Specification and DNA Barcoding of Thai Traditional Remedy for Chronic Kidney Disease: Pikad Tri-phol-sa-mut-than
Abstract
:1. Introduction
2. Results
2.1. Macroscopical and Microscopical Identification
2.2. Physiochemical Identification
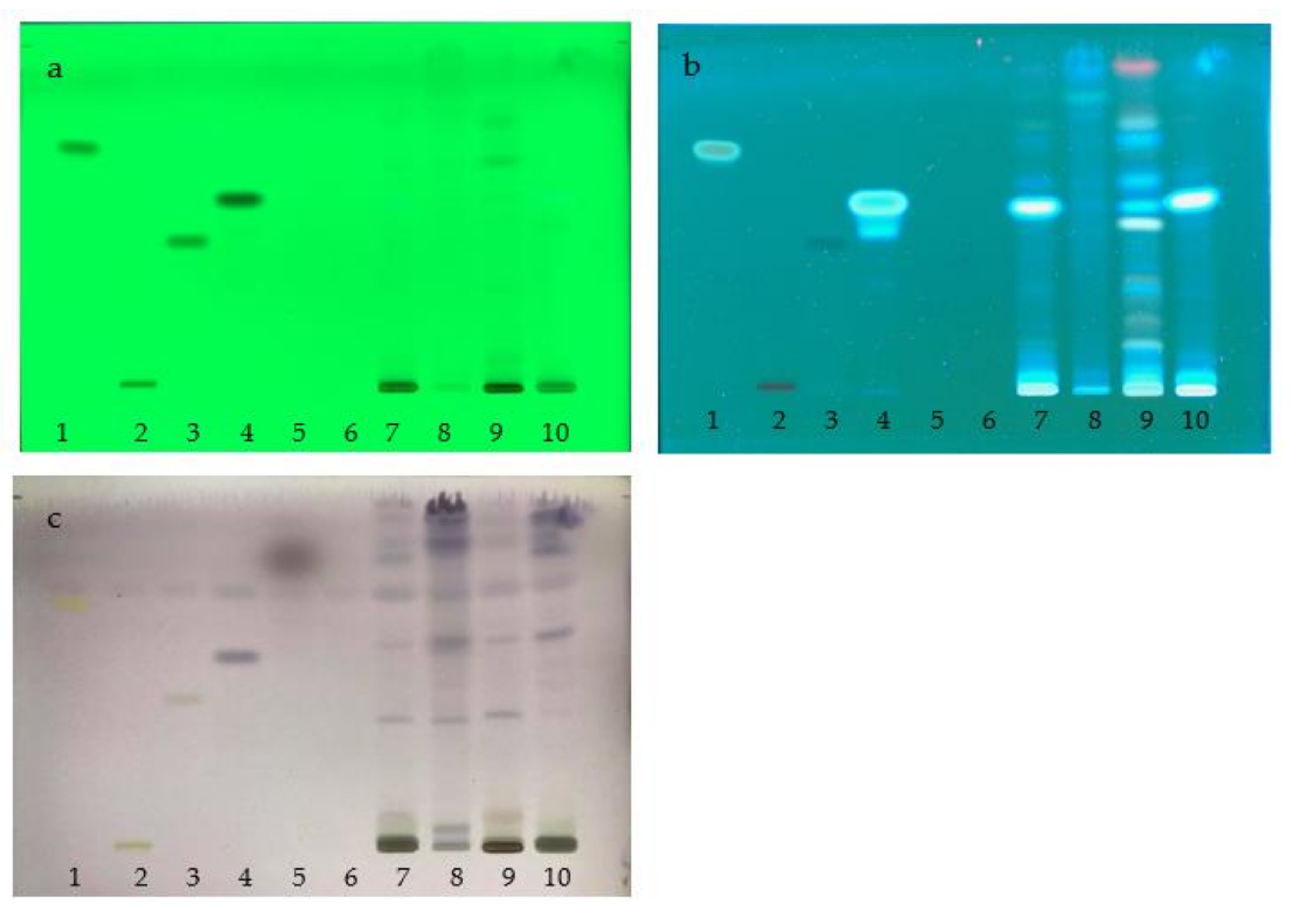
2.3. Chemical Constituents Determined by TLC
2.4. DNA Barcoding Analysis
2.5. Generation of the Predicted ITS2 Secondary Structure
2.6. Authentication of Crude Drugs of Aegle marmelos (L.), Coriandrum sativum L. Corrêa, and Morinda citrifolia L.
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.1.1. Collection of Crude Drugs for Specification Evaluation
4.1.2. Collection of Plant Materials for DNA Barcoding Analysis
4.2. Preparation of Crude Extracts
4.3. Specification of TS Recipe
4.3.1. Determination of Macroscopic and Microscopic Characteristics
4.3.2. Determination of Loss on Drying
4.3.3. Determination of Water Content
4.3.4. Determination of Ash Values
- Determination of Total Ash
- Determination of Acid-Insoluble Ash
4.3.5. Determination of Extractive Value
- Ethanol Soluble Extractive Value
- Water-Soluble Extractive Value
4.3.6. Determination of Volatile Oil
4.3.7. Thin Layer Chromatographic Fingerprint
4.3.8. DNA Extraction, Amplification, and Sequencing
4.3.9. Prediction of ITS2 Secondary Structure
4.3.10. Authentication of the Ingredients of TS Formulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phitsanuprasātwēt, P. Tamra Paetsart Sonkhrau Chabub Anurak—Textbooks of Thai Traditional Medicine, 1st ed.; Sarm Charoen Panich: Bangkok, Thailand, 1992; pp. 154–196. [Google Scholar]
- Ingsathit, A.; Thakkinstian, A.; Chaiprasert, A.; Sangthawan, P.; Gojaseni, P.; Kiattisunthorn, K.; Ongaiyooth, L.; Vanavanan, S.; Sirivongs, D.; Thirakhupt, P.; et al. Prevalence and risk factors of chronic kidney disease in the Thai adult population: Thai SEEK study. Nephrol. Dial. Transplant. 2010, 25, 1567–1575. [Google Scholar] [CrossRef] [Green Version]
- Stenvinkel, P. Chronic kidney disease: A public health priority and harbinger of premature cardiovascular disease. J. Intern. Med. 2010, 268, 456–467. [Google Scholar] [CrossRef]
- Yarnell, E.; Abascal, K. Herbs for Relieving Chronic Renal Failure. Altern. Complement. Ther. 2007, 13, 18–23. [Google Scholar] [CrossRef]
- Sahoo, N.; Manchikanti, P.; Dey, S. Herbal drugs: Standards and regulation. Fitoterapia 2010, 81, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cao, H.; But, P.P.-H.; Shaw, P.-C. Identification of herbal medicinal materials using DNA barcodes. J. Syst. Evol. 2011, 49, 271–283. [Google Scholar] [CrossRef]
- Zhang, W.; Yuan, Y.; Yang, S.; Huang, J.; Huang, L. ITS2 Secondary Structure Improves Discrimination between Medicinal “Mu Tong” Species when Using DNA Barcoding. PLoS ONE 2015, 10, e0131185. [Google Scholar] [CrossRef] [Green Version]
- Bain, J.; Jansen, R. A chloroplast DNA hairpin structure provides useful phylogenetic data within tribe Senecioneae (Asteraceae). Can. J. Bot. 2006, 84, 862–868. [Google Scholar] [CrossRef]
- Neamsuvan, O.; Komonhiran, P.; Boonming, K. Medicinal plants used for hypertension treatment by folk healers in Songkhla province, Thailand. J. Ethnopharmacol. 2018, 214, 58–70. [Google Scholar] [CrossRef]
- Wojcikowski, K.; Johnson, D.; Gobe, G. Herbs or natural substances as complementary therapies for chronic kidney disease: Ideas for future studies. J. Lab. Clin. Med. 2006, 147, 160–166. [Google Scholar] [CrossRef]
- Henrich, M. Ethnopharmacology in the 21st century-grand challenges. Front. Pharmacol. 2010, 8. [Google Scholar] [CrossRef] [Green Version]
- Raclariu, A.C.; Heinrich, M.; Ichim, M.C.; De Boer, H. Benefits and Limitations of DNA Barcoding and Metabarcoding in Herbal Product Authentication. Phytochem. Anal. 2017, 29, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Wang, M.; Zhang, L.; Wang, C. Application of Molecular Methods in the Identification of Ingredients in Chinese Herbal Medicines. Molecules 2018, 23, 2728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Liu, Y.; Wang, X.; Wei, X.; Han, J. DNA Mini-Barcoding: A Derived Barcoding Method for Herbal Molecular Identification. Front. Plant Sci. 2019, 10, 987. [Google Scholar] [CrossRef]
- Ahmad, S.; Singh, M.; Tamboli, E.; Kamal, Y.; Ahmad, W.; Ansari, S. Quality control and in vitro antioxidant potential of Coriandrum sativum Linn. J. Pharm. Bioallied Sci. 2015, 7, 280–283. [Google Scholar] [CrossRef]
- Ramesh, S.; Radhakrishnan, M.b.; Anburaj, R.; Elangomathavan, R.; Patharajan, S. Physicochemical, phytochemical and antimicrobial studies on Morinda citrifolia L. fruits at different maturity stages. Int. J. Pharm. Pharm. Sci. 2012, 4, 473–476. [Google Scholar]
- Thai Herbal Pharmacopoeia. Available online: https://bdn.go.th/thp/home (accessed on 24 June 2020).
- Hadjmohammadi, M.; Sharifi, V. Investigation of optimum extraction conditions for determination of quercetin and kaempferol in coriander (Conundrum sativum L.) by using experimental design and HPLC. J. Food Drug Anal. 2009, 17, 293–299. [Google Scholar] [CrossRef]
- Hussain, F.; Jahan, N.; Rahman, K.-U.; Sultana, B.; Jamil, S. Identification of hypotensive biofunctional compounds of coriandrum sativumand evaluation of their Angiotensin-Converting Enzyme (ACE) inhibition potential. Oxid. Med. Cell. Longev. 2018, 2018, 4643736. [Google Scholar] [CrossRef] [Green Version]
- Prachayasittikul, V.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Coriander (Coriandrum sativum): A promising functional food toward the well-being. Food Res. Int. 2018, 105, 305–323. [Google Scholar] [CrossRef]
- Oganesyan, E.T.; Nersesyan, Z.M.; Parkhomenko, A.Y. Chemical composition of the above-ground part of Coriandrum sativum. Pharm. Chem. J. 2007, 41, 149–153. [Google Scholar] [CrossRef]
- Rajeshwari, U.; Andallu, B. Medicinal benefits of coriander (Coriandrum Sativum L.). Spatula DD 2011, 1, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Wigati, D.; Anwar, K.; Sudarsono; Nugroho, A.E. Hypotensive activity of ethanolic extracts of Morinda citrifolia L. leaves and fruit in dexamethasone-induced hypertensive rat. J. Evid. Based Complement. Altern. Med. 2017, 22, 107–113. [Google Scholar] [CrossRef] [Green Version]
- De Moraes, G.P.; De Alencar, M.V.O.B.; Islam, T.; Araújo, L.D.S.; Sobral, A.L.P.; Machado, K.D.C.; De Aguiar, R.P.S.; Júnior, A.L.G.; Corrêa, D.; Paz, M.F.C.J.; et al. Cytogenotoxic and oxidative status evaluation of Morinda citrifolia. Int. Arch. Med. 2016, 8. [Google Scholar] [CrossRef] [Green Version]
- Deng, S.; Palu, A.K.; West, B.; Su, C.X.; Zhou, B.-N.; Jensen, J.C. Lipoxygenase inhibitory constituents of the fruits of noni (Morinda citrifolia) collected in Tahiti. J. Nat. Prod. 2007, 70, 859–862. [Google Scholar] [CrossRef]
- Charoensiddhi, S.; Anprung, P. Bioactive compounds and volatile compounds of Thai bael fruit (Aegle Marmelos (L.) Correa) as a valuable source for functional food ingredients. Int. Food Res. J. 2008, 15, 45–63. [Google Scholar]
- Abdullakasim, P.; Songchitsomboon, S.; Techagumpuch, M.; Balee, N.; Swatsitang, P.; Sungpuag, P. Antioxidant capacity, total phenolics and sugar content of selected Thai health beverages. Int. J. Food Sci. Nutr. 2007, 58, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.A.; Mohammed, M.M.; Aly, H.F.; Ali, S.A.; Al-Hady, D.-A. Efficiency of the leaves and fruits of Aegle marmelos methanol extract (L.) Correa and their relative hepatotoxicity induced by CCL4 and identification of their active constituents by using LC/MS/MS. Toxicol. Rep. 2018, 5, 1161–1168. [Google Scholar] [CrossRef]
- Garg, N.; Kumar, S.; Yadav, P. Indian goose berry fortified, anti-oxidant rich bael (Aegle marmelos) fermented beverage. J. Food Sci. Technol. 2021, 58, 4437–4441. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Pang, X.; Song, J.; Zhu, Y.; Xie, C.; Chen, S. Using DNA Barcoding to Identify Species within Euphorbiaceae. Planta Medica 2010, 76, 1784–1786. [Google Scholar] [CrossRef] [Green Version]
- Pang, X.; Liu, C.; Shi, L.; Liu, R.; Liang, D.; Li, H.; Cherny, S.S.; Chen, S. Utility of the trnH-psbA intergenic spacer region and its combination as plant DNA barcodes: A meta-analysis. PLoS ONE 2012, 7, e48833. [Google Scholar] [CrossRef]
- Gere, J.; Yessoufou, K.; Daru, B.H.; Mankga, L.T.; Maurin, O.; Bank, M.V. Incorporating trnH-psbA to the core DNA barcode improves significantly species discrimination with southern African Combretaceae. ZooKeys 2013, 365, 129–147. [Google Scholar] [CrossRef]
- Gao, T.; Ma, X.; Zhu, X. Use of the psbA-trnH region to authenticate medicinal species of Fabaceae. Biol. Pharm. Bull. 2013, 36, 1975–1979. [Google Scholar] [CrossRef] [Green Version]
- Michel, C.-I.; Meyer, R.S.; Taveras, Y.; Molina, J. The nuclear internal transcribed spacer (ITS2) as a practical plant DNA barcode for herbal medicines. J. Appl. Res. Med. Aromat. Plants 2016, 3, 94–100. [Google Scholar] [CrossRef]
- Parvathy, V.A.; Swetha, V.P.; Sheeja, T.E.; Leela, N.K.; Chempakam, B.; Sasikumar, B. DNA Barcoding to Detect Chilli Adulteration in Traded Black Pepper Powder. Food Biotechnol. 2014, 28, 25–40. [Google Scholar] [CrossRef]
- Han, J.; Zhu, Y.; Chen, X.; Liao, B.; Yao, H.; Song, J.; Chen, S.; Meng, F. The Short ITS2 Sequence Serves as an Efficient Taxonomic Sequence Tag in Comparison with the Full-Length ITS. BioMed Res. Int. 2013, 2013, 741476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.J.; Khan, I.A. DNA Barcoding for the Identification of Botanicals in Herbal Medicine and Dietary Supplements: Strengths and Limitations. Planta Medica 2016, 82, 1225–1235. [Google Scholar] [CrossRef] [Green Version]
- Gao, T.; Yao, H.; Song, J.; Liu, C.; Zhu, Y.; Ma, X.; Pang, X.; Xu, H.; Chen, S. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J. Ethnopharmacol. 2010, 130, 116–121. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, X.; Wang, P.; Huang, B.; Sun, W.; Xiong, C.; Hu, Z.; Chen, S. Investigation on Species Authenticity for Herbal Products of Celastrus Orbiculatus and Tripterygum Wilfordii from Markets Using ITS2 Barcoding. Molecules 2018, 23, 967. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Lv, Q.; Zhou, H.; Fang, J.; Cheng, W.; Jiang, C.; Cheng, K.; Yao, H. Identification of Traditional She Medicine Shi-Liang Tea Species and Closely Related Species Using the ITS2 Barcode. Appl. Sci. 2017, 7, 195. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Li, X.; Zhou, H.; Hu, H.; Zhang, H.; Sun, W.; Wang, Y.; Yao, H. Molecular identification of Chinese Materia Medica and its adulterants using ITS2 and psbA-trnH barcodes: A case study on Rhizoma Menispermi. Chin. Med. 2014, 5, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Vassou, S.L.; Kusuma, G.; Parani, M. DNA barcoding for species identification from dried and powdered plant parts: A case study with authentication of the raw drug market samples of Sida cordifolia. Gene 2015, 559, 86–93. [Google Scholar] [CrossRef]
- Intharuksa, A.; Sasaki, Y.; Ando, H.; Charoensup, W.; Suksathan, R.; Kertsawang, K.; Sirisa-Ard, P.; Mikage, M. The combination of ITS2 and psbA-trnH region is powerful DNA barcode markers for authentication of medicinal Terminalia plants from Thailand. J. Nat. Med. 2019, 74, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.N.; Yang, C.Y.; Shi, L.C.; Zhang, Z.L.; Xu, A.S.; Zhang, L.X.; Li, X.L.; Li, H.T. Identification of medicinal plants within the Apocynaceae family using ITS2 and trnH-psbA barcode. Chin. J. Nat. Med. 2020, 18, 594–605. [Google Scholar] [CrossRef]
- Liu, Z.W.; Gao, Y.Z.; Zhou, J. Molecular authentication of the medicinal species of Ligusticum (Ligustici Rhizoma et Radix, “Gao-ben”) by integrating non-coding Internal Trascribed Spacer 2 (ITS2) and its secondary structure. Front. Plant Sci. 2019, 10, 429. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.-W.; Li, Y.-C.; Zhong, D.-L.; Zhang, J.-Q. Establishment of the most comprehensive ITS2 barcode database to date of the traditional medicinal plant Rhodiola (Crassulaceae). Sci. Rep. 2017, 7, 10051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, N.; Wei, Y.L.; Zhang, X.; Zhu, N.; Wang, Y.L.; Zhu, Y.; Zhang, H.P.; Li, F.M.; Yang, L.; Sun, J.Q.; et al. Barcode ITS2: A useful tool for identifying Trachelospermu, jasminoides and a good monitor for medicine market. Sci. Rep. 2017, 7, 5037. [Google Scholar] [CrossRef]
- Consortium for the Barcode of Life Plant Working Group; Hollingsworth, P.; Forrest, L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- China Plant BOL Group; Li, D.-Z.; Gao, L.-M.; Li, H.T.; Wang, H.; Ge, X.-J.; Liu, J.-Q.; Chen, Z.-D.; Zhou, S.-L.; Chen, S.-L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Song, J.Y.; Ma, X.Y.; Liu, C.; Li, Y.; Xu, H.X.; Han, J.P.; Duan, L.S.; Chen, S.L. Identification of Dendrobium species by a candidate DNA barcode sequence: The chloroplast trnH-psbA intergenic region. Planta. Med. 2009, 75, 667–669. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Intharuksa, A.; Ando, H.; Miyake, K.; Sirisa-Ard, P.; Mikage, M.; Sasaki, Y. Molecular Analysis of Terminalia spp. Distributed in Thailand and Authentication of Crude Drugs from Terminalia Plants. Biol. Pharm. Bull. 2016, 39, 492–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitaoka, F.; Kakiuchi, N.; Long, C.; Itoga, M.; Mitsue, A.; Mouri, C.; Mikage, M. Molecular Characterization of Akebia Plants and the Derived Traditional Herbal Medicine. Biol. Pharm. Bull. 2009, 32, 665–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanford, A.M.; Harden, R.; Parks, C.R. Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS se-quence data. Am. J. Bot. 2000, 87, 872–882. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal Ribosomal RNA genes for phylogenetics. PCR Protoc. 1990, 315–322. [Google Scholar] [CrossRef]
- Hamilton, M.B. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol. EcoI. 1999, 8, 521–523. [Google Scholar]
- Kuzmina, M.; Johnson, K.L.; Barron, H.R.; Hebert, P.D. Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecol. 2012, 12, 25. [Google Scholar] [CrossRef] [Green Version]
- Fay, F.M.; Bayer, C.; Alverson, W.S.; Bruijin, A.Y.; Chase, M.W. Plastid rbcL sequence data indicate a close affinity between Diegodendron and Bixa. Taxon 1998, 47, 43–50. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. J. Bioinform. 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Selig, C.; Wolf, M.; Müller, T.; Dandekar, T.; Schultz, J. The ITS2 Database II: Homology modelling RNA structure for molecular systematics. Nucleic Acids Res. 2007, 36, D377–D380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundaresan, N.; Sahu, A.; Jagan, E.G.; Pandi, M. Evaluation of ITS2 molecular morphometric effectiveness in species delimitation of Ascomycota-A pilot study. Fungal Biol. 2019, 123, 517–527. [Google Scholar] [CrossRef] [PubMed]








| Specification | Content (%) | |||
|---|---|---|---|---|
| AM | CS | MC | TS | |
| Loss on drying | 8.42 ± 0.02 | - | - | - |
| Total ash | 3.95 ± 0.07 | 8.18 ± 0.05 | 6.77 ± 0.04 | 6.55 ± 0.03 |
| Acid-insoluble ash | 0.07 ± 0.02 | 0.06 ± 0.02 | 0.10 ± 0.01 | 0.07 ± 0.00 |
| Ethanol-soluble extractive value | 11.66 ± 0.04 | 17.19 ± 0.17 | 27.33 ± 0.13 | 21.75 ± 0.08 |
| Water-soluble extractive value | 48.95 ± 0.09 | 17.21 ± 0.24 | 48.95 ± 0.14 | 37.69 ± 0.21 |
| Water content | - | 4.33 ± 0.29 | 4.50 ± 0.00 | 5.50 ± 0.00 |
| Volatile oil content | - | 1.50 ± 0.00 | - | 0.50 ± 0.00 |
| Each sample analysis was performed in triplicate. | ||||
| Composition of TS Formulation | Voucher Numbers | Locality (District, Province) | Accession Numbers | |||
|---|---|---|---|---|---|---|
| ITS | matK | trnH-psbA | rbcL | |||
| Aegle marmelos (L.) Corrêa | Authentic Specimens | |||||
| AEM-PL06032021 | Mueang, Phitsanulok | |||||
| AEM-CM11032021 | Mueang, Chiang Mai | LC633819 | LC633825 | LC633828 | LC633822 | |
| AEM-AC24032021 | Mueang, Amnat Charoen | |||||
| AEM-UR24032021 | Det Udom, Ubon Ratchathani | |||||
| AEM-KP28032021 | Mueang, Kamphaeng Phet | |||||
| AEM-PL29032021 | Phrom Phiram, Phitsanulok | |||||
| AEM-PB30032021 | Prachantakham, Prachinburi | |||||
| AEM-KP31032021 | Phran Kratai, Kamphaeng Phet | |||||
| Crude drugs | ||||||
| MAT-NP08082018 | Mueang, Nakhon Pathom | - | - | - | - | |
| Coriandrum sativum L. | Authentic specimens | |||||
| COS-KP06032021 | Mueang, Kamphaeng Phet | |||||
| COS-KP08032021 | Mueang, Kamphaeng Phet | |||||
| COS-CM11032021 | Mueang, Chiang Mai | LC633820 | LC633826 | LC633829 | LC633823 | |
| COS-AC24032021 | Mueang, Amnat Charoen | |||||
| COS-UR24032021 | Det Udom, Ubon Ratchathani | |||||
| COS-NS28032021 | Mueang, Nakhon Sawan | |||||
| COS-PC30032021 | Wachirabarami, Phichit | |||||
| COS-PB08042021 | Lom Sak, Phetchabun | |||||
| Crude drugs | ||||||
| LPC-NP08082018 | Mueang, Nakhon Pathom | - | - | - | - | |
| Morinda citrifolia L. | Authentic specimens | |||||
| MOC-PL08032021 | Mueang, Phitsanulok | |||||
| MOC-KP06032021 | Mueang, Kamphaeng Phet | |||||
| MOC-CM11032021 | Mueang, Chiang Mai | LC633821 | LC633827 | LC633830 | LC633824 | |
| MOC-AC24032021 | Mueang, Amnat Charoen | |||||
| MOC-UR24032021 | Det Udom, Ubon Ratchathani | |||||
| MOC-PL29032021 | Phrom Phiram, Phitsanulok | |||||
| Crude drugs | ||||||
| YOC-CM07082019 | Mueang, Chiang Mai | - | - | - | - | |
| Regions | ITS2 | ITS | matK | psbA-trnH | rbcL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | AEM | COS | MOC | AEM | COS | MOC | AEM | COS | MOC | AEM | COS | MOC | AEM | COS | MOC |
| Length (bp) | 230 | 221 | 212 | 630 | 598 | 566 | 779 | 845 | 837 | 460 | 240 | 365 | 750 | 750 | 751 |
| %GC Content | 71.7 | 55.7 | 71.2 | 63.8 | 55.7 | 64.5 | 35.6 | 35.9 | 34.1 | 29.3 | 33.8 | 24.7 | 44.9 | 43.7 | 42.9 |
| Pairwise Distance | |||||||||||||||
| AEM | 0 | 0.070 | 0.057 | 0 | 0.037 | 0.034 | 0 | 0.022 | 0.023 | 0 | 0.061 | 0.078 | 0 | 0.012 | 0.013 |
| COS | 0.536 | 0 | 0.087 | 0.423 | 0 | 0.032 | 0.263 | 0 | 0.020 | 0.455 | 0 | 0.065 | 0.087 | 0 | 0.010 |
| MOC | 0.401 | 0.628 | 0 | 0.383 | 0.336 | 0 | 0.282 | 0.243 | 0 | 0.681 | 0.472 | 0 | 0.111 | 0.075 | 0 |
| Overall Mean Distance (S.E.) | 0.522 (0.06) | 0.381 (0.03) | 0.263 (0.02) | 0.536 (0.05) | 0.091 (0.01) | ||||||||||
| No. | Scientific Name | Family | Part Used | |
|---|---|---|---|---|
| TS Recipe | 1. | Aegle marmelos (L.) Corrêa | Rutaceae | Fruit |
| 2. | Coriandrum sativum L. | Apiaceae | Fruit | |
| 3. | Morinda citrifoli L. | Rubiaceae | Fruit |
| Primers | Sequences 5′ → 3′ | Tm (°C) | Reference |
|---|---|---|---|
| ITS2 intergenic region | |||
| Un.3F | CGA CTC TCG GCA AGG GAT AT | 56.73 | [57] |
| Akebi-26SR | GTA AGT TTC TTC TCC TCC GC | 52.89 | [58] |
| Internal transcribed spacer (ITS) | |||
| ITS5A | CCT TAT CAT TTA GAG GAA GGA G | 49.22 | [59] |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | 51.67 | [60] |
| maturase K (matK) | |||
| matK-1RKIM-f | ACC CAG TCC ATC TGG AAA TCT TGG TTC | 60.74 | [61] |
| matK-3FKIM-r | CGT ACA GTA CTT TTG TGT TTA CGA G | 53.29 | |
| trnH-psbA intergenic spacer | |||
| trnH-psbAF * | ACT GCC TTG ATC CAC TTG GC | 58.31 | [62] |
| trnH-psbAR * | CGA AGC TCC ATC TAC AAA TGG | 53.32 | |
| MOC-psbAF ** | GCGCATGATGGATTCACAAT | 53.50 | This study |
| MOC-psbAR ** | GAAGTTATGCACGAACGTAAT | 50.01 | |
| ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) | |||
| rbcL 1F | ATC TCA CCA CAA ACA GAA AC | 50.22 | [63] |
| rbcL 724R | TCG CAT GTA CCT GCA GTA GC | 57.64 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thariwong, S.; Intharuksa, A.; Sirisa-ard, P.; Charoensup, W.; Chansakaow, S. Specification and DNA Barcoding of Thai Traditional Remedy for Chronic Kidney Disease: Pikad Tri-phol-sa-mut-than. Plants 2021, 10, 2023. https://doi.org/10.3390/plants10102023
Thariwong S, Intharuksa A, Sirisa-ard P, Charoensup W, Chansakaow S. Specification and DNA Barcoding of Thai Traditional Remedy for Chronic Kidney Disease: Pikad Tri-phol-sa-mut-than. Plants. 2021; 10(10):2023. https://doi.org/10.3390/plants10102023
Chicago/Turabian StyleThariwong, Suwimol, Aekkhaluck Intharuksa, Panee Sirisa-ard, Wannaree Charoensup, and Sunee Chansakaow. 2021. "Specification and DNA Barcoding of Thai Traditional Remedy for Chronic Kidney Disease: Pikad Tri-phol-sa-mut-than" Plants 10, no. 10: 2023. https://doi.org/10.3390/plants10102023
APA StyleThariwong, S., Intharuksa, A., Sirisa-ard, P., Charoensup, W., & Chansakaow, S. (2021). Specification and DNA Barcoding of Thai Traditional Remedy for Chronic Kidney Disease: Pikad Tri-phol-sa-mut-than. Plants, 10(10), 2023. https://doi.org/10.3390/plants10102023






