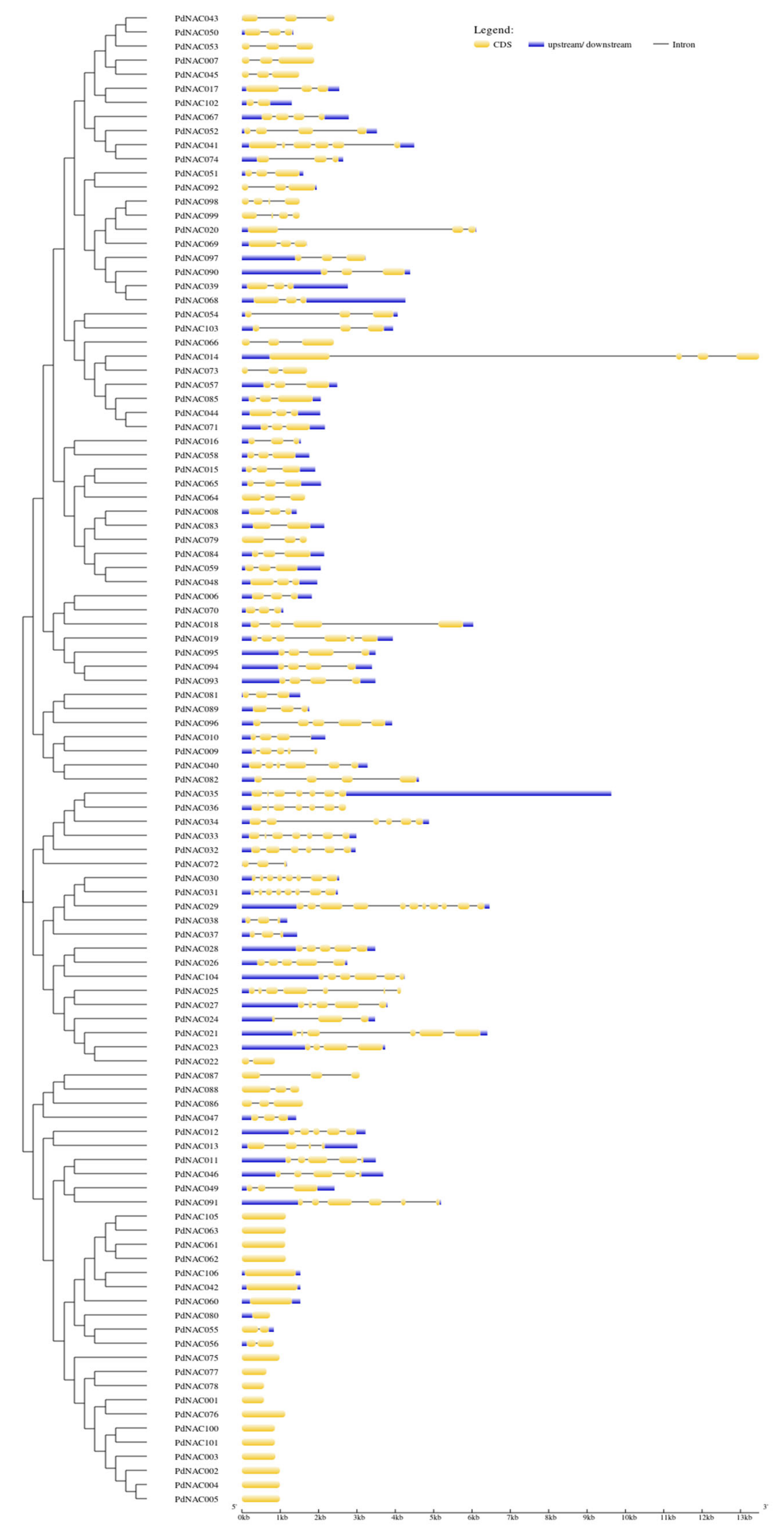
2.4. Gene Structure, Motif Composition and Domain Analyses of PdNAC
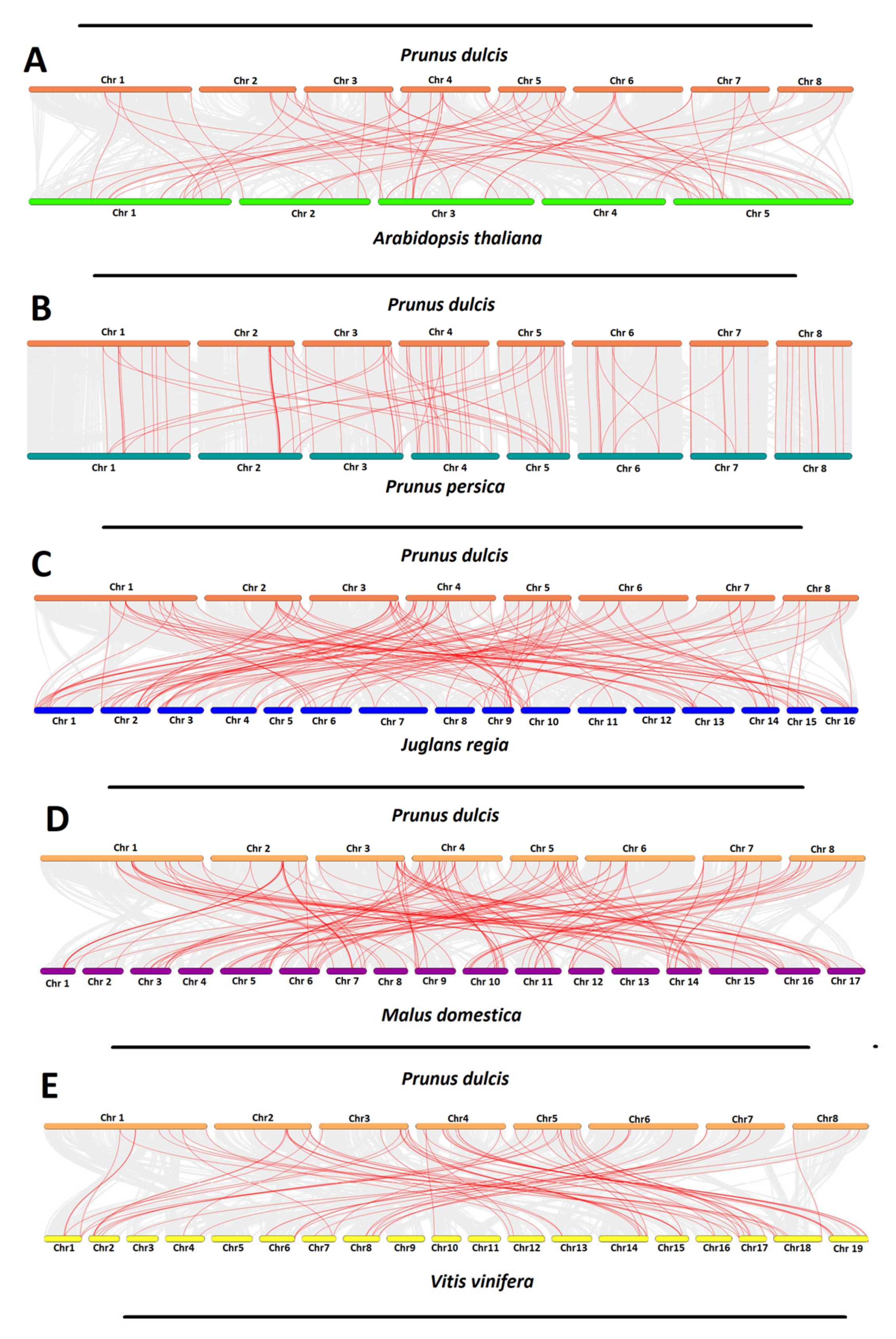
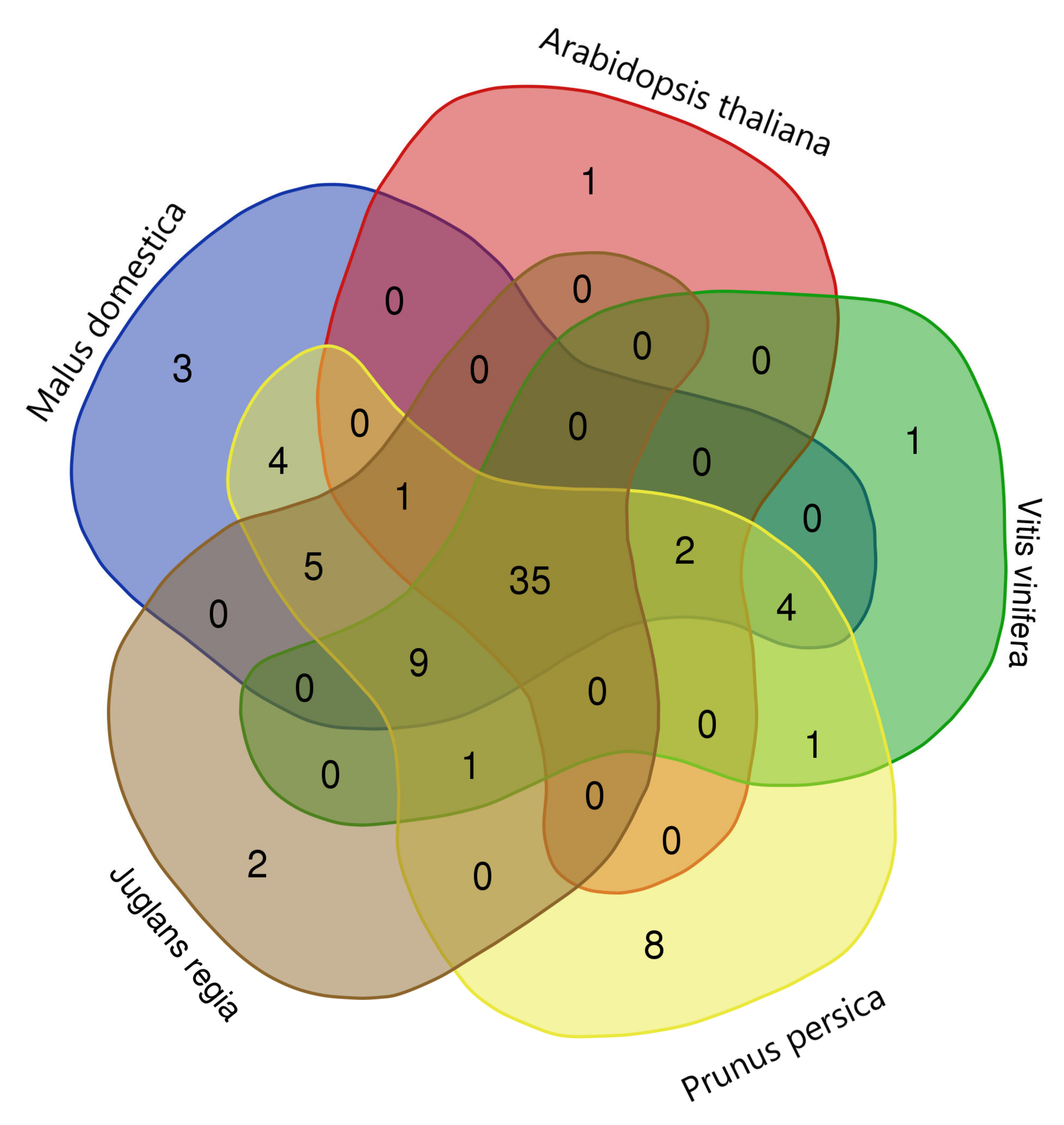
2.5. Gene Duplication and Syntenic Analyses of P. dulcis NAC Genes
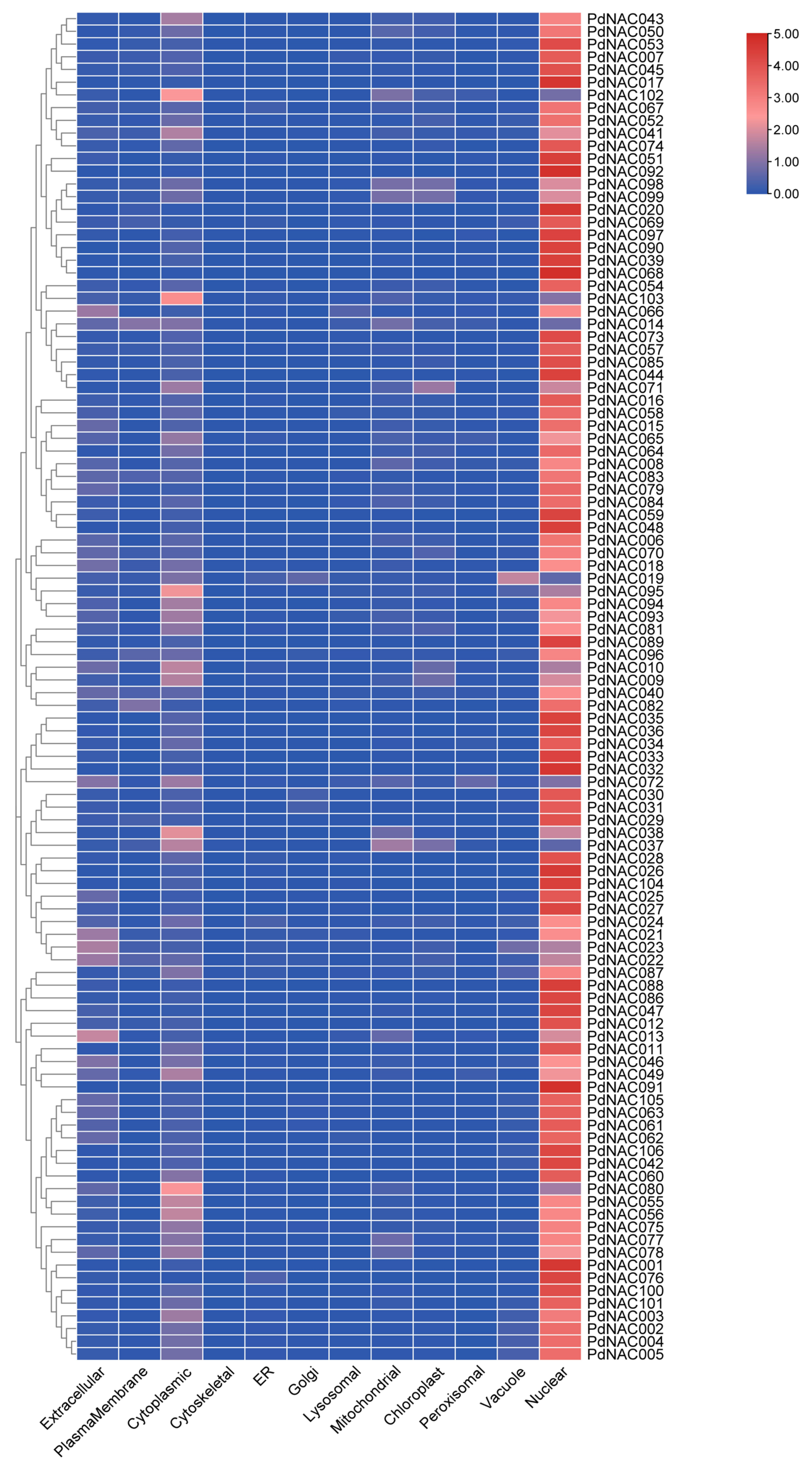
2.6. Physicochemical Properties and Subcellular Localization Prediction
2.7. Gene Ontology Analysis
2.8. Differential Gene Expression of NAC Genes in Normal and Abnormal Fruitlets
3. Discussion
4. Materials and Methods
4.1. Identification of PdNAC Gene Family
4.2. Chromosomal Mapping and Cis-Acting Regulatory Analysis of NAC Genes
4.3. Gene Structure and Motif Analyses of PdNAC
4.4. Physicochemical Properties and Subcellular Localization of PdNAC
4.5. Multiple Sequence Alignment and Phylogenetic Analysis of PdNAC
4.6. Analysis of Gene Ontology
4.7. Gene Duplication and Synteny Analyses
4.8. Differential Expression Analysis of PdNAC in Fruits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.-S. Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaño-Pachón, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Jin, J.; Tang, L.; Zhao, Y.; Gu, X.; Gao, G.; Luo, J. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011, 39, D1114–D1117. [Google Scholar] [CrossRef] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef]
- Sun, H.; Hu, M.; Li, J.; Chen, L.; Li, M.; Zhang, S.; Zhang, X.; Yang, X. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 2017, 120, 61–74. [Google Scholar] [CrossRef]
- Chai, M.; Bellizzi, M.; Wan, C.; Cui, Z.; Li, Y.; Wang, G.-L. The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 2015, 58, 44–51. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Q.; Chen, F.; Wang, M.; Dixon, R.A. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 2011, 68, 1104–1114. [Google Scholar] [CrossRef]
- Ye, Y.; Wu, K.; Chen, J.; Liu, Q.; Wu, Y.; Liu, B.; Fu, X. OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Cheng, H.-Y.; Chen, C.; Yu, X.-W.; Yang, J.-N.; Gao, W.-R.; Shi, Q.-H.; Zhang, H.; Li, J.-G.; Ma, H. A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 2009, 166, 1934–1945. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.-S.; van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Alshareef, N.O.; Wang, J.Y.; Ali, S.; Al-Babili, S.; Tester, M.; Schmöckel, S.M. Overexpression of the NAC transcription factor JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato. Plant Physiol. Biochem. 2019, 140, 113–121. [Google Scholar] [CrossRef]
- Liu, X.; Lyu, Y.; Yang, W.; Yang, Z.; Lu, S.; Liu, J. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2019, 18, 1317–1329. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Li, G.; Yan, C.; Liu, L.; Zhang, Q.; Han, Z.; Li, B. DRL1, encoding a NAC transcription factor, is involved in leaf senescence in grapevine. Int. J. Mol. Sci. 2019, 20, 2678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.-Y.; Kim, S.Y.; Hyeon, D.Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.-H.; Kim, S.-K.; Hong, J.C.; et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, L.-S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Zhang, S.; Yuan, X.; Chen, C.; Wang, X.-F.; Hao, Y.-J. Genome-wide analysis and identification of stress-responsive genes of the NAM–ATAF1,2–CUC2 transcription factor family in apple. Plant Physiol. Biochem. 2013, 71, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Shiriga, K.; Sharma, R.; Kumar, K.; Yadav, S.K.; Hossain, F.; Thirunavukkarasu, N. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2014, 2, 407–417. [Google Scholar] [CrossRef]
- Singh, A.K.; Sharma, V.; Pal, A.K.; Acharya, V.; Ahuja, P.S. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 2013, 20, 403–423. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Ouyang, Z.; Zhang, Y.; Li, X.; Hong, Y.; Huang, L.; Liu, S.; Zhang, H.; Li, D.; Song, F. Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS ONE 2014, 9, e102067. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, H.; Su, C.; Qi, Y.; Liu, X.; Pu, J. Genome-wide identification and expression profile analysis of the NAC transcription factor family during abiotic and biotic stress in woodland strawberry. PLoS ONE 2018, 13, e0197892. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2013, 55, 119–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, N.; Feng, H.; Meng, X.; Li, D.; Yang, D.; Wu, C.; Meng, Q. Overexpression of tomato SlNAC1transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tigchelaar, E.C.; Tomes, M.; Kerr, E.; Barman, R. A new ripening mutant, non-ripening (nor). Rep. Tomato Genet. Coop. 1973, 35, 20. [Google Scholar]
- Carrasco-Orellana, C.; Stappung, Y.; Mendez-Yañez, A.; Allan, A.C.; Espley, R.; Plunkett, B.J.; Moya-Leon, M.A.; Herrera, R. Characterization of a ripening-related transcription factor FcNAC1 from Fragaria chiloensis fruit. Sci. Rep. 2018, 8, 10524. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, T.; Bartholomew, E.; Black, K.; Dong, M.; Zhang, Y.; Yang, S.; Cai, Y.; Xue, S.; Weng, Y.; et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.). Hortic. Res. 2018, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Watkins, C.B.; Gan, S.-S. Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 2012, 63, 6139–6147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-Z.; Baig, M.N.R.; Fan, R.; Ye, J.; Cao, Y.-C.; Deng, X.-X. Identification and expression pattern of a novel NAM, ATAF, and CUC-like gene from Citrus sinensis Osbeck. Plant Mol. Biol. Rep. 2009, 27, 292–297. [Google Scholar] [CrossRef]
- Grasselly, C.H. Origine et évolution de l’amandier cultivé. Cah. Options Méditerranéennes 1976, 32, 44–49. [Google Scholar]
- Küden, A. Crop situation and research in almond. In Proceedings of the 5th International Symposium on Pistachios and Almonds, Sanliurfa, Turkey, 13 October 2009; Volume 912, pp. 515–521. [Google Scholar]
- Kester, D.E.; Gradziel, T.M.; Grasselly, C. Almonds (Prunus). Genet. Resour. Temp. Fruit Nut Crops 1991, 290, 701–760. [Google Scholar] [CrossRef]
- Barreca, D.; Nabavi, S.M.; Sureda, A.; Rasekhian, M.; Raciti, R.; Silva, A.S.; Annunziata, G.; Arnone, A.; Tenore, G.C.; Süntar, İ.; et al. Almonds (Prunus dulcis Mill. DA webb): A source of nutrients and health-promoting compounds. Nutrients 2020, 12, 672. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Yuan, Z.; Zhang, P.; Liu, Z.; Wang, T.; Wei, L. Genome-wide analysis of NAC transcription factor family in maize under drought stress and rewatering. Physiol. Mol. Biol. Plants 2020, 26, 705–717. [Google Scholar] [CrossRef]
- Le, D.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Shinozaki, K.; Tran, L.-S.P. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Kjaersgaard, T.; Jensen, M.K.; Christiansen, M.W.; Gregersen, P.; Kragelund, B.; Skriver, K. Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem. 2011, 286, 35418–35429. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Zhang, S.; Yin, Y.; Zhu, D.; Han, L. Genome-wide analysis of NAM-ATAF1,2-CUC2 transcription factor family in Solanum lycopersicum. J. Plant Biochem. Biotechnol. 2014, 24, 176–183. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, Q.; Shi, Y.; Sun, Z.; Zhang, Q.; Hou, S.; Wu, R.; Jiang, L.; Zhao, X.; Guo, Y. Genome-wide analysis of the NAC domain transcription factor gene family in Theobroma cacao. Genes 2019, 11, 35. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chang, Y.; Ma, S.; Shen, J.; Hu, H.; Xiong, L. Genome-wide identification of SNAC1-targeted genes involved in drought response in rice. Front. Plant Sci. 2019, 10, 982. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Khurana, P.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct. Integr. Genom. 2007, 8, 69–78. [Google Scholar] [CrossRef]
- Heyn, P.; Kalinka, A.T.; Tomancak, P.; Neugebauer, K.M. Introns and gene expression: Cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 2015, 37, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Li, T.; Zhang, L.; Dong, W.; Wang, A.; Qijing, Z.; Tong, L.; Lijie, Z.; Wenxuan, D.; Aide, W. Expression analysis of NAC genes during the growth and ripening of apples. Hortic. Sci. 2018, 45, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Moyano, E.; Martínez-Rivas, F.J.; Blanco-Portales, R.; Molina-Hidalgo, F.J.; Ric-Varas, P.; Arroyo, A.J.M.; Caballero, J.L.; Muñoz-Blanco, J.; Rodríguez-Franco, A. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PLoS ONE 2018, 13, e0196953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, M.; Gu, C.; Zhang, S. Genome-wide identification and expression analysis of genes associated with peach (Prunus persica) fruit ripening. Sci. Hortic. 2019, 246, 317–327. [Google Scholar] [CrossRef]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Wei, Y.; Yang, B.; Ayup, M.; Li, N.; Liu, J.; Liao, K.; Wang, H. Developmental transcriptome profiling uncovered carbon signaling genes associated with almond fruit drop. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef] [Green Version]
- Andrew, S. A quality control tool for high throughput sequence data. FastQC 2016, 532, 1. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafar, Z.; Fatima, S.; Bhatti, M.F. Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development. Plants 2021, 10, 2200. https://doi.org/10.3390/plants10102200
Zafar Z, Fatima S, Bhatti MF. Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development. Plants. 2021; 10(10):2200. https://doi.org/10.3390/plants10102200
Chicago/Turabian StyleZafar, Zeeshan, Sidra Fatima, and Muhammad Faraz Bhatti. 2021. "Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development" Plants 10, no. 10: 2200. https://doi.org/10.3390/plants10102200
APA StyleZafar, Z., Fatima, S., & Bhatti, M. F. (2021). Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development. Plants, 10(10), 2200. https://doi.org/10.3390/plants10102200





