Genetic and Epigenetic Regulation of Drosophila Oocyte Determination
Abstract
1. Introduction
Germ Line Cyst and Oocyte Determination
2. Drosophila Melanogaster Oogenesis
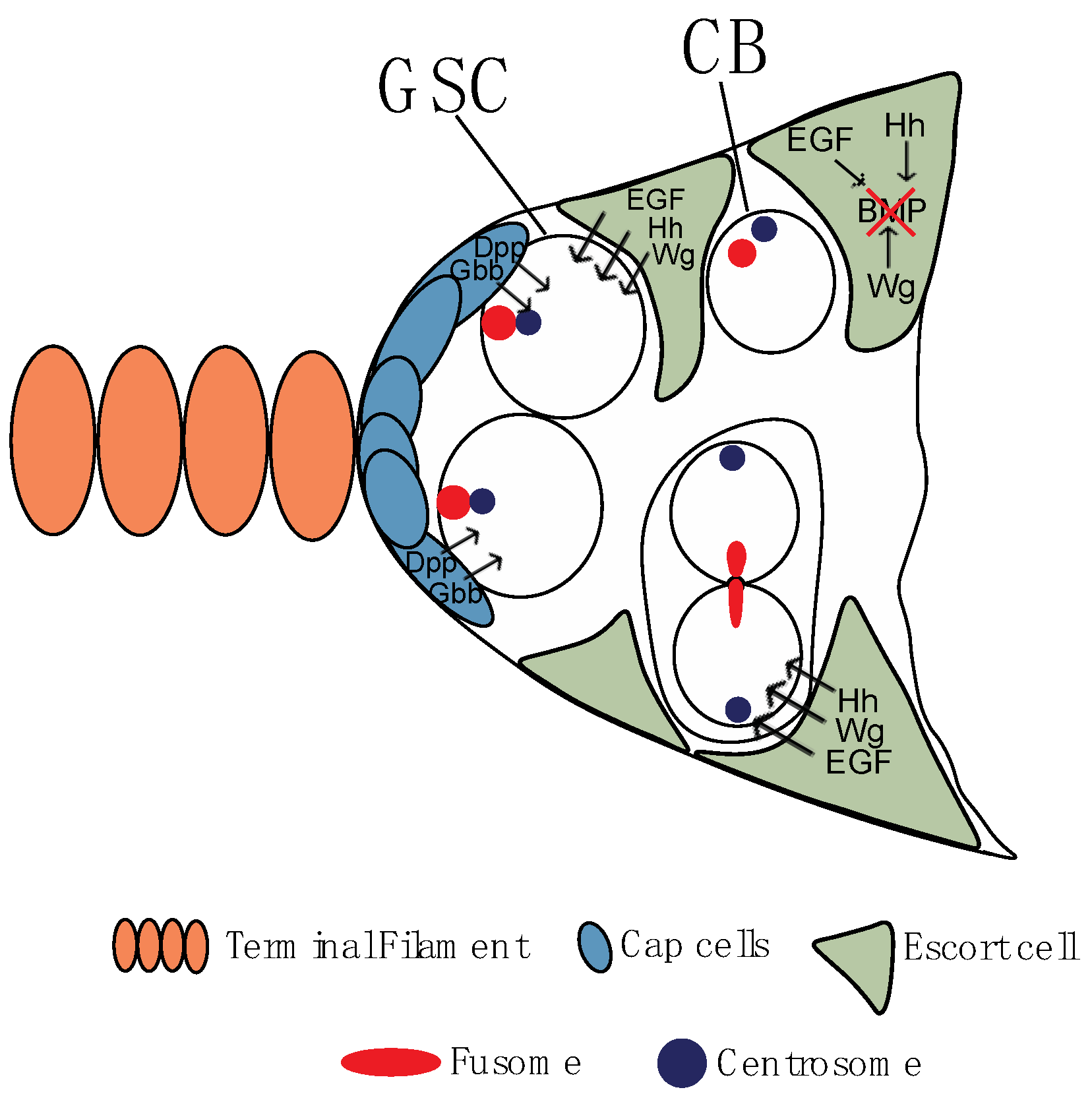
2.1. Cystoblast, Fusome, and Germ Line Cyst Formation
2.2. First among Equals: Oocyte Determination
2.3. Oocyte-Specific Factors
2.4. Shall I Stay or Shall I Go: Entry into Meiosis
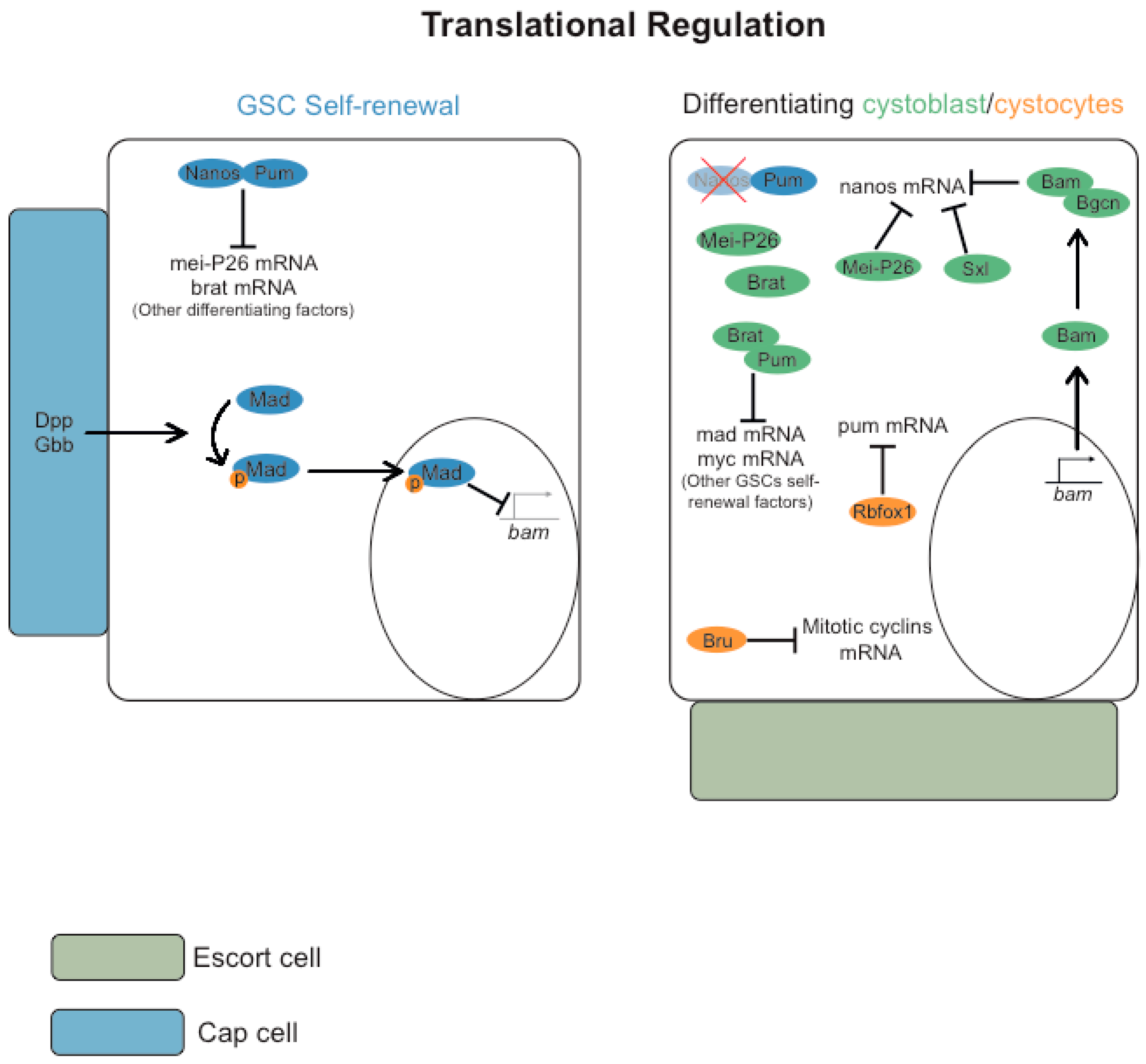
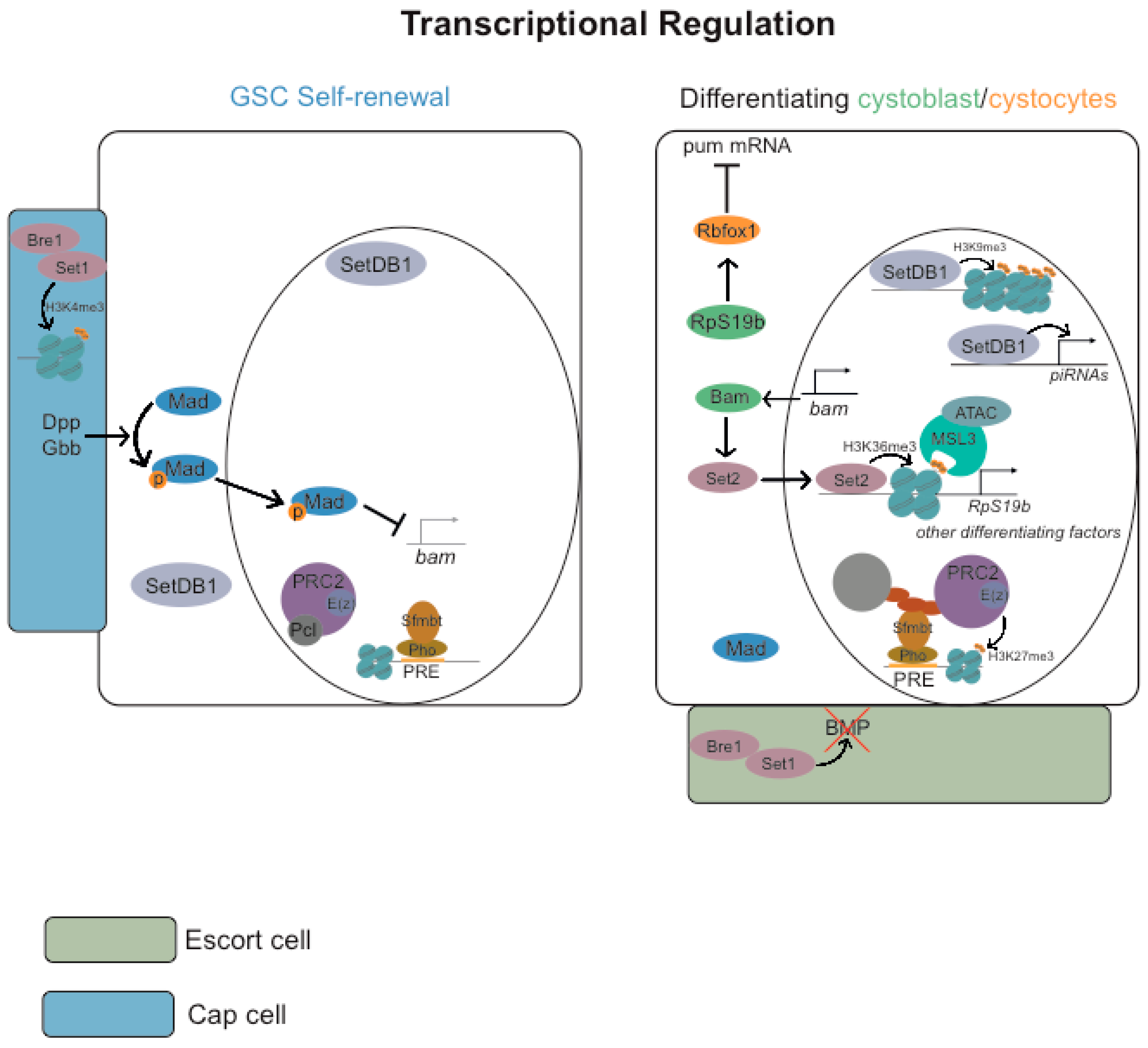
3. Germ Cells Differentiation: Regulation of Gene Expression
3.1. Post-Transcriptional Regulation
3.2. Transcriptional Regulation
3.2.1. Repressive Histone Marks (H3K9me3 and H3K27me3)
3.2.2. Active Histone Marks (H2B Monoubiquitylation H3K4me3, H3K36me3 and H3K79me3)
3.2.3. ATP-Dependent Chromatin-Remodeling
4. Drosophila and Mammalian Oogenesis
5. Final Remarks and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peterson, N.G.; Fox, D.T. Communal living: The role of polyploidy and syncytia in tissue biology. Chromosome Res. 2021, 29, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Spradling, A.C.; Niu, W.; Yin, Q.; Pathak, M.; Maurya, B. Conservation of oocyte development in germline cysts from Drosophila to mouse. eLife 2022, 11, e83230. [Google Scholar] [CrossRef] [PubMed]
- Spradling, A.C. Developmental genetics of oogenesis. In The Development of Drosophila Melanogaster; Bate, M., Martinez Arias, A., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993; Volume 1. [Google Scholar]
- Mahajan-Miklos, S.; Cooley, L. Intercellular cytoplasm transport during Drosophila oogenesis. Dev. Biol. 1994, 165, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.R.; St Johnston, D. The origin of asymmetry: Early polarisation of the Drosophila germline cyst and oocyte. Curr. Biol. 2004, 14, R438–R449. [Google Scholar] [CrossRef]
- Lei, L.; Spradling, A.C. Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 2016, 352, 95–99. [Google Scholar] [CrossRef]
- Niu, W.; Spradling, A.C. Mouse oocytes develop in cysts with the help of nurse cells. Cell 2022, 185, 2576–2590.e12. [Google Scholar] [CrossRef]
- Matova, N.; Cooley, L. Comparative aspects of animal oogenesis. Dev. Biol. 2001, 231, 291–320. [Google Scholar] [CrossRef]
- Buning, J. Germ cell cluster formation in insect ovaries. Int. J. Insect Morphol. Embryol. 1993, 22, 237–253. [Google Scholar] [CrossRef]
- Jezierska, M.; Miernik, A.; Sojka, J.; Student, S.; Sliwinska, M.A.; Gross, V.; Poprawa, I. Oogenesis in the tardigrade Hypsibius exemplaris Gasiorek, Stec, Morek & Michalczyk, 2018 (Eutardigrada, Hypsibiidae). Micron 2021, 150, 103126. [Google Scholar] [CrossRef]
- Kloc, M.; Bilinski, S.; Dougherty, M.T.; Brey, E.M.; Etkin, L.D. Formation, architecture and polarity of female germline cyst in Xenopus. Dev. Biol. 2004, 266, 43–61. [Google Scholar] [CrossRef]
- Nakamura, S.; Kobayashi, K.; Nishimura, T.; Tanaka, M. Ovarian germline stem cells in the teleost fish, medaka (Oryzias latipes). Int. J. Biol. Sci. 2011, 7, 403–409. [Google Scholar] [CrossRef]
- Bertho, S.; Clapp, M.; Banisch, T.U.; Bandemer, J.; Raz, E.; Marlow, F.L. Zebrafish dazl regulates cystogenesis and germline stem cell specification during the primordial germ cell to germline stem cell transition. Development 2021, 148, dev187773. [Google Scholar] [CrossRef]
- Hughes, S.E.; Miller, D.E.; Miller, A.L.; Hawley, R.S. Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila melanogaster. Genetics 2018, 208, 875–908. [Google Scholar] [CrossRef]
- Hinnant, T.D.; Merkle, J.A.; Ables, E.T. Coordinating Proliferation, Polarity, and Cell Fate in the Drosophila Female Germline. Front. Cell Dev. Biol. 2020, 8, 19. [Google Scholar] [CrossRef]
- Kahney, E.W.; Snedeker, J.C.; Chen, X. Regulation of Drosophila germline stem cells. Curr. Opin. Cell Biol. 2019, 60, 27–35. [Google Scholar] [CrossRef]
- Xie, T.; Spradling, A.C. Decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 1998, 94, 251–260. [Google Scholar] [CrossRef]
- Song, X.; Wong, M.D.; Kawase, E.; Xi, R.; Ding, B.C.; McCarthy, J.J.; Xie, T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 2004, 131, 1353–1364. [Google Scholar] [CrossRef]
- Wilcockson, S.G.; Ashe, H.L. Drosophila Ovarian Germline Stem Cell Cytocensor Projections Dynamically Receive and Attenuate BMP Signaling. Dev. Cell 2019, 50, 296–312.e5. [Google Scholar] [CrossRef]
- Chen, D.; McKearin, D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 2003, 13, 1786–1791. [Google Scholar] [CrossRef]
- Morris, L.X.; Spradling, A.C. Steroid signaling within Drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PLoS ONE 2012, 7, e46109. [Google Scholar] [CrossRef]
- Eliazer, S.; Palacios, V.; Wang, Z.; Kollipara, R.K.; Kittler, R.; Buszczak, M. Lsd1 restricts the number of germline stem cells by regulating multiple targets in escort cells. PLoS Genet. 2014, 10, e1004200. [Google Scholar] [CrossRef] [PubMed]
- Konig, A.; Shcherbata, H.R. Soma influences GSC progeny differentiation via the cell adhesion-mediated steroid-let-7-Wingless signaling cascade that regulates chromatin dynamics. Biol. Open 2015, 4, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhong, G.; Chai, P.C.; Luo, L.; Liu, S.; Yang, Y.; Baeg, G.H.; Cai, Y. Coordinated niche-associated signals promote germline homeostasis in the Drosophila ovary. J. Cell Biol. 2015, 211, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Wang, S.; Gao, Y.; Mao, Y.; Yang, Z.; Liu, L.; Song, X.; Ni, J.; Xie, T. COP9-Hedgehog axis regulates the function of the germline stem cell progeny differentiation niche in the Drosophila ovary. Development 2015, 142, 4242–4252. [Google Scholar] [CrossRef]
- Mottier-Pavie, V.I.; Palacios, V.; Eliazer, S.; Scoggin, S.; Buszczak, M. The Wnt pathway limits BMP signaling outside of the germline stem cell niche in Drosophila ovaries. Dev. Biol. 2016, 417, 50–62. [Google Scholar] [CrossRef]
- Su, Y.H.; Rastegri, E.; Kao, S.H.; Lai, C.M.; Lin, K.Y.; Liao, H.Y.; Wang, M.H.; Hsu, H.J. Diet regulates membrane extension and survival of niche escort cells for germline homeostasis via insulin signaling. Development 2018, 145, dev159186. [Google Scholar] [CrossRef]
- Wang, X.; Page-McCaw, A. Wnt6 maintains anterior escort cells as an integral component of the germline stem cell niche. Development 2018, 145, dev158527. [Google Scholar] [CrossRef]
- Morris, L.X.; Spradling, A.C. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development 2011, 138, 2207–2215. [Google Scholar] [CrossRef]
- Hsu, H.J.; LaFever, L.; Drummond-Barbosa, D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 2008, 313, 700–712. [Google Scholar] [CrossRef]
- Ables, E.T.; Drummond-Barbosa, D. Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development 2013, 140, 530–540. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, H. The division of Drosophila germline stem cells and their precursors requires a specific cyclin. Curr. Biol. 2005, 15, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, I.; Lilly, M.A. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev. Cell 2006, 10, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Lilly, M.A.; de Cuevas, M.; Spradling, A.C. Cyclin A associates with the fusome during germline cyst formation in the Drosophila ovary. Dev. Biol. 2000, 218, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.; Tan, C. Germline cyst formation and incomplete cytokinesis during Drosophila melanogaster oogenesis. Dev. Biol. 2010, 337, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.N.; Cooley, L. Stable intercellular bridges in development: The cytoskeleton lining the tunnel. Trends Cell Biol. 1996, 6, 474–479. [Google Scholar] [CrossRef]
- Carpenter, A.T. Egalitarian and the choice of cell fates in Drosophila melanogaster oogenesis. Ciba Found. Symp. 1994, 182, 223–246; discussion 246–254. [Google Scholar]
- Huynh, J.R.; St Johnston, D. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development 2000, 127, 2785–2794. [Google Scholar] [CrossRef]
- Huynh, J.R. Fusome as a Cell-Cell Communication Channel of Drosophila Ovarian Cyst. In Cell-Cell Channels; Baluska, F., Volkmann, D., Barlow, P.W., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar]
- de Cuevas, M.; Spradling, A.C. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 1998, 125, 2781–2789. [Google Scholar] [CrossRef]
- Lin, H.; Yue, L.; Spradling, A.C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 1994, 120, 947–956. [Google Scholar] [CrossRef]
- Lin, H.; Spradling, A.C. Fusome asymmetry and oocyte determination in Drosophila. Dev. Genet. 1995, 16, 6–12. [Google Scholar] [CrossRef]
- Nashchekin, D.; Busby, L.; Jakobs, M.; Squires, I.; St Johnston, D. Symmetry breaking in the female germline cyst. Science 2021, 374, 874–879. [Google Scholar] [CrossRef]
- Röper, K.; Brown, N.H. A Spectraplakin Is Enriched on the Fusome and Organizes Microtubules during Oocyte Specification in Drosophila. Curr. Biol. 2004, 14, 99–110. [Google Scholar] [CrossRef]
- Roper, K. Rtnl1 is enriched in a specialized germline ER that associates with ribonucleoprotein granule components. J. Cell Sci. 2007, 120 Pt 6, 1081–1092. [Google Scholar] [CrossRef]
- Snapp, E.L.; Iida, T.; Frescas, D.; Lippincott-Schwartz, J.; Lilly, M.A. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol. Biol. Cell 2004, 15, 4512–4521. [Google Scholar] [CrossRef]
- Lighthouse, D.V.; Buszczak, M.; Spradling, A.C. New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev. Biol. 2008, 317, 59–71. [Google Scholar] [CrossRef]
- Pielage, J.; Bulat, V.; Zuchero, J.B.; Fetter, R.D.; Davis, G.W. Hts/Adducin controls synaptic elaboration and elimination. Neuron 2011, 69, 1114–1131. [Google Scholar] [CrossRef]
- Yue, L.; Spradling, A.C. hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev. 1992, 6, 2443–2454. [Google Scholar] [CrossRef]
- Petrella, L.N.; Smith-Leiker, T.; Cooley, L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development 2007, 134, 703–712. [Google Scholar] [CrossRef]
- Huynh, J.R.; Shulman, J.M.; Benton, R.; St Johnston, D. PAR-1 is required for the maintenance of oocyte fate in Drosophila. Development 2001, 128, 1201–1209. [Google Scholar] [CrossRef]
- Bolivar, J.; Huynh, J.R.; Lopez-Schier, H.; Gonzalez, C.; St Johnston, D.; Gonzalez-Reyes, A. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development 2001, 128, 1889–1897. [Google Scholar] [CrossRef]
- Cox, R.T.; Spradling, A.C. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 2003, 130, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Wharton, R.P.; Struhl, G. Structure of the Drosophila BicaudalD protein and its role in localizing the posterior determinant nanos. Cell 1989, 59, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Suter, B.; Romberg, L.M.; Steward, R. Bicaudal-D, a Drosophila gene involved in developmental asymmetry: Localized transcript accumulation in ovaries and sequence similarity to myosin heavy chain tail domains. Genes Dev. 1989, 3, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Mach, J.M.; Lehmann, R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 1997, 11, 423–435. [Google Scholar] [CrossRef]
- Navarro, C.; Puthalakath, H.; Adams, J.M.; Strasser, A.; Lehmann, R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 2004, 6, 427–435. [Google Scholar] [CrossRef]
- Ephrussi, A.; Dickinson, L.K.; Lehmann, R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991, 66, 37–50. [Google Scholar] [CrossRef]
- Lantz, V.; Chang, J.S.; Horabin, J.I.; Bopp, D.; Schedl, P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 1994, 8, 598–613. [Google Scholar] [CrossRef]
- Wong, L.C.; Schedl, P. Cup blocks the precocious activation of the orb autoregulatory loop. PLoS ONE 2011, 6, e28261. [Google Scholar] [CrossRef]
- Christerson, L.B.; McKearin, D.M. orb is required for anteroposterior and dorsoventral patterning during Drosophila oogenesis. Genes Dev. 1994, 8, 614–628. [Google Scholar] [CrossRef]
- Chang, J.S.; Tan, L.; Schedl, P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev. Biol. 1999, 215, 91–106. [Google Scholar] [CrossRef]
- Chang, J.S.; Tan, L.; Wolf, M.R.; Schedl, P. Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 2001, 128, 3169–3177. [Google Scholar] [CrossRef]
- Castagnetti, S.; Ephrussi, A. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development 2003, 130, 835–843. [Google Scholar] [CrossRef]
- Tan, L.; Chang, J.S.; Costa, A.; Schedl, P. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development 2001, 128, 1159–1169. [Google Scholar] [CrossRef]
- Barr, J.; Gilmutdinov, R.; Wang, L.; Shidlovskii, Y.; Schedl, P. The Drosophila CPEB Protein Orb Specifies Oocyte Fate by a 3’UTR-Dependent Autoregulatory Loop. Genetics 2019, 213, 1431–1446. [Google Scholar] [CrossRef]
- Rubin, T.; Christophorou, N.; Huynh, J.R. How to pre-pair chromosomes for meiosis. Cell Cycle 2016, 15, 609–610. [Google Scholar] [CrossRef]
- Christophorou, N.; Rubin, T.; Huynh, J.R. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 2013, 9, e1004012. [Google Scholar] [CrossRef]
- Rust, K.; Byrnes, L.E.; Yu, K.S.; Park, J.S.; Sneddon, J.B.; Tward, A.D.; Nystul, T.G. A single-cell atlas and lineage analysis of the adult Drosophila ovary. Nat. Commun. 2020, 11, 5628. [Google Scholar] [CrossRef]
- Slaidina, M.; Lehmann, R. Translational control in germline stem cell development. J. Cell Biol. 2014, 207, 13–21. [Google Scholar] [CrossRef]
- Lin, H.; Spradling, A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 1997, 124, 2463–2476. [Google Scholar] [CrossRef]
- Forbes, A.; Lehmann, R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 1998, 125, 679–690. [Google Scholar] [CrossRef]
- Gilboa, L.; Lehmann, R. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 2004, 14, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Joly, W.; Chartier, A.; Rojas-Rios, P.; Busseau, I.; Simonelig, M. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 2013, 1, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Neumuller, R.A.; Betschinger, J.; Fischer, A.; Bushati, N.; Poernbacher, I.; Mechtler, K.; Cohen, S.M.; Knoblich, J.A. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 2008, 454, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.E.; Pargett, M.; Sutcliffe, C.; Umulis, D.; Ashe, H.L. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell 2011, 20, 72–83. [Google Scholar] [CrossRef]
- Li, Y.; Minor, N.T.; Park, J.K.; McKearin, D.M.; Maines, J.Z. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. USA 2009, 106, 9304–9309. [Google Scholar] [CrossRef]
- Chau, J.; Kulnane, L.S.; Salz, H.K. Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 9465–9470. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Carreira-Rosario, A.; Maines, J.Z.; McKearin, D.M.; Buszczak, M. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS ONE 2013, 8, e58301. [Google Scholar] [CrossRef]
- Carreira-Rosario, A.; Bhargava, V.; Hillebrand, J.; Kollipara, R.K.; Ramaswami, M.; Buszczak, M. Repression of Pumilio Protein Expression by Rbfox1 Promotes Germ Cell Differentiation. Dev. Cell 2016, 36, 562–571. [Google Scholar] [CrossRef]
- Navarro-Costa, P.; McCarthy, A.; Prudencio, P.; Greer, C.; Guilgur, L.G.; Becker, J.D.; Secombe, J.; Rangan, P.; Martinho, R.G. Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling. Nat. Commun. 2016, 7, 12331. [Google Scholar] [CrossRef]
- Vidaurre, V.; Chen, X. Epigenetic regulation of Drosophila germline stem cell maintenance and differentiation. Dev. Biol. 2021, 473, 105–118. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Kassis, J.A.; Kennison, J.A.; Tamkun, J.W. Polycomb and Trithorax Group Genes in Drosophila. Genetics 2017, 206, 1699–1725. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833. [Google Scholar] [CrossRef]
- Li, X.Y.; Harrison, M.M.; Villalta, J.E.; Kaplan, T.; Eisen, M.B. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 2014, 3, e03737. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef]
- DeLuca, S.Z.; Ghildiyal, M.; Pang, L.Y.; Spradling, A.C. Differentiating Drosophila female germ cells initiate Polycomb silencing by regulating PRC2-interacting proteins. eLife 2020, 9, e56922. [Google Scholar] [CrossRef] [PubMed]
- Klymenko, T.; Papp, B.; Fischle, W.; Kocher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Muller, J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef]
- Alfieri, C.; Gambetta, M.C.; Matos, R.; Glatt, S.; Sehr, P.; Fraterman, S.; Wilm, M.; Muller, J.; Muller, C.W. Structural basis for targeting the chromatin repressor Sfmbt to Polycomb response elements. Genes Dev. 2013, 27, 2367–2379. [Google Scholar] [CrossRef]
- Kang, H.; McElroy, K.A.; Jung, Y.L.; Alekseyenko, A.A.; Zee, B.M.; Park, P.J.; Kuroda, M.I. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev. 2015, 29, 1136–1150. [Google Scholar] [CrossRef]
- Frey, F.; Sheahan, T.; Finkl, K.; Stoehr, G.; Mann, M.; Benda, C.; Muller, J. Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev. 2016, 30, 1116–1127. [Google Scholar] [CrossRef]
- Birve, A.; Sengupta, A.K.; Beuchle, D.; Larsson, J.; Kennison, J.A.; Rasmuson-Lestander, A.; Muller, J. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 2001, 128, 3371–3379. [Google Scholar] [CrossRef] [PubMed]
- Iovino, N.; Ciabrelli, F.; Cavalli, G. PRC2 controls Drosophila oocyte cell fate by repressing cell cycle genes. Dev. Cell 2013, 26, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Feijao, T.; Marques, B.; Silva, R.D.; Carvalho, C.; Sobral, D.; Matos, R.; Tan, T.; Pereira, A.; Morais-de-Sa, E.; Maiato, H.; et al. Polycomb group (PcG) proteins prevent the assembly of abnormal synaptonemal complex structures during meiosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2204701119. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Moon, W.; Wang, S.; Smith, K.; Hazelrigg, T. Histone methylation is required for oogenesis in Drosophila. Development 2007, 134, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Lee, K.S.; Park, J.S.; Yu, K.; Paik, S.G.; Kang, Y.K. dSETDB1 and SU(VAR)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS ONE 2008, 3, e2234. [Google Scholar] [CrossRef]
- Rangan, P.; Malone, C.D.; Navarro, C.; Newbold, S.P.; Hayes, P.S.; Sachidanandam, R.; Hannon, G.J.; Lehmann, R. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol. 2011, 21, 1373–1379. [Google Scholar] [CrossRef]
- Shokri, L.; Inukai, S.; Hafner, A.; Weinand, K.; Hens, K.; Vedenko, A.; Gisselbrecht, S.S.; Dainese, R.; Bischof, J.; Furger, E.; et al. A Comprehensive Drosophila melanogaster Transcription Factor Interactome. Cell Rep. 2019, 27, 955–970.e7. [Google Scholar] [CrossRef]
- Yi, X.; de Vries, H.I.; Siudeja, K.; Rana, A.; Lemstra, W.; Brunsting, J.F.; Kok, R.M.; Smulders, Y.M.; Schaefer, M.; Dijk, F.; et al. Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol. Biol. Cell 2009, 20, 983–994. [Google Scholar] [CrossRef]
- Maines, J.Z.; Park, J.K.; Williams, M.; McKearin, D.M. Stonewalling Drosophila stem cell differentiation by epigenetic controls. Development 2007, 134, 1471–1479. [Google Scholar] [CrossRef]
- Zinshteyn, D.; Barbash, D.A. Stonewall prevents expression of ectopic genes in the ovary and accumulates at insulator elements in D. melanogaster. PLoS Genet. 2022, 18, e1010110. [Google Scholar] [CrossRef]
- Wood, A.; Krogan, N.J.; Dover, J.; Schneider, J.; Heidt, J.; Boateng, M.A.; Dean, K.; Golshani, A.; Zhang, Y.; Greenblatt, J.F.; et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 2003, 11, 267–274. [Google Scholar] [CrossRef]
- Bray, S.; Musisi, H.; Bienz, M. Bre1 is required for Notch signaling and histone modification. Dev. Cell 2005, 8, 279–286. [Google Scholar] [CrossRef]
- Mohan, M.; Herz, H.M.; Smith, E.R.; Zhang, Y.; Jackson, J.; Washburn, M.P.; Florens, L.; Eissenberg, J.C.; Shilatifard, A. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 2011, 31, 4310–4318. [Google Scholar] [CrossRef]
- Xuan, T.; Xin, T.; He, J.; Tan, J.; Gao, Y.; Feng, S.; He, L.; Zhao, G.; Li, M. dBre1/dSet1-dependent pathway for histone H3K4 trimethylation has essential roles in controlling germline stem cell maintenance and germ cell differentiation in the Drosophila ovary. Dev. Biol. 2013, 379, 167–181. [Google Scholar] [CrossRef]
- Prudencio, P.; Guilgur, L.G.; Sobral, J.; Becker, J.D.; Martinho, R.G.; Navarro-Costa, P. The Trithorax group protein dMLL3/4 instructs the assembly of the zygotic genome at fertilization. EMBO Rep. 2018, 19, e45728. [Google Scholar] [CrossRef]
- Navarro-Costa, P.; Martinho, R.G. The emerging role of transcriptional regulation in the oocyte-to-zygote transition. PLoS Genet. 2020, 16, e1008602. [Google Scholar] [CrossRef]
- Mukai, M.; Hira, S.; Nakamura, K.; Nakamura, S.; Kimura, H.; Sato, M.; Kobayashi, S. H3K36 Trimethylation-Mediated Epigenetic Regulation is Activated by Bam and Promotes Germ Cell Differentiation During Early Oogenesis in Drosophila. Biol. Open 2015, 4, 119–124. [Google Scholar] [CrossRef]
- McCarthy, A.; Sarkar, K.; Martin, E.T.; Upadhyay, M.; Jang, S.; Williams, N.D.; Forni, P.E.; Buszczak, M.; Rangan, P. Msl3 promotes germline stem cell differentiation in female Drosophila. Development 2022, 149, dev199625. [Google Scholar] [CrossRef]
- Stabell, M.; Larsson, J.; Aalen, R.B.; Lambertsson, A. Drosophila dSet2 functions in H3-K36 methylation and is required for development. Biochem. Biophys. Res. Commun. 2007, 359, 784–789. [Google Scholar] [CrossRef]
- Larschan, E.; Alekseyenko, A.A.; Gortchakov, A.A.; Peng, S.; Li, B.; Yang, P.; Workman, J.L.; Park, P.J.; Kuroda, M.I. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell 2007, 28, 121–133. [Google Scholar] [CrossRef]
- Drelon, C.; Belalcazar, H.M.; Secombe, J. The Histone Demethylase KDM5 Is Essential for Larval Growth in Drosophila. Genetics 2018, 209, 773–787. [Google Scholar] [CrossRef] [PubMed]
- Drelon, C.; Rogers, M.F.; Belalcazar, H.M.; Secombe, J. The histone demethylase KDM5 controls developmental timing in Drosophila by promoting prothoracic gland endocycles. Development 2019, 146, dev182568. [Google Scholar] [CrossRef] [PubMed]
- Lloret-Llinares, M.; Perez-Lluch, S.; Rossell, D.; Moran, T.; Ponsa-Cobas, J.; Auer, H.; Corominas, M.; Azorin, F. dKDM5/LID regulates H3K4me3 dynamics at the transcription-start site (TSS) of actively transcribed developmental genes. Nucleic Acids Res. 2012, 40, 9493–9505. [Google Scholar] [CrossRef] [PubMed]
- Zhaunova, L.; Ohkura, H.; Breuer, M. Kdm5/Lid Regulates Chromosome Architecture in Meiotic Prophase I Independently of Its Histone Demethylase Activity. PLoS Genet. 2016, 12, e1006241. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Secombe, J. The Histone Demethylase KDM5 Activates Gene Expression by Recognizing Chromatin Context through Its PHD Reader Motif. Cell Rep. 2015, 13, 2219–2231. [Google Scholar] [CrossRef]
- Liu, X.; Greer, C.; Secombe, J. KDM5 interacts with Foxo to modulate cellular levels of oxidative stress. PLoS Genet. 2014, 10, e1004676. [Google Scholar] [CrossRef]
- Gajan, A.; Barnes, V.L.; Liu, M.; Saha, N.; Pile, L.A. The histone demethylase dKDM5/LID interacts with the SIN3 histone deacetylase complex and shares functional similarities with SIN3. Epigenet. Chromatin 2016, 9, 4. [Google Scholar] [CrossRef]
- Torres-Campana, D.; Kimura, S.; Orsi, G.A.; Horard, B.; Benoit, G.; Loppin, B. The Lid/KDM5 histone demethylase complex activates a critical effector of the oocyte-to-zygote transition. PLoS Genet. 2020, 16, e1008543. [Google Scholar] [CrossRef]
- Nieken, K.J.; O’Brien, K.; McDonnell, A.; Zhaunova, L.; Ohkura, H. A large-scale RNAi screen reveals that mitochondrial function is important for meiotic chromosome organization in oocytes. Chromosoma 2023, 132, 1–18. [Google Scholar] [CrossRef]
- Xi, R.; Xie, T. Stem cell self-renewal controlled by chromatin remodeling factors. Science 2005, 310, 1487–1489. [Google Scholar] [CrossRef]
- Buszczak, M.; Paterno, S.; Spradling, A.C. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 2009, 323, 248–251. [Google Scholar] [CrossRef]
- Fuchs, G.; Hollander, D.; Voichek, Y.; Ast, G.; Oren, M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014, 24, 1572–1583. [Google Scholar] [CrossRef]
- Neumuller, R.A.; Wirtz-Peitz, F.; Lee, S.; Kwon, Y.; Buckner, M.; Hoskins, R.A.; Venken, K.J.; Bellen, H.J.; Mohr, S.E.; Perrimon, N. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics 2012, 190, 931–940. [Google Scholar] [CrossRef]
- Ardehali, M.B.; Yao, J.; Adelman, K.; Fuda, N.J.; Petesch, S.J.; Webb, W.W.; Lis, J.T. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009, 28, 1067–1077. [Google Scholar] [CrossRef]
- He, J.; Xuan, T.; Xin, T.; An, H.; Wang, J.; Zhao, G.; Li, M. Evidence for chromatin-remodeling complex PBAP-controlled maintenance of the Drosophila ovarian germline stem cells. PLoS ONE 2014, 9, e103473. [Google Scholar] [CrossRef]
- Mondragon, A.A.; Yalonetskaya, A.; Ortega, A.J.; Zhang, Y.; Naranjo, O.; Elguero, J.; Chung, W.S.; McCall, K. Lysosomal Machinery Drives Extracellular Acidification to Direct Non-apoptotic Cell Death. Cell Rep. 2019, 27, 11–19.e3. [Google Scholar] [CrossRef]
- Seki, Y.; Yamaji, M.; Yabuta, Y.; Sano, M.; Shigeta, M.; Matsui, Y.; Saga, Y.; Tachibana, M.; Shinkai, Y.; Saitou, M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 2007, 134, 2627–2638. [Google Scholar] [CrossRef]
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 2012, 48, 849–862. [Google Scholar] [CrossRef]
- Seki, Y.; Hayashi, K.; Itoh, K.; Mizugaki, M.; Saitou, M.; Matsui, Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005, 278, 440–458. [Google Scholar] [CrossRef]
- Staubli, A.; Peters, A.H. Mechanisms of maternal intergenerational epigenetic inheritance. Curr. Opin. Genet. Dev. 2021, 67, 151–162. [Google Scholar] [CrossRef]
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017, 547, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yin, Q.; Inoue, A.; Zhang, C.; Zhang, Y. Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci. Adv. 2019, 5, eaay7246. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.; Kozuka, C.; Hayashi, R.; Kumon, M.; Koseki, H.; Inoue, A. H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat. Genet. 2021, 53, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Zenk, F.; Loeser, E.; Schiavo, R.; Kilpert, F.; Bogdanovic, O.; Iovino, N. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 2017, 357, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Samata, M.; Alexiadis, A.; Richard, G.; Georgiev, P.; Nuebler, J.; Kulkarni, T.; Renschler, G.; Basilicata, M.F.; Zenk, F.L.; Shvedunova, M.; et al. Intergenerationally Maintained Histone H4 Lysine 16 Acetylation Is Instructive for Future Gene Activation. Cell 2020, 182, 127–144.e23. [Google Scholar] [CrossRef]
- Lyko, F.; Ramsahoye, B.H.; Jaenisch, R. DNA methylation in Drosophila melanogaster. Nature 2000, 408, 538–540. [Google Scholar] [CrossRef]
- Church, S.H.; de Medeiros, B.A.S.; Donoughe, S.; Márquez Reyes, N.L.; Extavour, C.G. Repeated loss of variation in insect ovary morphology highlights the role of development in life-history evolution. Proc. R. Soc. B Biol. Sci. 2021, 288, 20210150. [Google Scholar] [CrossRef]
- Munro, C.; Cadis, H.; Pagnotta, S.; Houliston, E.; Huynh, J.R. Conserved meiotic mechanisms in the cnidarian Clytia hemisphaerica revealed by Spo11 knockout. Sci. Adv. 2023, 9, eadd2873. [Google Scholar] [CrossRef]
- Miramon-Puertolas, P.; Steinmetz, P.R.H. An adult stem-like cell population generates germline and neurons in the sea anemone Nematostella vectensis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Saitou, M.; Hayashi, K. Mammalian in vitro gametogenesis. Science 2021, 374, eaaz6830. [Google Scholar] [CrossRef]
- Aizawa, E.; Ozonov, E.A.; Kawamura, Y.K.; Dumeau, C.-E.; Nagaoka, S.; Saitou, M.; Peters, A.H.F.M.; Wutz, A. Comprehensive comparison of female germ cell development in vitro and in vivo identifies epigenetic gene regulation crucial for oocyte development and embryonic competence. bioRxiv 2023. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabrita, B.; Martinho, R.G. Genetic and Epigenetic Regulation of Drosophila Oocyte Determination. J. Dev. Biol. 2023, 11, 21. https://doi.org/10.3390/jdb11020021
Cabrita B, Martinho RG. Genetic and Epigenetic Regulation of Drosophila Oocyte Determination. Journal of Developmental Biology. 2023; 11(2):21. https://doi.org/10.3390/jdb11020021
Chicago/Turabian StyleCabrita, Brigite, and Rui Gonçalo Martinho. 2023. "Genetic and Epigenetic Regulation of Drosophila Oocyte Determination" Journal of Developmental Biology 11, no. 2: 21. https://doi.org/10.3390/jdb11020021
APA StyleCabrita, B., & Martinho, R. G. (2023). Genetic and Epigenetic Regulation of Drosophila Oocyte Determination. Journal of Developmental Biology, 11(2), 21. https://doi.org/10.3390/jdb11020021






