Lysosomal Function Impacts the Skeletal Muscle Extracellular Matrix
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry/Mutant/Transgenic Lines
2.2. Itga7 Mutant Construction
2.3. Morpholino Injections
2.4. Cyclopamine (CyA)/Aurintricarboxylic Acid (ATA) Treatment
2.5. Evans Blue Dye (EBD) Injection
2.6. Phalloidin Staining and Immunohistochemistry
2.7. Comparative qRT-PCR
2.8. Imaging
2.9. Birefringence
2.10. Analysis and Statistics
3. Results
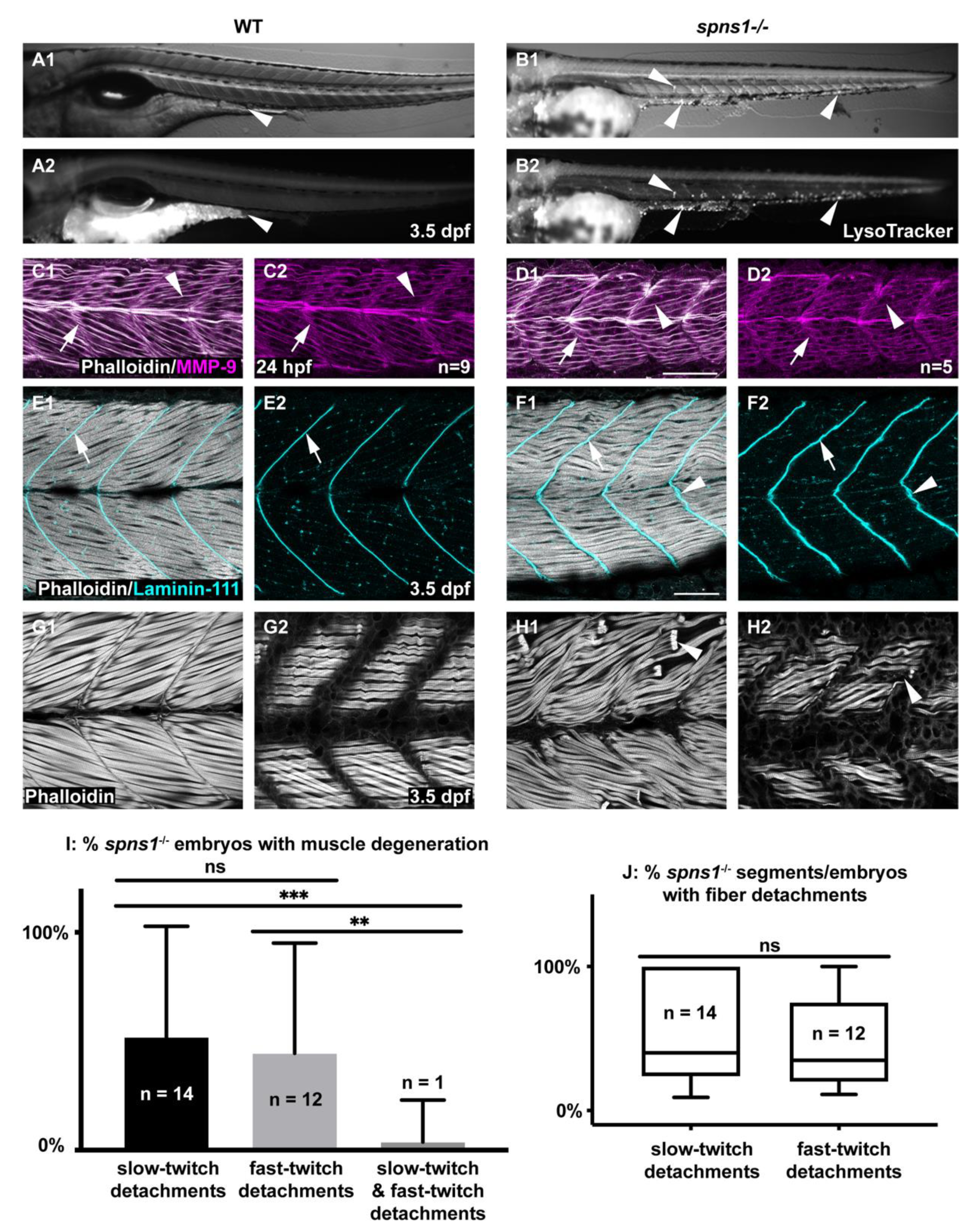
3.1. Initial Muscle Development Proceeded Normally But Degeneration Began at 3.5 Days Post-Fertilization
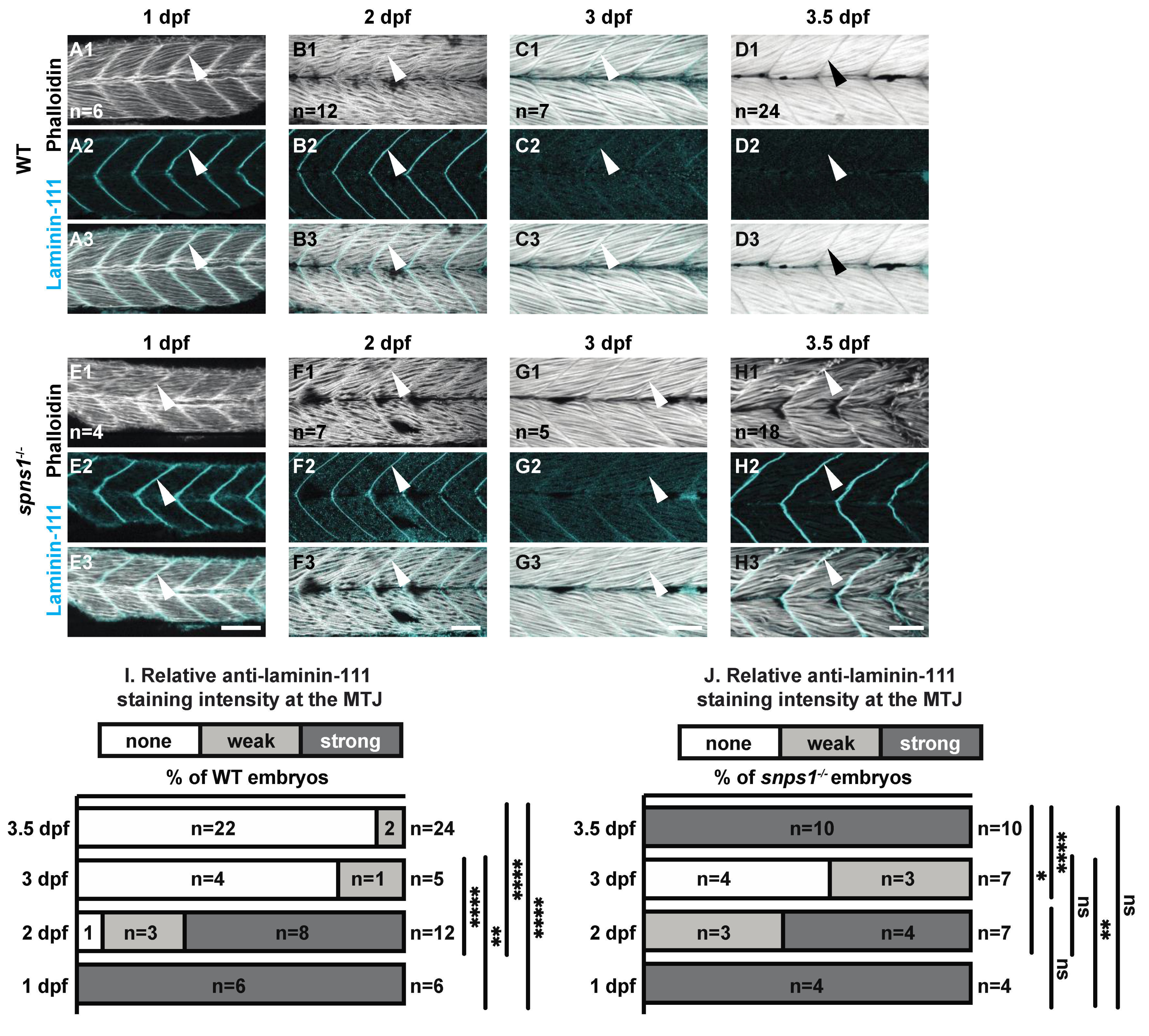
3.2. Aberrant Upregulation of the Extracellular Matrix Protein Laminin 111 at the MTJ Coincided with the Onset of Muscle Degeneration
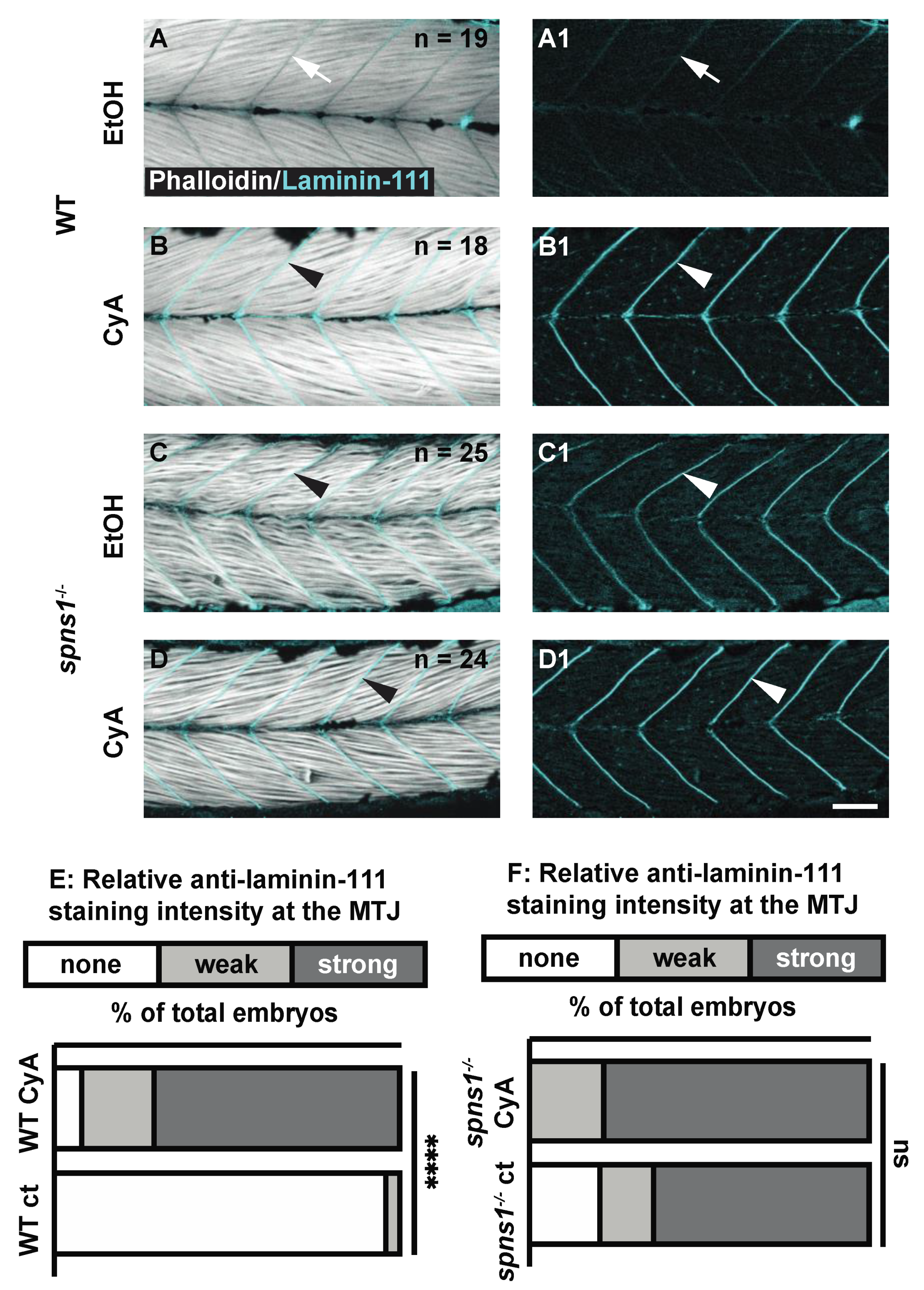
3.3. A Different Mechanism Underlies Laminin 111 Re-Expression Than Underlies Laminin 111 Expression during Initial MTJ Development
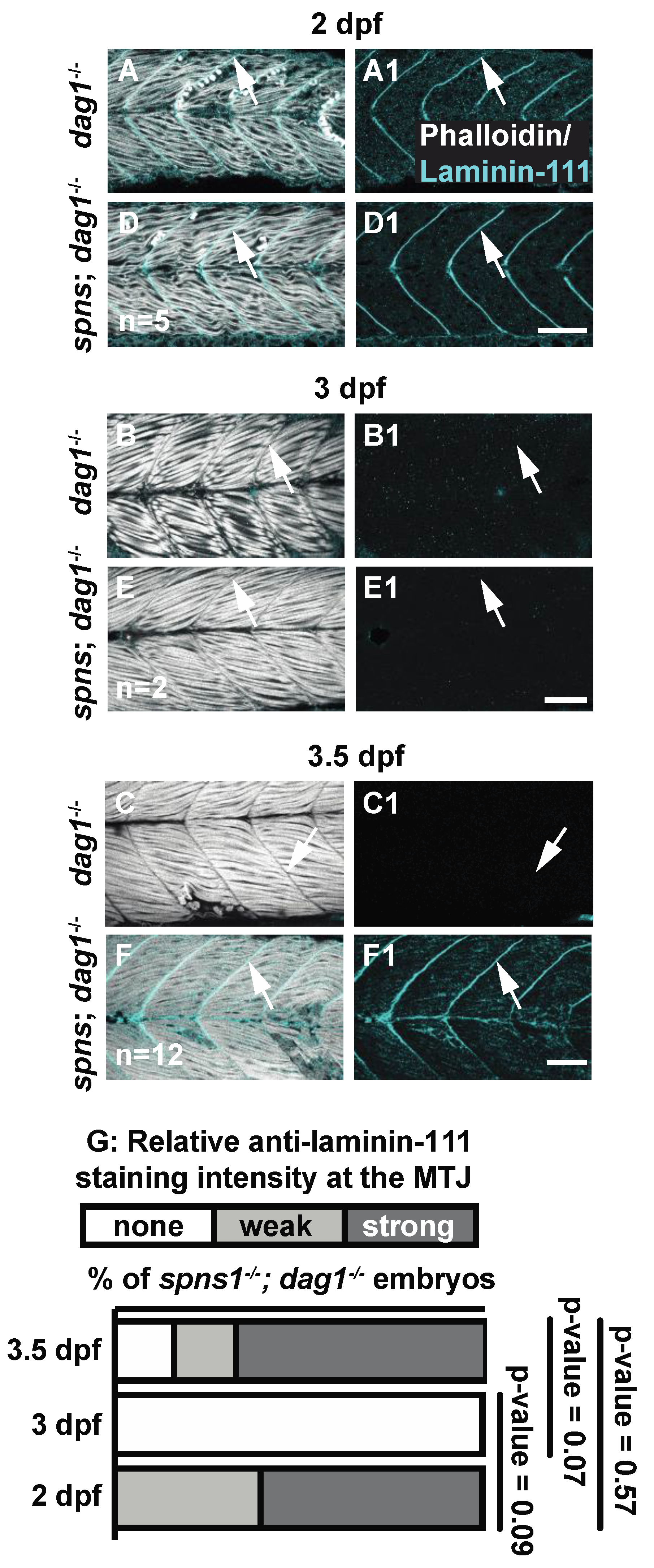
3.4. The Laminin Receptor Dystroglycan Did Not Genetically Interact with Spinster
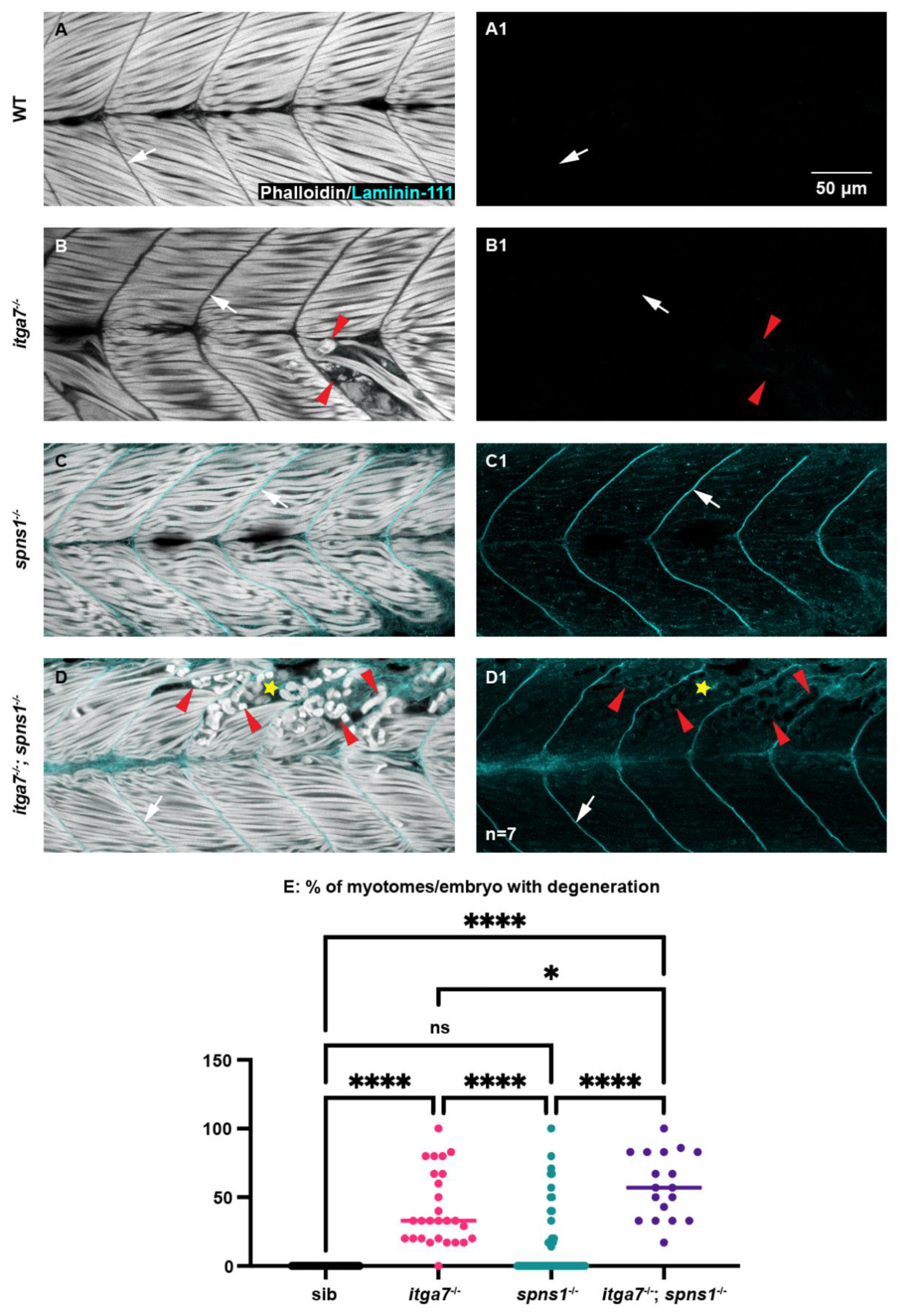
3.5. The Laminin Receptor Integrin α7 Contributed to Muscle Homeostasis in spns1−/− Mutants
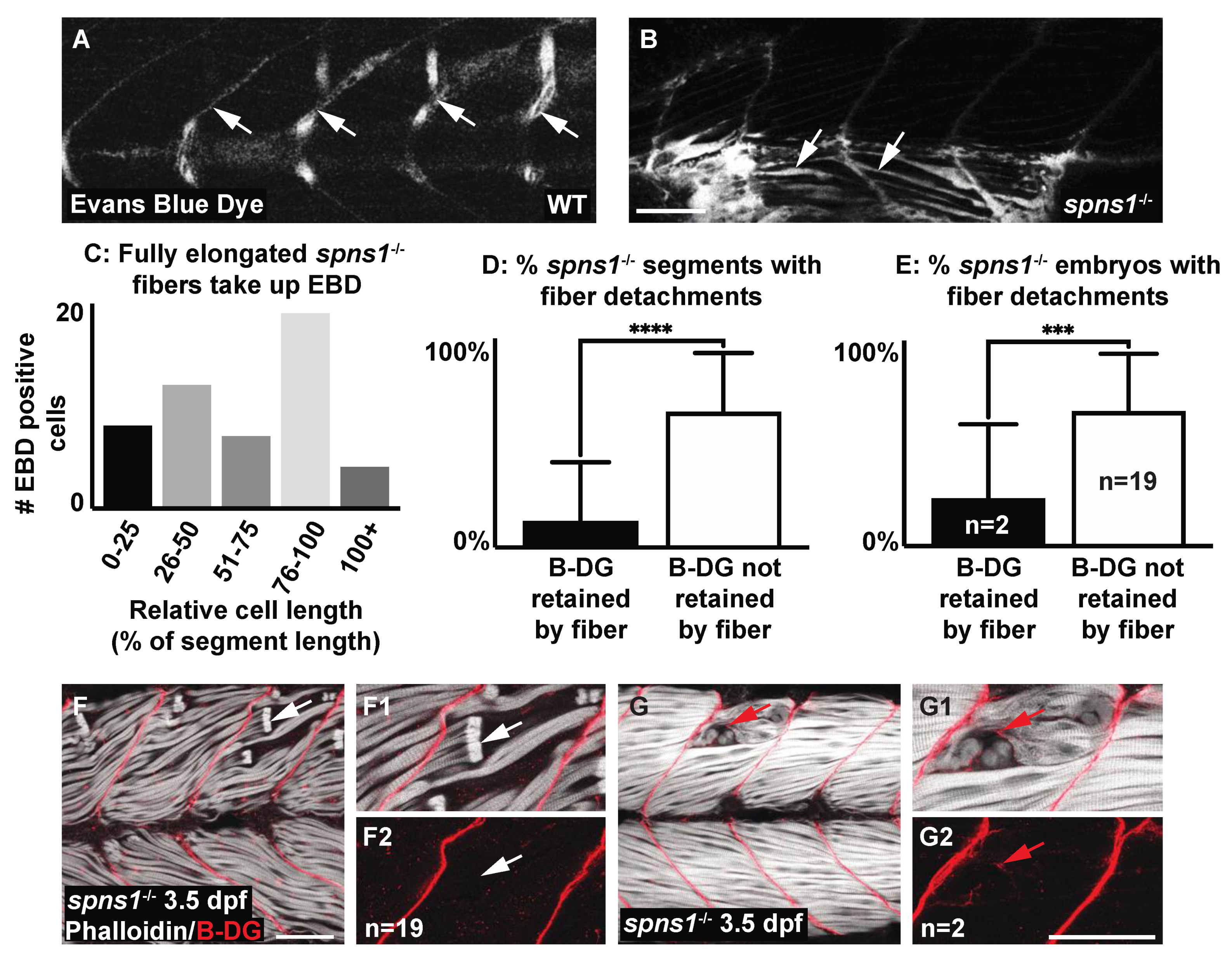
3.6. Sarcolemmal Instability in spns1 Muscle Fibers
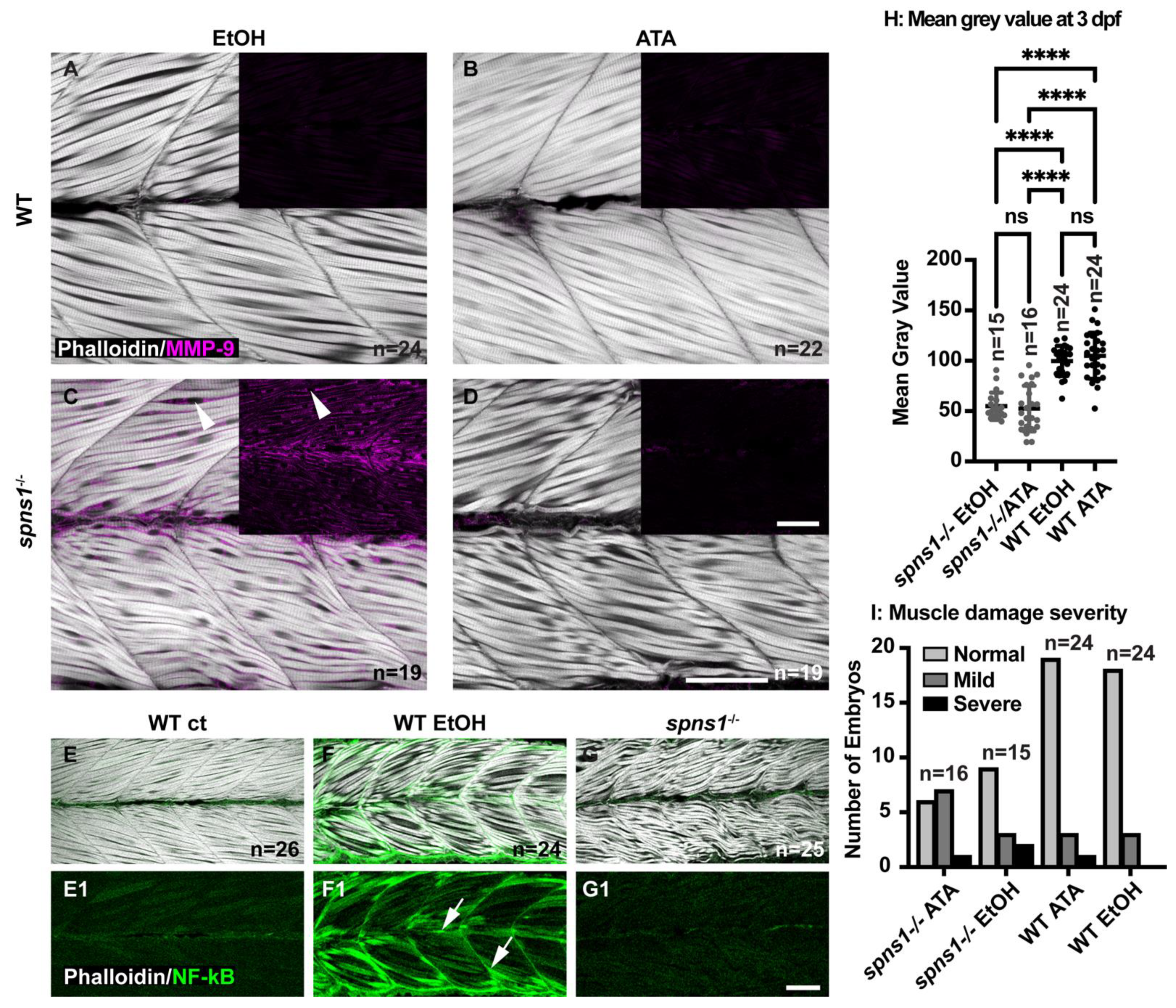
3.7. MMP-9 Was Upregulated in spns1−/− Mutants
3.8. Tnfsd12-Fn14 Signaling Axis Regulated Increased MMP-9 Expression
3.9. Reduced MMP-9 Expression Did Not Impact Muscle Degeneration
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tesseraud, S.; Chagneau, A.M.; Grizard, J. Muscle Protein Turnover during Early Development in Chickens Divergently Selected for Growth Rate. Poult. Sci. 2000, 79, 1465–1471. [Google Scholar] [CrossRef] [PubMed]
- Nemova, N.N.; Lysenko, L.A.; Kantserova, N.P. [Degradation of skeletal muscle protein during growth and development of salmonid fish]. Ontogenez 2016, 47, 197–208. [Google Scholar] [CrossRef]
- Rajawat, Y.S.; Hilioti, Z.; Bossis, I. Aging: Central Role for Autophagy and the Lysosomal Degradative System. Ageing Res. Rev. 2009, 8, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.L.; Escaleira, R.C.; Jazenko, F.; Mermelstein, C.S. Cell Adhesion in Zebrafish Myogenesis: Distribution of Intermediate Filaments, Microfilaments, Intracellular Adhesion Structures and Extracellular Matrix. Cell Motil. 2008, 65, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, D.; Velling, T.; Sjöberg, G.; Salmivirta, K.; Gaggero, B.; Tiger, C.F.; Edström, L.; Sejersen, T. Tenascin-C Expression Correlates with Macrophage Invasion in Duchenne Muscular Dystrophy and in Myositis. Neuromuscul. Disord. 1997, 7, 39–54. [Google Scholar] [CrossRef]
- Hall, T.E.; Bryson-Richardson, R.J.; Berger, S.; Jacoby, A.S.; Cole, N.J.; Hollway, G.E.; Berger, J.; Currie, P.D. The Zebrafish Candyfloss Mutant Implicates Extracellular Matrix Adhesion Failure in Laminin A2-Deficient Congenital Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2007, 104, 7092–7097. [Google Scholar] [CrossRef] [Green Version]
- Leonoudakis, D.; Huang, G.; Akhavan, A.; Fata, J.E.; Singh, M.; Gray, J.W.; Muschler, J.L. Endocytic Trafficking of Laminin Is Controlled by Dystroglycan and Is Disrupted in Cancers. J. Cell Sci. 2014, 127, 4894–4903. [Google Scholar] [CrossRef] [Green Version]
- Carmignac, V.; Svensson, M.; Körner, Z.; Elowsson, L.; Matsumura, C.; Gawlik, K.I.; Allamand, V.; Durbeej, M. Autophagy Is Increased in Laminin A2 Chain-Deficient Muscle and Its Inhibition Improves Muscle Morphology in a Mouse Model of MDC1A. Hum. Mol. Genet. 2011, 20, 4891–4902. [Google Scholar] [CrossRef]
- Grumati, P.; Coletto, L.; Sabatelli, P.; Cescon, M.; Angelin, A.; Bertaggia, E.; Blaauw, B.; Urciuolo, A.; Tiepolo, T.; Merlini, L.; et al. Autophagy Is Defective in Collagen VI Muscular Dystrophies, and Its Reactivation Rescues Myofiber Degeneration. Nat. Med. 2010, 16, 1313–1320. [Google Scholar] [CrossRef]
- Graves, A.R.; Curran, P.K.; Smith, C.L.; Mindell, J.A. The Cl-/H+ Antiporter ClC-7 Is the Primary Chloride Permeation Pathway in Lysosomes. Nature 2008, 453, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Sagné, C.; Agulhon, C.; Ravassard, P.; Darmon, M.; Hamon, M.; El Mestikawy, S.; Gasnier, B.; Giros, B. Identification and Characterization of a Lysosomal Transporter for Small Neutral Amino Acids. Proc. Natl. Acad. Sci. USA 2001, 98, 7206–7211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, T.; Lian, S.; Khan, A.; Llop, J.R.; Samuelson, A.V.; Chen, W.; Klionsky, D.J.; Kishi, S. Autolysosome Biogenesis and Developmental Senescence Are Regulated by Both Spns1 and V-ATPase. Autophagy 2017, 13, 386–403. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; McPhee, C.K.; McPhee, C.; Deng, S.; Huang, L.; Chen, L.; Liu, M.; Tracy, K.; Baehrecke, E.H.; Baehreck, E.H.; et al. Spinster Is Required for Autophagic Lysosome Reformation and MTOR Reactivation Following Starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 7826–7831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadarajah, V.D.; van Putten, M.; Chaouch, A.; Garrood, P.; Straub, V.; Lochmüller, H.; Ginjaar, H.B.; Aartsma-Rus, A.M.; van Ommen, G.J.B.; den Dunnen, J.T.; et al. Serum Matrix Metalloproteinase-9 (MMP-9) as a Biomarker for Monitoring Disease Progression in Duchenne Muscular Dystrophy (DMD). Neuromuscul. Disord. 2011, 21, 569–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiba, N.; Miyazaki, D.; Yoshizawa, T.; Fukushima, K.; Shiba, Y.; Inaba, Y.; Imamura, M.; Takeda, S.; Koike, K.; Nakamura, A. Differential Roles of MMP-9 in Early and Late Stages of Dystrophic Muscles in a Mouse Model of Duchenne Muscular Dystrophy. Biochim. Biophys. Acta 2015, 1852, 2170–2182. [Google Scholar] [CrossRef] [Green Version]
- Kishi, S.; Bayliss, P.E.; Uchiyama, J.; Koshimizu, E.; Qi, J.; Nanjappa, P.; Imamura, S.; Islam, A.; Neuberg, D.; Amsterdam, A.; et al. The Identification of Zebrafish Mutants Showing Alterations in Senescence-Associated Biomarkers. PLoS Genet. 2008, 4, e1000152. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-Y.; White, R.J.; Torelli, S.; Cirak, S.; Muntoni, F.; Stemple, D.L. Zebrafish Fukutin Family Proteins Link the Unfolded Protein Response with Dystroglycanopathies. Hum. Mol. Genet. 2011, 20, 1763–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanther, M.; Sun, X.; Mühlbauer, M.; Mackey, L.C.; Flynn, E.J.; Bagnat, M.; Jobin, C.; Rawls, J.F. Microbial Colonization Induces Dynamic Temporal and Spatial Patterns of NF-ΚB Activation in the Zebrafish Digestive Tract. Gastroenterology 2011, 141, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Akhmetova, L.; Ahkmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; et al. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef] [PubMed]
- Barresi, M.J.F.; Hutson, L.D.; Chien, C.-B.; Karlstrom, R.O. Hedgehog Regulated Slit Expression Determines Commissure and Glial Cell Position in the Zebrafish Forebrain. Development 2005, 132, 3643–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurusamy, S.; López-Maderuelo, D.; Little, R.; Cadagan, D.; Savage, A.M.; Ihugba, J.C.; Baggott, R.R.; Rowther, F.B.; Martínez-Martínez, S.; Arco, P.G.-D.; et al. Selective Inhibition of Plasma Membrane Calcium ATPase 4 Improves Angiogenesis and Vascular Reperfusion. J. Mol. Cell Cardiol. 2017, 109, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Sztal, T.; Currie, P.D. Quantification of Birefringence Readily Measures the Level of Muscle Damage in Zebrafish. Biochem. Biophys. Res. Commun. 2012, 423, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.H.F.; Zhang, H.; Jiang, A.; Zai, C.C.; Liu, F. Specific Alterations in Astrocyte Properties via the GluA2-GAPDH Complex Associated with Multiple Sclerosis. Sci Rep. 2018, 8, 12856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, T.; Lian, S.; Qi, J.; Bayliss, P.E.; Carr, C.E.; Johnson, J.L.; Guha, S.; Kobler, P.; Catz, S.D.; Gill, M.; et al. Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and P53 in Vertebrate Spns1 Deficiency. PLoS Genet. 2014, 10, e1004409. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Fahad, T.M.; Akther, T.; Zaman, T.; Hasan, M.F.; Islam Khan, M.R.; Islam, M.S.; Kishi, S. Carbofuran Accelerates the Cellular Senescence and Declines the Life Span of Spns1 Mutant Zebrafish. J. Cell Mol. Med. 2021, 25, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.; Maves, L. Skeletal Muscle Fiber Type: Using Insights from Muscle Developmental Biology to Dissect Targets for Susceptibility and Resistance to Muscle Disease. Wiley Interdiscip Rev. Dev. Biol. 2016, 5, 518–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jessen, J.R. Recent Advances in the Study of Zebrafish Extracellular Matrix Proteins. Dev. Biol. 2015, 401, 110–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, M.T.; Henry, C.A. Hedgehog Signaling and Laminin Play Unique and Synergistic Roles in Muscle Development. Dev. Dyn. 2010, 239, 905–913. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.; Thorsteinsdóttir, S.; Borycki, A.-G. Sonic Hedgehog-Dependent Synthesis of Laminin A1 Controls Basement Membrane Assembly in the Myotome. Development 2009, 136, 3495–3504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.K.; Taipale, J.; Cooper, M.K.; Beachy, P.A. Inhibition of Hedgehog Signaling by Direct Binding of Cyclopamine to Smoothened. Genes Dev. 2002, 16, 2743–2748. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Adiarte, E.G.; Devoto, S.H. Hedgehog Acts Directly on the Zebrafish Dermomyotome to Promote Myogenic Differentiation. Dev. Biol. 2006, 300, 736–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamill, K.J.; Kligys, K.; Hopkinson, S.B.; Jones, J.C.R. Laminin Deposition in the Extracellular Matrix: A Complex Picture Emerges. J. Cell Sci. 2009, 122, 4409–4417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, Y.K.; Chou, F.L.; Engvall, E.; Ogawa, M.; Matsuda, C.; Hirabayashi, S.; Yokochi, K.; Ziober, B.L.; Kramer, R.H.; Kaufman, S.J.; et al. Mutations in the Integrin Alpha7 Gene Cause Congenital Myopathy. Nat. Genet. 1998, 19, 94–97. [Google Scholar] [CrossRef]
- Yurchenco, P.D.; McKee, K.K.; Reinhard, J.R.; Rüegg, M.A. Laminin-Deficient Muscular Dystrophy: Molecular Pathogenesis and Structural Repair Strategies. Matrix Biol. 2018, 71–72, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Van Ry, P.M.; Minogue, P.; Hodges, B.L.; Burkin, D.J. Laminin-111 Improves Muscle Repair in a Mouse Model of Merosin-Deficient Congenital Muscular Dystrophy. Hum. Mol. Genet. 2014, 23, 383–396. [Google Scholar] [CrossRef] [Green Version]
- Postel, R.; Vakeel, P.; Topczewski, J.; Knöll, R.; Bakkers, J. Zebrafish Integrin-Linked Kinase Is Required in Skeletal Muscles for Strengthening the Integrin–ECM Adhesion Complex. Dev. Biol. 2008, 318, 92–101. [Google Scholar] [CrossRef] [Green Version]
- Bassett, D.I. Dystrophin Is Required for the Formation of Stable Muscle Attachments in the Zebrafish Embryo. Development 2003, 130, 5851–5860. [Google Scholar] [CrossRef] [Green Version]
- Fukushima, K.; Nakamura, A.; Ueda, H.; Yuasa, K.; Yoshida, K.; Takeda, S.; Ikeda, S. Activation and Localization of Matrix Metalloproteinase-2 and -9 in the Skeletal Muscle of the Muscular Dystrophy Dog (CXMDJ). BMC Musculoskelet Disord. 2007, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Kherif, S.; Lafuma, C.; Dehaupas, M.; Lachkar, S.; Fournier, J.G.; Verdière-Sahuqué, M.; Fardeau, M.; Alameddine, H.S. Expression of Matrix Metalloproteinases 2 and 9 in Regenerating Skeletal Muscle: A Study in Experimentally Injured and Mdx Muscles. Dev. Biol. 1999, 205, 158–170. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, M.L.; Kibbey, M.C.; Kleinman, H.K.; Wahl, L.M. Laminin SIKVAV Peptide Induction of Monocyte/Macrophage Prostaglandin E2 and Matrix Metalloproteinases. J. Biol. Chem. 1995, 270, 10365–10368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuratomi, Y.; Nomizu, M.; Tanaka, K.; Ponce, M.L.; Komiyama, S.; Kleinman, H.K.; Yamada, Y. Laminin Gamma 1 Chain Peptide, C-16 (KAFDITYVRLKF), Promotes Migration, MMP-9 Secretion, and Pulmonary Metastasis of B16-F10 Mouse Melanoma Cells. Br. J. Cancer 2002, 86, 1169–1173. [Google Scholar] [CrossRef] [PubMed]
- Roos, A.; Dhruv, H.D.; Mathews, I.T.; Inge, L.J.; Tuncali, S.; Hartman, L.K.; Chow, D.; Millard, N.; Yin, H.H.; Kloss, J.; et al. Identification of Aurintricarboxylic Acid as a Selective Inhibitor of the TWEAK-Fn14 Signaling Pathway in Glioblastoma Cells. Oncotarget 2017, 8, 12234–12246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Mittal, A.; Paul, P.K.; Kumar, M.; Srivastava, D.S.; Tyagi, S.C.; Kumar, A. Tumor Necrosis Factor-Related Weak Inducer of Apoptosis Augments Matrix Metalloproteinase 9 (MMP-9) Production in Skeletal Muscle through the Activation of Nuclear Factor-KappaB-Inducing Kinase and P38 Mitogen-Activated Protein Kinase: A Potential Role of MMP-9 in Myopathy. J. Biol. Chem. 2009, 284, 4439–4450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffey, E.C.; Pasquarella, M.E.; Goody, M.F.; Henry, C.A. Ethanol Exposure Causes Muscle Degeneration in Zebrafish. J. Dev. Biol. 2018, 6, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wibo, M.; Poole, B. Protein degradation in cultured cells. J.Cell Biol. 1974, 63, 430–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Follo, C.; Ozzano, M.; Montalenti, C.; Santoro, M.M.; Isidoro, C. Knockdown of Cathepsin D in Zebrafish Fertilized Eggs Determines Congenital Myopathy. Biosci. Rep. 2013, 33, e00034. [Google Scholar] [CrossRef]
- Hersheson, J.; Burke, D.; Clayton, R.; Anderson, G.; Jacques, T.S.; Mills, P.; Wood, N.W.; Gissen, P.; Clayton, P.; Fearnley, J.; et al. Cathepsin D Deficiency Causes Juvenile-Onset Ataxia and Distinctive Muscle Pathology. Neurology 2014, 83, 1873–1875. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, H.; Ishii, T.; Endo, K.; Kawakami, E.; Nagao, K.; Miyashita, T.; Akiyama, K.; Watabe, K.; Komatsu, M.; Yamamoto, D.; et al. L-Leucine and SPNS1 Coordinately Ameliorate Dysfunction of Autophagy in Mouse and Human Niemann-Pick Type C Disease. Sci. Rep. 2017, 7, 15944. [Google Scholar] [CrossRef]
- Kharbanda, K.K.; McVicker, D.L.; Zetterman, R.K.; MacDonald, R.G.; Donohue, T.M. Flow Cytometric Analysis of Vesicular PH in Rat Hepatocytes after Ethanol Administration. Hepatology 1997, 26, 929–934. [Google Scholar] [CrossRef]
- Gullberg, D.; Tiger, C.F.; Velling, T. Laminins during Muscle Development and in Muscular Dystrophies. Cell Mol. Life Sci. 1999, 56, 442–460. [Google Scholar] [CrossRef] [PubMed]
- Goody, M.F.; Sher, R.B.; Henry, C.A. Hanging on for the Ride: Adhesion to the Extracellular Matrix Mediates Cellular Responses in Skeletal Muscle Morphogenesis and Disease. Dev. Biol. 2015, 401, 75–91. [Google Scholar] [CrossRef] [Green Version]
- Rooney, J.E.; Knapp, J.R.; Hodges, B.L.; Wuebbles, R.D.; Burkin, D.J. Laminin-111 Protein Therapy Reduces Muscle Pathology and Improves Viability of a Mouse Model of Merosin-Deficient Congenital Muscular Dystrophy. Am. J. Pathol. 2012, 180, 1593–1602. [Google Scholar] [CrossRef] [Green Version]
- Brkic, M.; Balusu, S.; Libert, C.; Vandenbroucke, R.E. Friends or Foes: Matrix Metalloproteinases and Their Multifaceted Roles in Neurodegenerative Diseases. Mediat. Inflamm. 2015, 2015, e620581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahiya, S.; Givvimani, S.; Bhatnagar, S.; Qipshidze, N.; Tyagi, S.C.; Kumar, A. Osteopontin-Stimulated Expression of Matrix Metalloproteinase-9 Causes Cardiomyopathy in the Mdx Model of Duchenne Muscular Dystrophy. J. Immunol. 2011, 187, 2723–2731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogra, C.; Changoua, H.; Wedhas, N.; Qin, X.; Wergedal, J.E.; Kumar, A. TNF-Related Weak Inducer of Apoptosis (TWEAK) Is a Potent Skeletal Muscle-Wasting Cytokine. FASEB J. 2007, 21, 1857–1869. [Google Scholar] [CrossRef] [Green Version]
- Mittal, A.; Bhatnagar, S.; Kumar, A.; Lach-Trifilieff, E.; Wauters, S.; Li, H.; Makonchuk, D.Y.; Glass, D.J.; Kumar, A. The TWEAK-Fn14 System Is a Critical Regulator of Denervation-Induced Skeletal Muscle Atrophy in Mice. J. Cell Biol. 2010, 188, 833–849. [Google Scholar] [CrossRef] [Green Version]
- Tajrishi, M.M.; Sato, S.; Shin, J.; Zheng, T.S.; Burkly, L.C.; Kumar, A. The TWEAK-Fn14 Dyad Is Involved in Age-Associated Pathological Changes in Skeletal Muscle. Biochem. Biophys. Res. Commun. 2014, 446, 1219–1224. [Google Scholar] [CrossRef] [Green Version]
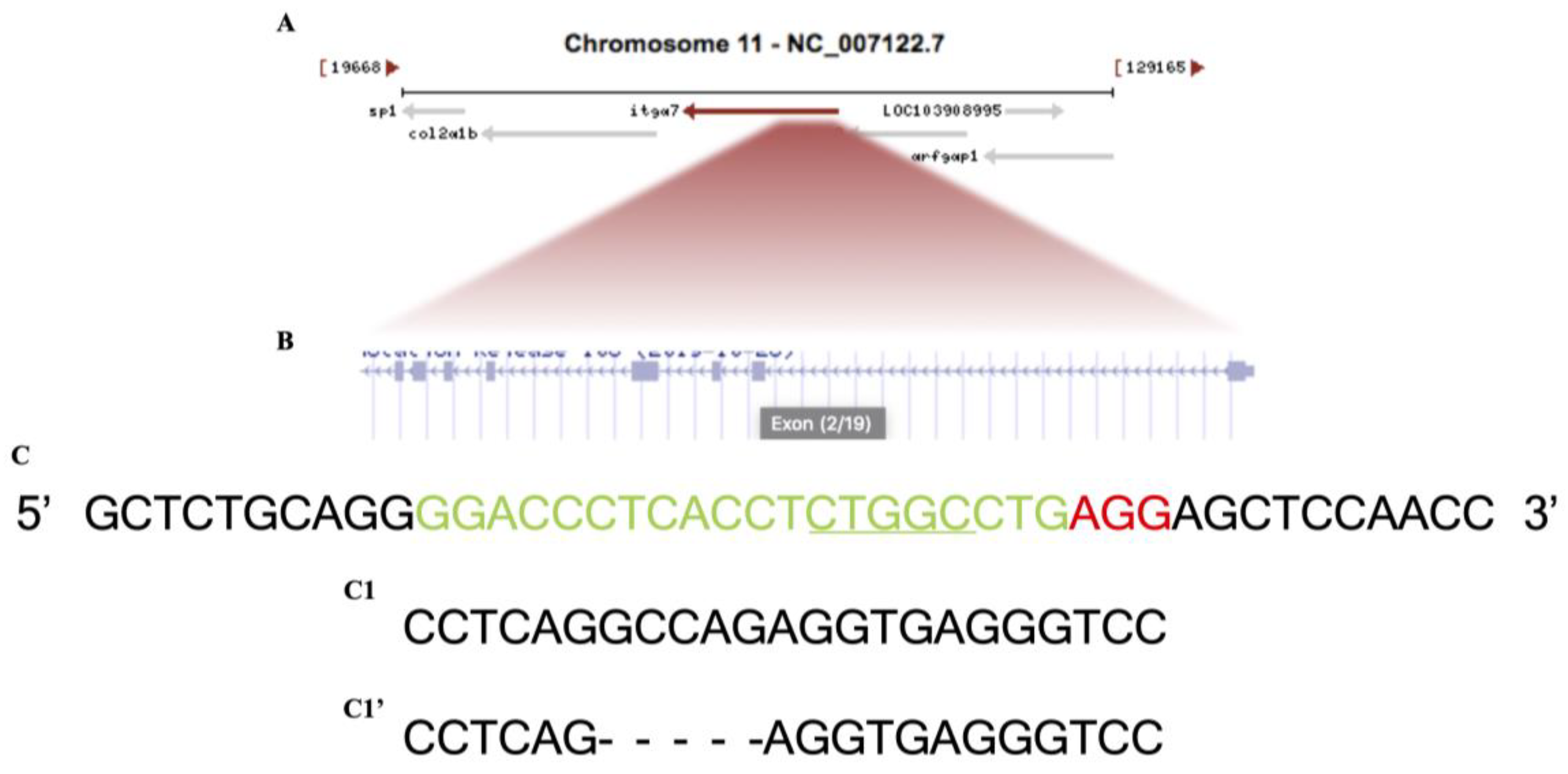

| Name | Sequence (5′→3′) |
|---|---|
| Itga7 target site (PAM) | GGACCCTCACCTCTGGCCTG(AGG) |
| Itga7 specific oligonuclutide (sp6 + target-last 3 letters + overlap region) | ATTTAGGTGACACTATAGGACCCTCACCTCTGGCCTGGTTTTAGAGCTAGAAATAGCAAG |
| Itga7 stop-Cassette oligonuclutide (left homology + stop cassette + right homology) | TTCTGGTTGGAGCTCCTCAGGTCATGGCGTTTAAACCTTAATTAAGCTGTTGTAGGCCAGAGGTGAGGGTCCCCT |
| Left Homology | TTCTGGTTGGAGCTCCTCAG |
| Right Homology | GCCAGAGGTGAGGGTCCCCT |
| Primer | Sequence | Product Size | Final Conc. |
|---|---|---|---|
| tweak forward | 5′-TGAATATGAGCAGGGCAGT-3′ | 168 bp | 100 uM |
| tweak reverse | 5′-TTCAATGCACTGGAGCAAAG-3′ | ||
| mmp9 forward | 5′-TGATGTGCTTGGACCACGTAA-3′ | 102 bp | 100 uM |
| mmp9 reverse | 5′-ACAGGAGCACCTTGCCTTTTC-3′ |
| Primer | ∆∆ Cq | Exponential Change |
|---|---|---|
| tweak | −5.09 | 34.12 |
| mmp9 | −3.92 | 15.12 |
| Marker Being Assessed | Wildtype (WT) 3.5 dpf | spns1−/− 3.5 dpf | dag1; spns1−/− 3.5 dpf | itga7; spns1−/− 3.5 dpf |
|---|---|---|---|---|
| Lysotracker | No puncta | Positive puncta | N/A | N/A |
| MMP9 mRNA | N/A | +15.1 fold compared to WT | N/A | N/A |
| MMP9 antibody staining compared to 1 dpf | MMP9 absent compared to 1 dpf WT p < 0.0001 | MMP9 returns to 1 dpf spns1−/− level | N/A | N/A |
| MMP9 antibody staining after 100 µM ATA (Tnfsf12 inhibitor) | MMP9 absent in ATA and in 0.1% EtOH vehicle control treated embryos | MMP9 absent in ATA but present in 0.1% EtOH vehicle control treated embryos | N/A | N/A |
| NF-κB:EGFP relative levels | Lower in negative controls than EtOH treated p-value < 0.0001 | Less than positive controls same as negative controls p-values < 0.0001 | N/A | N/A |
| Sarcolemma instability: beta-dystroglycan (b-DG) at the myotendinous junction(MTJ) versus at fiber ends | N/A | More embryos with b-DG at MTJ p-value <0.001 More segments with MTJ staining p-value < 0.0001 | N/A | N/A |
| Laminin-111 staining at 3.5 dpf compared to 1 dpf | Laminin-111 absent compared to 1 dpf WT p < 0.0001 | Laminin-111 returns to 1 dpf spns−/− level NS p-value > 0.9999 | Laminin-111 Expression returns to 1 dpf dag1; spns1−/− level NS p-value = 0.057 | Laminin-111 expression returns to 1 dpf itga7; spns1−/− level NS |
| Laminin-111 staining at 3.5 dpf after 50 µM cyclopamine (sonic hedgehog signaling blocker) | Stronger compared to WT vehicle controls p-value < 0.0001 | No change compared to spns1−/− vehicle controls NS p-value = 0.1838 | N/A | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coffey, E.C.; Astumian, M.; Alrowaished, S.S.; Schaffer, C.; Henry, C.A. Lysosomal Function Impacts the Skeletal Muscle Extracellular Matrix. J. Dev. Biol. 2021, 9, 52. https://doi.org/10.3390/jdb9040052
Coffey EC, Astumian M, Alrowaished SS, Schaffer C, Henry CA. Lysosomal Function Impacts the Skeletal Muscle Extracellular Matrix. Journal of Developmental Biology. 2021; 9(4):52. https://doi.org/10.3390/jdb9040052
Chicago/Turabian StyleCoffey, Elizabeth C., Mary Astumian, Sarah S. Alrowaished, Claire Schaffer, and Clarissa A. Henry. 2021. "Lysosomal Function Impacts the Skeletal Muscle Extracellular Matrix" Journal of Developmental Biology 9, no. 4: 52. https://doi.org/10.3390/jdb9040052
APA StyleCoffey, E. C., Astumian, M., Alrowaished, S. S., Schaffer, C., & Henry, C. A. (2021). Lysosomal Function Impacts the Skeletal Muscle Extracellular Matrix. Journal of Developmental Biology, 9(4), 52. https://doi.org/10.3390/jdb9040052







