Genome-Wide Analysis and the Expression Pattern of the ERF Gene Family in Hypericum perforatum
Abstract
1. Introduction
2. Results
2.1. Identification of HpERFs and Their Sequence Features
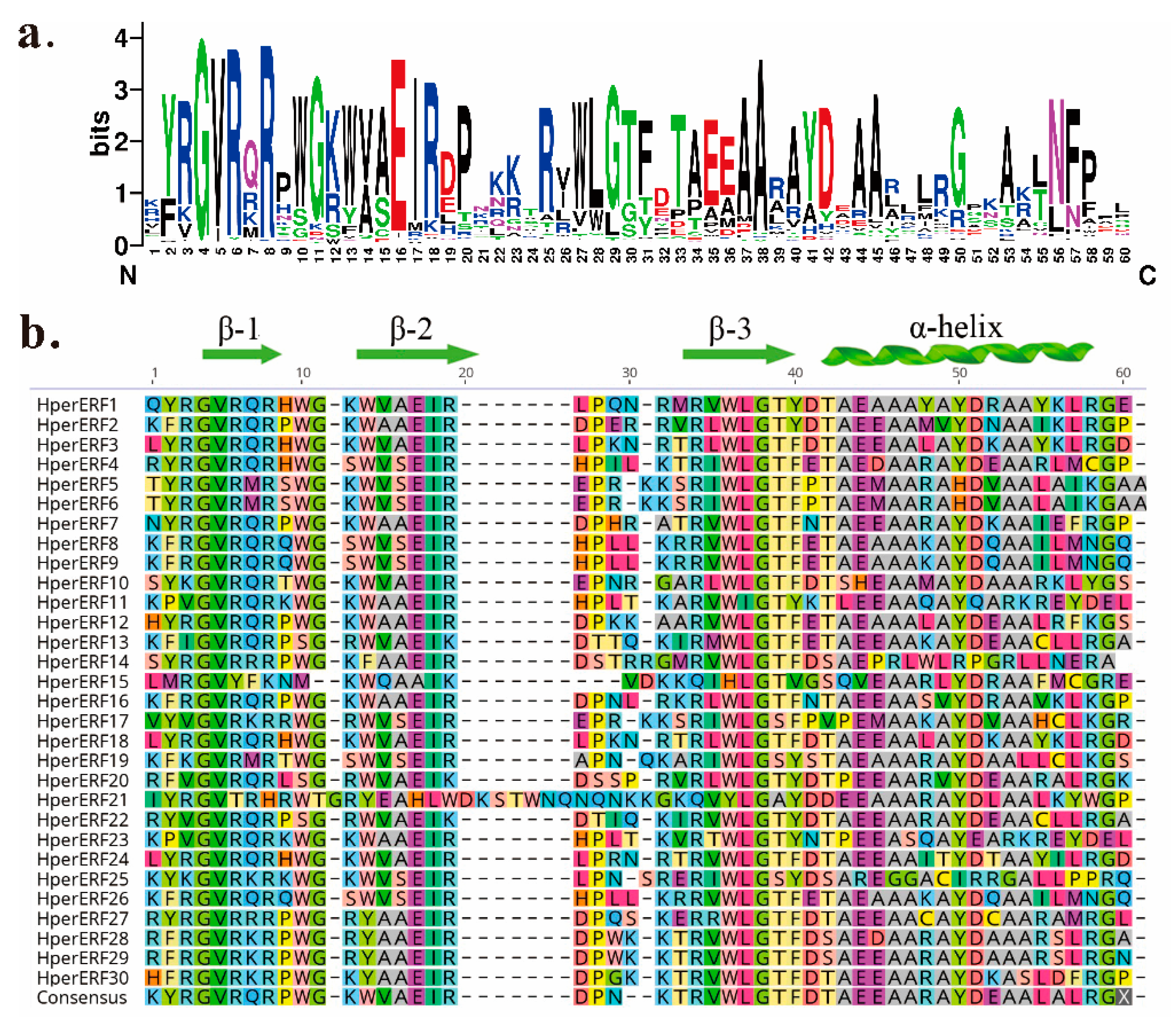
2.2. Analysis of Multiple Sequence Alignments and Cis-Acting Elements
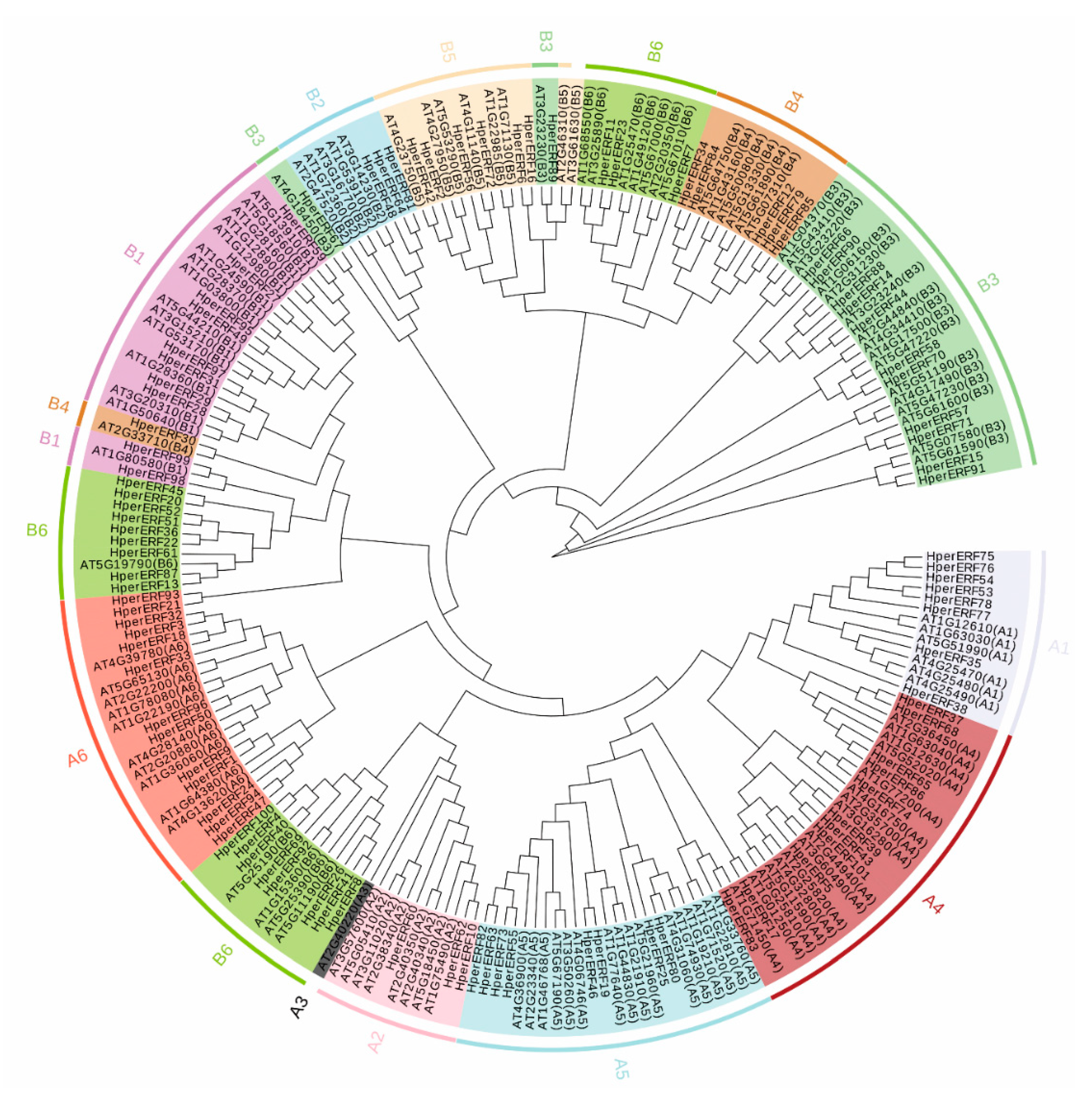
2.3. Phylogenetic Analysis of the ERF Family between H. perforatum and Arabidopsis
2.4. Gene Structure, Motif Composition, and Ka/Ks Analysis of HpERFs
2.5. Transcript Abundance Profiling by RNA-Seq
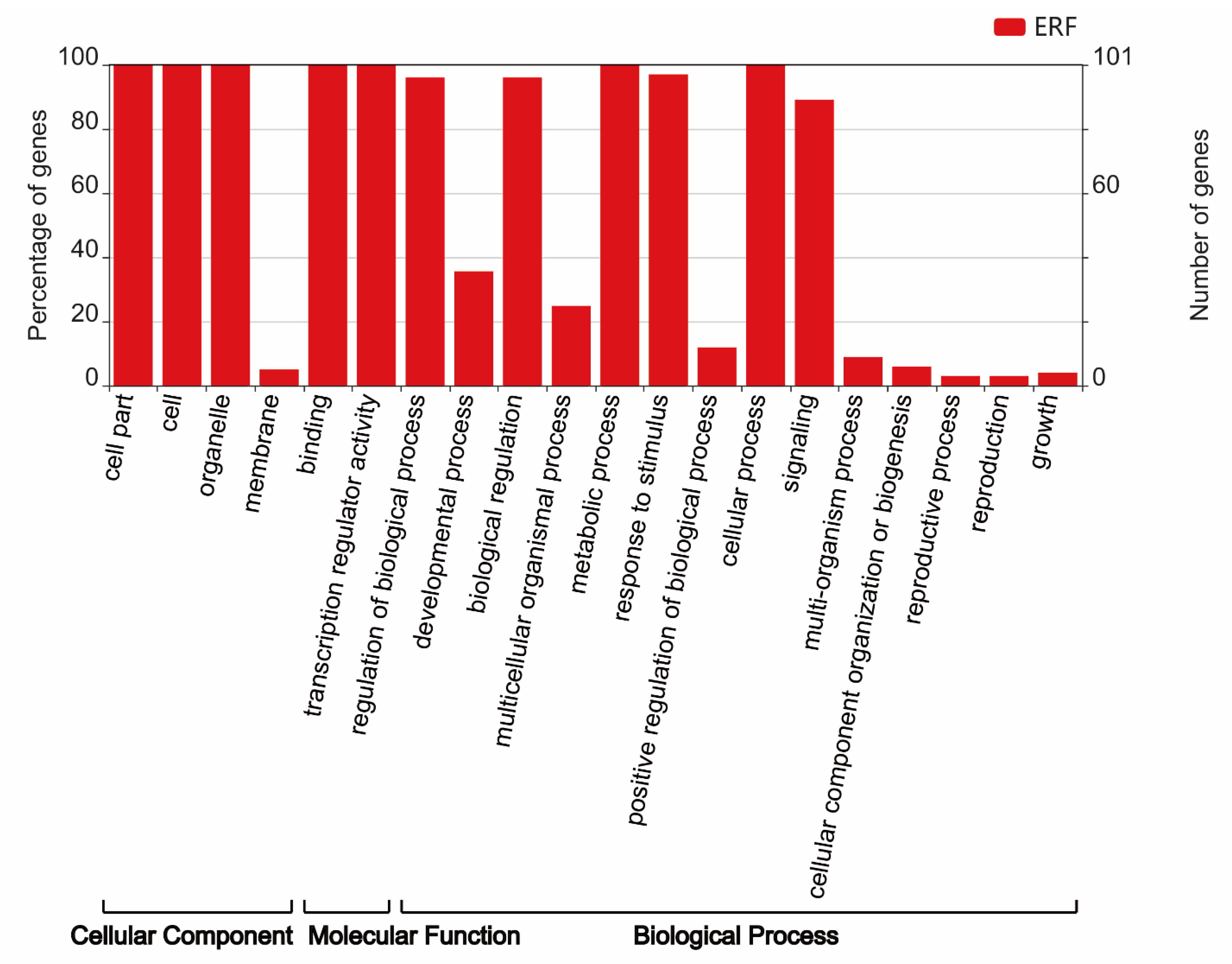
2.6. Gene Ontology (GO) Annotation
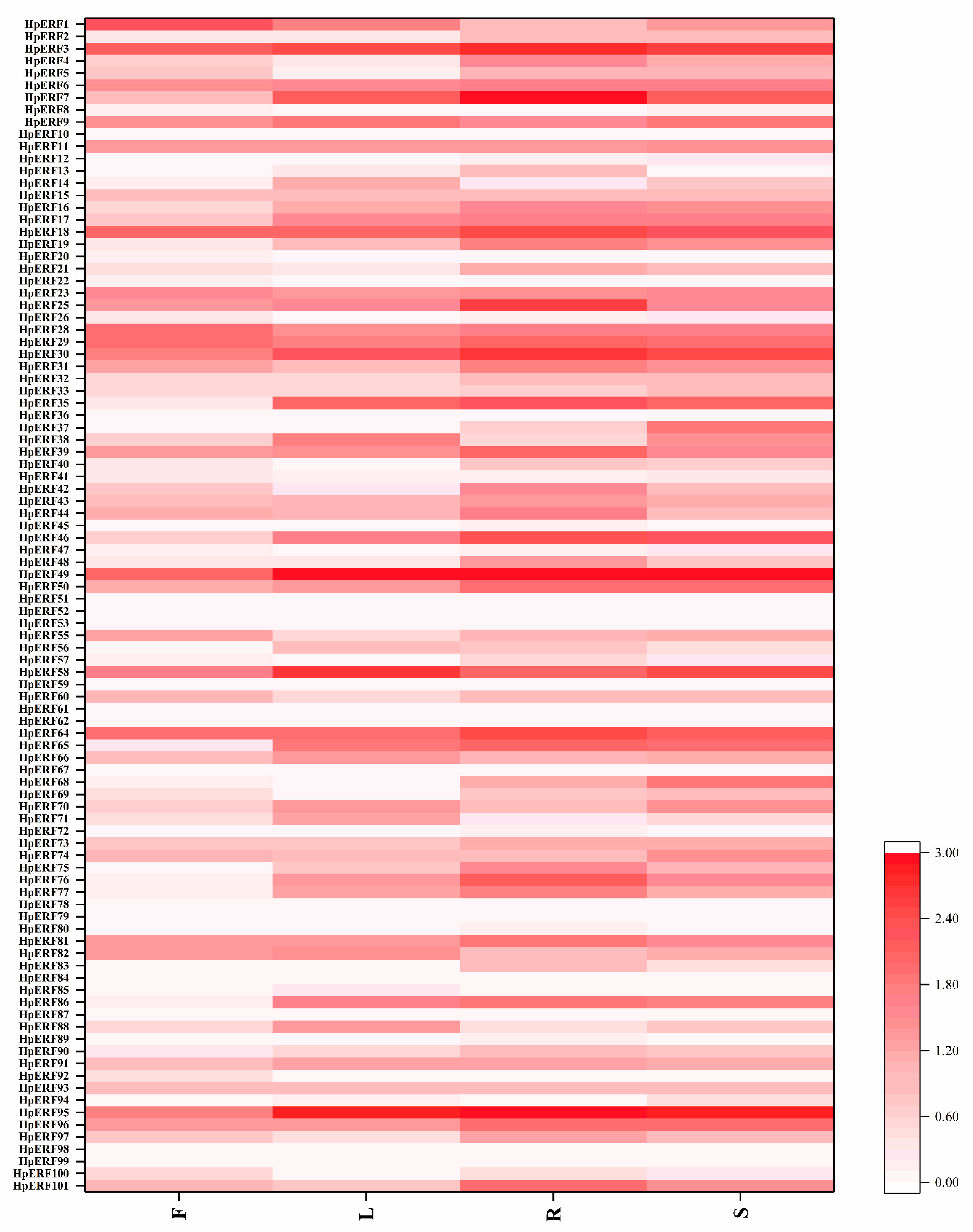
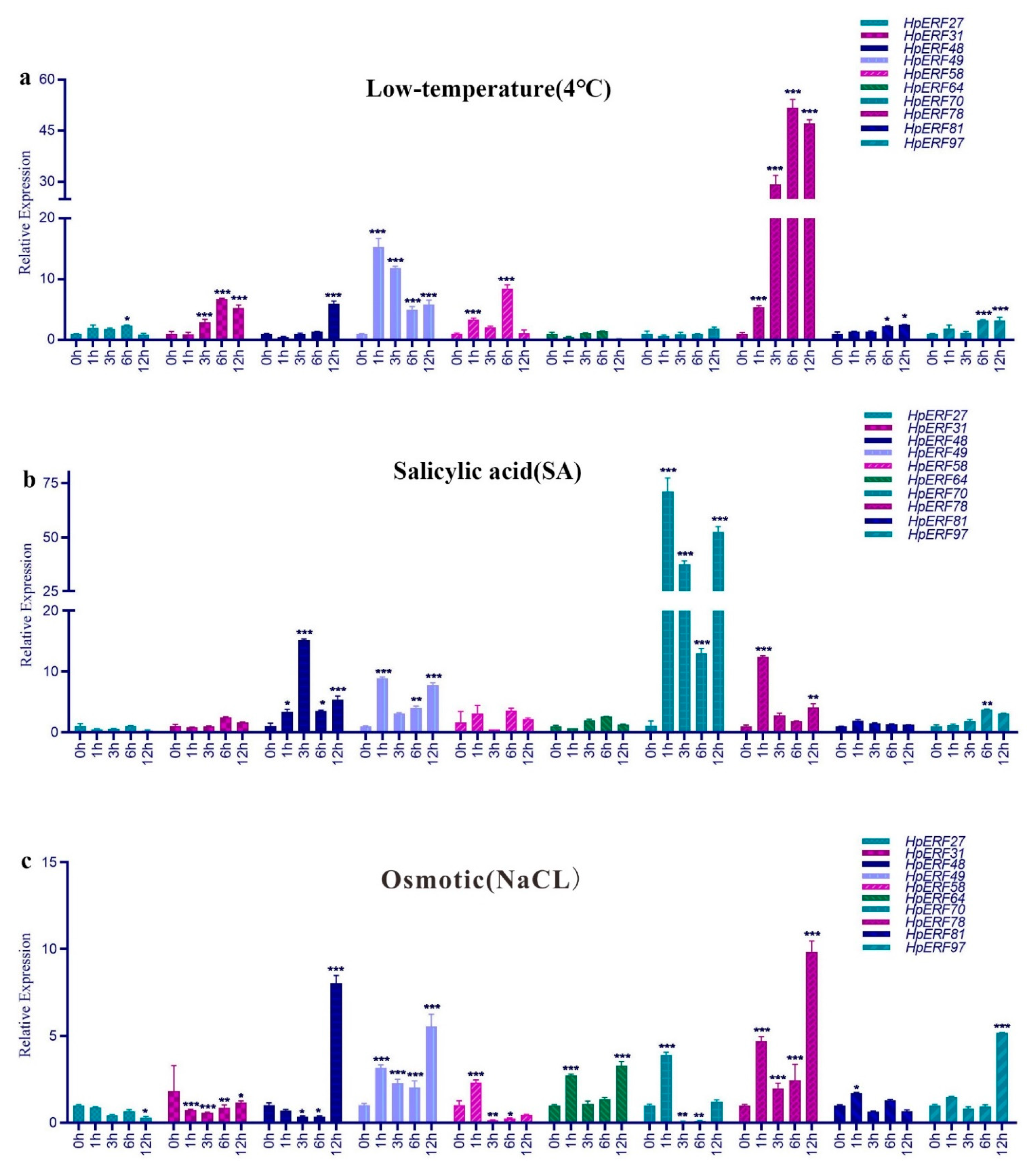
2.7. Analysis of Expression Patterns under Different Stress Treatments
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Database Search for Sequences and the Prediction of the Subcellular Locations
4.3. Multiple Alignments and Phylogenetic Tree Analysis
4.4. The Prediction of Conserved Motifs and Gene Structure Analysis
4.5. Cis-Elements and Ka/Ks Analysis
4.6. Gene Ontology Annotation Analysis
4.7. Isolation of RNA and cDNA Preparation
4.8. RNA-Seq Expression and qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sirvent, T.M.; Krasnoff, S.B.; Gibson, D.M. Induction of Hypericins and Hyperforins in Hypericum perforatum in Response to Damage by Herbivores. J. Chem. Ecol. 2003, 29, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Weina, H.; Preeti, S.; Gregory, F. A Perspective on Hypericum perforatum Genetic Transformation. Front. Plant Sci. 2016, 7, 879–890. [Google Scholar]
- Zdunić, G.; Gođevac, D.; Milenković, M.; Šavikin, K.; Menković, N.; Petrović, S. Anti-inflammatory and Gastroprotective Properties of Hypericum richeri Oil Extracts. Nat. Prod. Commun. 2010, 5, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Palo, J. Herbal Medicines: The Complete German Commission E Monographs: Therapeutic Guide to Herbal Medicines. Am. Med. Assoc. 1999, 281, 1851-1194. [Google Scholar]
- Butterweck, V. Mechanism of Action of St John’s Wort in Depression: What is Known? CNS Drugs 2003, 17, 539–562. [Google Scholar] [CrossRef]
- Butterweck, V.; Wall, A.; Liefländer-Wulf, U.; Winterhoff, H.; Nahrstedt, A. Effects of the total extract and fractions of Hypericum perforatum in animal assays for antidepressant activity. Pharmacopsychiatry 1997, 30, 117–124. [Google Scholar] [CrossRef]
- Group, H.D.T.S.; Davidson, J.R.T.; Gadde, K.M.; Fairbank, J.A.; Weisler, R.H. Effect of Hypericum perforatum (St John’s Wort) in Major Depressive Disorder: A Randomized Controlled Trial. JAMA 2002, 287, 1807–1814. [Google Scholar]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Ito, T.M.; Polido, P.B.; Rampim, M.C.; Kaschuk, G.; Souza, S.G.H. Genome-wide identification and phylogenetic analysis of the AP2/ERF gene superfamily in sweet orange (Citrus sinensis). Genet. Mol. Res. 2014, 13, 7839–7851. [Google Scholar] [CrossRef]
- Zhuang, J.; Peng, R.H.; Cheng, Z.M.; Zhang, J.; Cai, B.; Zhang, Z.; Gao, F.; Zhu, B.; Fu, X.Y.; Jin, X.F. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Entia Hortic. 2009, 123, 73–81. [Google Scholar] [CrossRef]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-Inducible DNA Binding Proteins That Interact with an Ethylene-Responsive Element. Plant Cell 1995, 7, 173–182. [Google Scholar] [PubMed]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Dong, Y.; Qiao, D.; Wang, Q.; Ma, Z.; Zhang, F.; Zhou, Q.; Xu, H.; Deng, F.; Li, Y. Isolation and characterization of ZmERF1encoding ethylene responsive factor-like protein 1 in popcorn (Zea mays L.). Plant Cell Tissue Org. 2015, 120, 747–756. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Banno, H.; Ikeda, Y.; Niu, Q.W.; Chua, N.H. Overexpression of Arabidopsis ESR1 Induces Initiation of Shoot Regeneration. Plant Cell 2001, 13, 2609–2618. [Google Scholar] [CrossRef]
- Deng, B.; Huang, Z.J.; Ge, F.; Liu, D.Q.; Lu, R.J.; Chen, C.Y. An AP2/ERF Family Transcription Factor PnERF1 Raised the Biosynthesis of Saponins in Panax notoginseng. J. Plant Growth Regul. 2017, 36, 566–576. [Google Scholar] [CrossRef]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The Jasmonate-Responsive AP2/ERF Transcription Factors AaERF1 and AaERF2 Positively Regulate Artemisinin Biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Du, H.W.; Huang, M.; Zhang, Z.X.; Cheng, S.Y. Genome-wide analysis of the AP2/ERF gene family in maize waterlogging stress response. Euphytica 2014, 198, 115–126. [Google Scholar] [CrossRef]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Quan, R.; Pan, X.; Wan, L.; Huang, R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 2013, 237, 1443–1451. [Google Scholar] [CrossRef]
- Guodong, R.; Jinkai, S.; Yanfei, Z.; He, C.; Zhang, J. Genome-wide analysis of the AP2/ERF gene family in Salix arbutifolia. FEBS Open Bio 2015, 5, 132–137. [Google Scholar]
- Du, D.L.; Hao, R.J.; Cheng, T.R.; Pan, H.T.; Yang, W.R.; Wang, J.; Zhang, Q.X. Genome-Wide Analysis of the AP2/ERF Gene Family in Prunus mume. Plant Mol. Biol. Rep. 2013, 31, 741–750. [Google Scholar] [CrossRef]
- Wang, S.; Yang, S.; Yin, Y.; Xi, J.; Li, S.; Hao, D. Molecular dynamics simulations reveal the disparity in specific recognition of GCC-box by AtERFs transcription factors super family in Arabidopsis. J. Mol. Recognit. 2010, 22, 474–479. [Google Scholar] [CrossRef]
- Wang, L.; Qin, L.; Liu, W.; Zhang, D.; Wang, Y. A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol. Plant 2014, 152, 84–97. [Google Scholar] [CrossRef]
- Lee, S.B.; Lee, S.J.; Kim, S.Y. AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 2015, 34, 71–81. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, L.; Dai, Y.; Liu, S.; Hong, Y.; Tian, L.; Huang, L.; Cao, Z.; Li, D.; Song, F. Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 2015, 6, 686–698. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, L.; Yu, Y.; Quan, R.; Zhang, Z.; Zhang, H.; Huang, R. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 2011, 68, 88–99. [Google Scholar] [CrossRef]
- Wei, L.; Umuhoza, K.N.J.; Wu, T.; Yang, Y.; Zhang, X.; Xu, X.; Yi, W.; Han, Z.; Wu, K. The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana. PLoS ONE 2017, 12, e0186580. [Google Scholar]
- Lee, J.H.; Hong, J.P.; Oh, S.K.; Lee, S.; Choi, D.; Kim, W. The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: Possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol. Biol. 2004, 55, 61–81. [Google Scholar] [PubMed]
- Yi, S.Y.; Kim, J.H.; Joung, Y.H.; Lee, S.; Kim, W.T.; Yu, S.H.; Choi, D. The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol. 2004, 136, 2862–2874. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.H.; Yin, L.R.; Hong, G.E.; Huang, R.F.; Xiao, Q.M.; Xie, B.Y. Overexpression of a Novel Member of ERF Transcription Factor JERF3 in Lily Enhances the Tolerance to Salt. Acta Hortic. Sin. 2007, 34, 1485–1490. [Google Scholar]
- Gao, S.; Zhang, H.; Tian, Y.; Li, F.; Zhang, Z.; Lu, X.; Chen, X.; Huang, R. Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep. 2008, 27, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.G.; Li, H.; Liu, J.Y. Molecular characterization of three novel ethylene responsive 2 element binding factor genes from cotton. J. Integr. Plant Biol. 2010, 52, 485–495. [Google Scholar]
- Oñate-Sánchez, L.; Singh, K.B. Identification of Arabidopsis Ethylene-Responsive Element Binding Factors with Distinct Induction Kinetics after Pathogen Infection. Plant Physiol. 2002, 128, 1313–1322. [Google Scholar] [CrossRef]
- Shinwari, Z.K.; Nakashima, K.; Miura, S.; Kasuga, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. An Arabidopsis Gene Family Encoding DRE/CRT Binding Proteins Involved in Low-Temperature-Responsive Gene Expression. Biochem. Biophys. Res. Commun. 1998, 250, 161–170. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Qin, S.; Guo, Y.; Chen, Y.; Wu, G. Genome-Wide Analysis of the AP2/ERF Gene Family in Physic Nut and Overexpression of the JcERF011 Gene in Rice Increased Its Sensitivity to Salinity Stress. PLoS ONE 2016, 11, e0150879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Shangguan, L.F.; Ma, R.J.; Sun, X.; Tao, R.; Guo, L.; Korir, N.K.; Yu, M.L. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet. Mol. Res. 2012, 11, 4789–4809. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.M.; Tang, Y.X.; Wu, Y.M. Genome-Wide Analysis of AP2/ERF Transcription Factor Family in Zea Mays. Curr. Bioinform. 2012, 7, 324–332. [Google Scholar] [CrossRef]
- Yao, W.; Zhang, X.; Zhou, B.; Zhao, K.; Li, R.; Jiang, T. Expression Pattern of ERF Gene Family under Multiple Abiotic Stresses in Populus simonii × P. nigra. Front. Plant Sci. 2017, 8, 181–189. [Google Scholar] [CrossRef]
- Lynch, M. Genomics. Gene duplication and evolution. Science 2002, 297, 945–947. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Wang, F.; Jiang, Q.; Li, R.; Ma, J. Genome-wide Analysis of the Distribution of AP2/ERF Transcription Factors Reveals Duplication and Elucidates their Potential Function in Chinese Cabbage (Brassica rapa ssp. pekinensis). Plant Mol. Biol. Rep. 2013, 31, 1002–1011. [Google Scholar] [CrossRef]
- Lata, C.; Mishra, A.K.; Muthamilarasan, M.; Bonthala, V.S.; Khan, Y.; Prasad, M. Genome-Wide Investigation and Expression Profiling of AP2/ERF Transcription Factor Superfamily in Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e113092. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Bi, S.D.; Li, F. Genome-wide analysis of expansion and contraction of gene families in parasitic wasps. J. Anhui Agric. Univ. 2018, 45, 945–950. [Google Scholar]
- Ken, W.; Ó’hUigín, C. Significance of positive selection and gene duplication in adaptive evolution: In memory of Austin L. Hughes. Immunogenetics 2016, 68, 749–753. [Google Scholar]
- Papdi, C.; Pérez-Salamó, I.; Joseph, M.P.; Giuntoli, B.; Szabados, L. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015, 82, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Robert, C. Edgar. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed]
- Lifang, H.; Shiqiang, L. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 2011, 34, 624–633. [Google Scholar]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N.; et al. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.K. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 2010, 25, 131–139. [Google Scholar] [CrossRef]
- Wang, Z.; Xiong, L.; Li, W.; Zhu, J.; Zhu, J. The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 2011, 23, 1971–1984. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Li, B.; Petijová, L.; Hu, S.; Zhang, Q.; Niu, J.; Wang, D.; Wang, S.; Dong, Y.; et al. Whole-genome sequence data of Hypericum perforatum and functional characterization of melatonin biosynthesis by N-acetylserotonin O-methyltransferase. J. Pineal Res. 2020, e12709. [Google Scholar]
- Bailey, T.L.; Nadya, W.; Chris, M.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P.; Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; GÖtz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) Database and Informatics Resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar]
- Carbon, S.; Ireland, A.; Christopher, J.M.; Shu, S.; Marshall, B.; Lewis, S.; AmiGO, Hub. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, S.Q.; Yang, L.; Sun, Y.; Zhang, Q.; Li, B.; Wang, B.; Li, L.; Wang, D.; Wang, Z. Reference genes for qRT-PCR normalisation in different tissues, developmental stages, and stress conditions of Hypericum perforatum. PeerJ 2019, 7, e7133. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int. J. Biol. Macromol. 2020, 145, 341–354. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhou, W.; Li, B.; Li, L.; Fu, M.; Zhou, L.; Yu, X.; Wang, D.; Wang, Z. Genome-Wide Analysis and the Expression Pattern of the ERF Gene Family in Hypericum perforatum. Plants 2021, 10, 133. https://doi.org/10.3390/plants10010133
Zhang Q, Zhou W, Li B, Li L, Fu M, Zhou L, Yu X, Wang D, Wang Z. Genome-Wide Analysis and the Expression Pattern of the ERF Gene Family in Hypericum perforatum. Plants. 2021; 10(1):133. https://doi.org/10.3390/plants10010133
Chicago/Turabian StyleZhang, Qian, Wen Zhou, Bin Li, Lin Li, Meng Fu, Li Zhou, Xiaoding Yu, Donghao Wang, and Zhezhi Wang. 2021. "Genome-Wide Analysis and the Expression Pattern of the ERF Gene Family in Hypericum perforatum" Plants 10, no. 1: 133. https://doi.org/10.3390/plants10010133
APA StyleZhang, Q., Zhou, W., Li, B., Li, L., Fu, M., Zhou, L., Yu, X., Wang, D., & Wang, Z. (2021). Genome-Wide Analysis and the Expression Pattern of the ERF Gene Family in Hypericum perforatum. Plants, 10(1), 133. https://doi.org/10.3390/plants10010133




