Seeking Sense in the Hox Gene Cluster
Abstract
1. Introduction
2. Seeking Sense in the Hox Gene Cluster
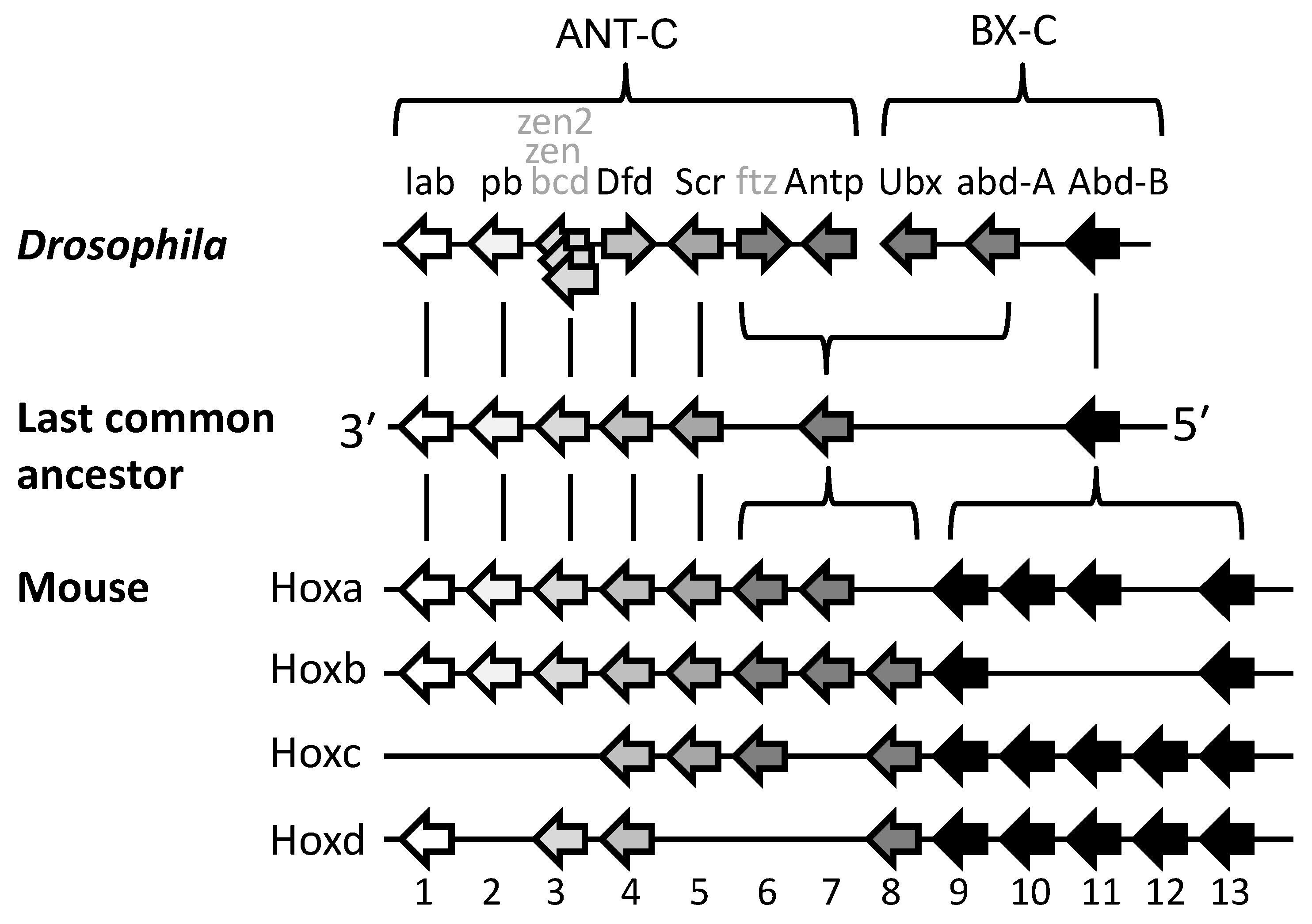
2.1. Seeking Sense in the Evolutionary Origin of Hox Clustering and Transcriptional Direction
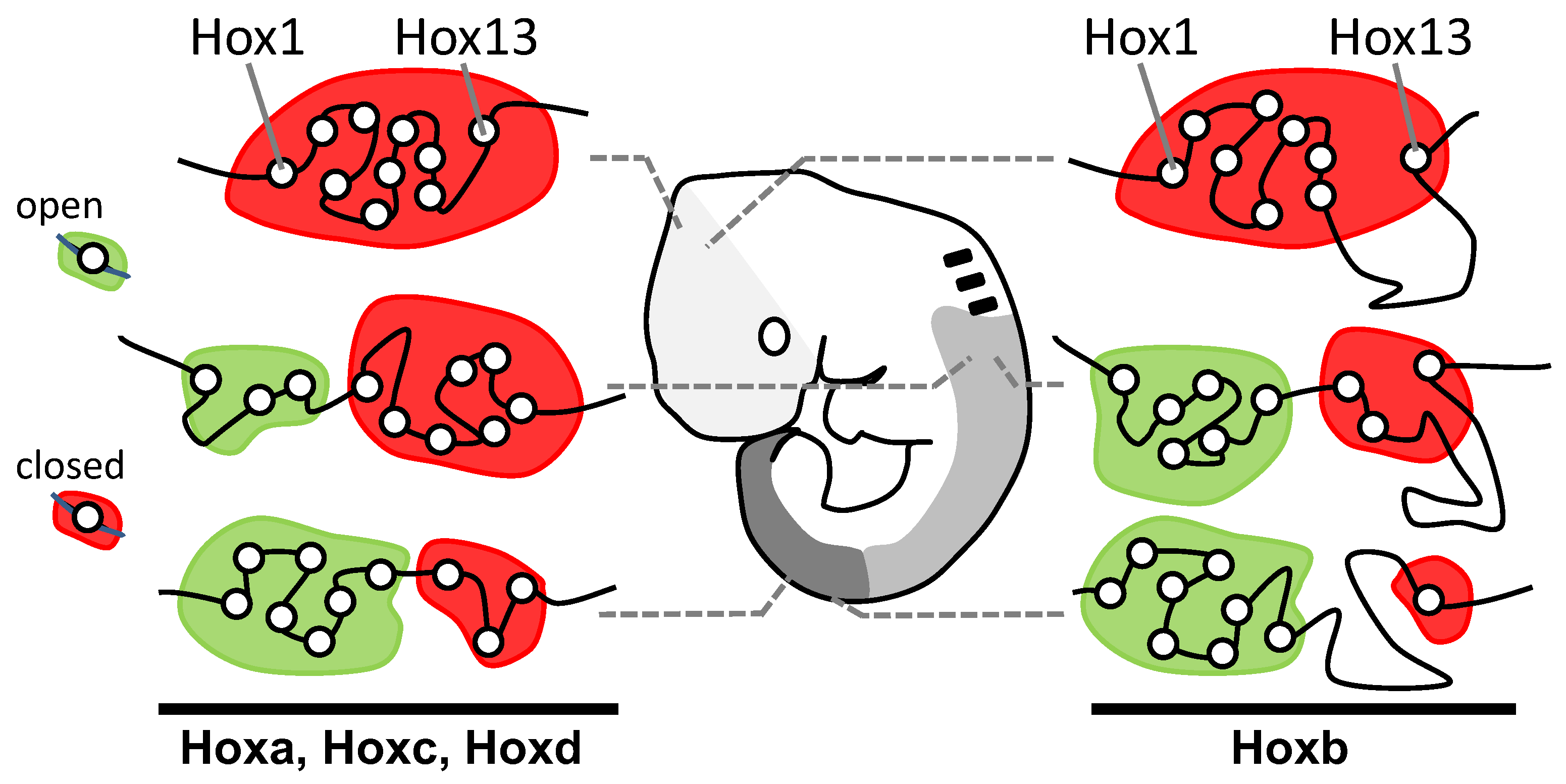
2.2. Seeking Sense in the Maintenance and Compactness of Clusters
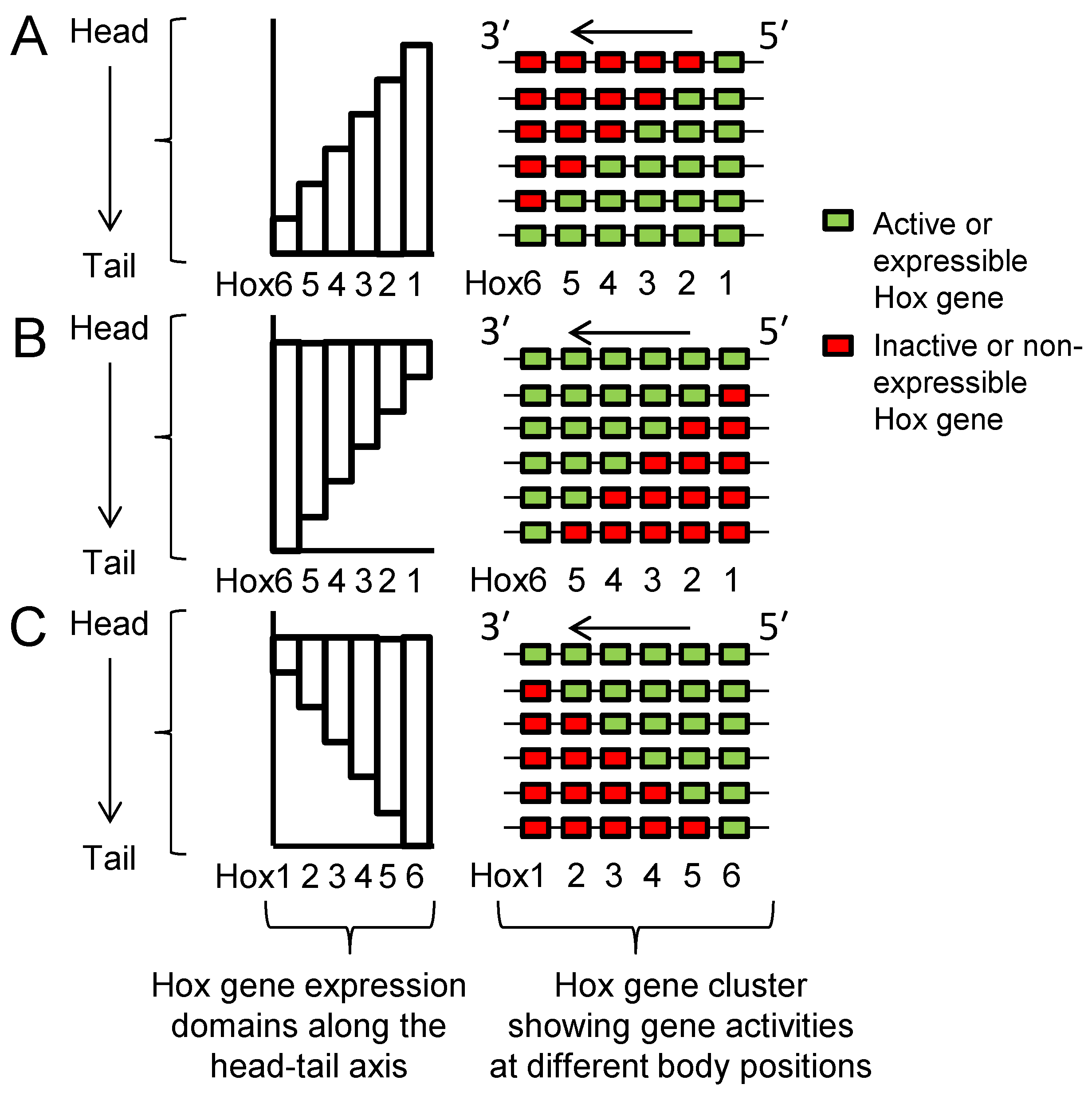
2.3. Seeking Sense in Spatial and Temporal Collinearities
2.4. Seeking Sense in the Partially Overlapping Nature of Hox Gene Expression Patterns
2.5. Seeking Sense in the Head–Tail Polarity of Hox Expression Domains
2.6. Seeking Sense in the Duplication of Hox Genes and Clusters
2.7. Seeking Sense in the Phenotype of Hox Gene Knockouts
2.8. Seeking Sense in the Evolution of Axial Morphologies by Shifts in Hox Expression

2.9. Seeking Sense in the Hox Clustering of Pre-Bilaterians
3. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boncinelli, E.; Somma, R.; Acampora, D.; Pannese, M.; D’Esposito, M.; Faiella, A.; Simeone, A. Organization of human homeobox genes. Hum. Reprod. 1988, 3, 880–886. [Google Scholar] [CrossRef]
- Duboule, D.; Dolle, P. The structural and functional organization of the murine hox gene family resembles that of drosophila homeotic genes. EMBO J. 1989, 8, 1497–1505. [Google Scholar] [CrossRef]
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378. [Google Scholar] [PubMed]
- Balavoine, G.; de Rosa, R.; Adoutte, A. Hox clusters and bilaterian phylogeny. Mol. Phylogenet. Evol. 2002, 24, 366–373. [Google Scholar] [PubMed]
- Gaunt, S.J. Hox cluster genes and collinearities throughout the tree of animal life. Int. J. Dev. Biol. 2018, 62, 673–683. [Google Scholar] [CrossRef]
- Gaunt, S.J. Made in the Image of a Fly; Amazon Books and Ebooks, 2019. [Google Scholar]
- Lewis, E.B. A gene complex controlling segmentation in drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, S.J. The significance of hox gene collinearity. Int. J. Dev. Biol. 2015, 59, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Soshnikova, N.; Duboule, D. Epigenetic temporal control of mouse hox genes in vivo. Science 2009, 324, 1320–1323. [Google Scholar]
- Noordermeer, D.; Leleu, M.; Splinter, E.; Rougemont, J.; De Laat, W.; Duboule, D. The dynamic architecture of hox gene clusters. Science 2011, 334, 222–225. [Google Scholar]
- Bowman, S.K.; Deaton, A.M.; Domingues, H.; Wang, P.I.; Sadreyev, R.I.; Kingston, R.E.; Bender, W. H3k27 modifications define segmental regulatory domains in the drosophila bithorax complex. eLife 2014, 3, e02833. [Google Scholar]
- Akam, M.; Dawson, I.; Tear, G. Homeotic genes and the control of segment diversity. Dev. Suppl. 1988, 104, 123–133. [Google Scholar] [CrossRef]
- Maeda, R.K.; Karch, F. The open for business model of the bithorax complex in drosophila. Chromosoma 2015, 124, 293–307. [Google Scholar]
- Izpisua-Belmonte, J.C.; Falkenstein, H.; Dolle, P.; Renucci, A.; Duboule, D. Murine genes related to the drosophila abdb homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991, 10, 2279–2289. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Anaya, J.; Adachi, N.; Alvarez, S.; Kuratani, S.; D’Aniello, S.; Garcia-Fernandez, J. Broken colinearity of the amphioxus hox cluster. EvoDevo 2012, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Frobius, A.C.; Matus, D.Q.; Seaver, E.C. Genomic organization and expression demonstrate spatial and temporal hox gene colinearity in the lophotrochozoan capitella sp. I. PLoS ONE 2008, 3, e4004. [Google Scholar]
- Serano, J.M.; Martin, A.; Liubicich, D.M.; Jarvis, E.; Bruce, H.S.; La, K.; Browne, W.E.; Grimwood, J.; Patel, N.H. Comprehensive analysis of hox gene expression in the amphipod crustacean parhyale hawaiensis. Dev. Biol. 2016, 409, 297–309. [Google Scholar]
- Aboobaker, A.; Blaxter, M. Hox gene evolution in nematodes: Novelty conserved. Curr. Opin. Genet. Dev. 2003, 13, 593–598. [Google Scholar]
- Seo, H.C.; Edvardsen, R.B.; Maeland, A.D.; Bjordal, M.; Jensen, M.F.; Hansen, A.; Flaat, M.; Weissenbach, J.; Lehrach, H.; Wincker, P.; et al. Hox cluster disintegration with persistent anteroposterior order of expression in oikopleura dioica. Nature 2004, 431, 67–71. [Google Scholar] [CrossRef]
- Gehring, W.J.; Kloter, U.; Suga, H. Evolution of the hox gene complex from an evolutionary ground state. Curr. Top. Dev. Biol. 2009, 88, 35–61. [Google Scholar]
- Lewis, E.B. The bithorax complex: The first fifty years. Int. J. Dev. Biol. 1998, 42, 403–415. [Google Scholar]
- Tarailo-Graovac, M.; Chen, N. Gene Clustering in Eukaryotes. In Els; John Wiley Sons Ltd.: Chichester, UK, 2013. [Google Scholar]
- Gaunt, S.J.; Gaunt, A.L. Possible rules for the ancestral origin of hox gene collinearity. J. Theor. Biol. 2016, 410, 1–8. [Google Scholar]
- Holland, P.W.H. Evolution of homeobox genes. Wires Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Hui, J.H.L.; McDougall, C.; Monteiro, A.S.; Holland, P.W.H.; Arendt, D.; Balavoine, G.; Ferrier, D.E.K. Extensive chordate and annelid macrosynteny reveals ancestral homeobox gene organization. Mol. Biol. Evol. 2012, 29, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Pollard, S.L.; Holland, P.W.H. Evidence for 14 homeobox gene clusters in human genome ancestry. Curr. Biol. 2000, 10, 1059–1062. [Google Scholar]
- Garcia-Fernandez, J. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 2005, 6, 881–892. [Google Scholar]
- Ferrier, D.E. The origin of the hox/parahox genes, the ghost locus hypothesis and the complexity of the first animal. Brief. Funct. Genom. 2016, 15, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Derelle, R.; Lopez, P.; Le Guyader, H.; Manuel, M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol. Dev. 2007, 9, 212–219. [Google Scholar]
- Negre, B.; Ruiz, A. Hom-c evolution in drosophila: Is there a need for hox gene clustering? Trends Genet. TIG 2007, 23, 55–59. [Google Scholar]
- Shippy, T.D.; Hosmani, P.S.; Florez-Gonzalez, M.; Mueller, L.A.; Hunter, W.B.; Brown, S.J.; D’Elia, T.; Saha, S. Annotation of hox cluster and hox cofactor genes in the asian citrus psyllid, diaphorina citri, reveals novel features. Gigabyte 2022. [Google Scholar] [CrossRef]
- Celniker, S.E.; Sharma, S.; Keelan, D.J.; Lewis, E.B. The molecular genetics of the bithorax complex of drosophila: Cis-regulation in the abdominal-b domain. EMBO J. 1990, 9, 4277–4286. [Google Scholar] [CrossRef]
- Gould, A.; Morrison, A.; Sproat, G.; White, R.A.; Krumlauf, R. Positive cross-regulation and enhancer sharing: Two mechanisms for specifying overlapping hox expression patterns. Genes Dev. 1997, 11, 900–913. [Google Scholar]
- Kwan, C.T.; Tsang, S.L.; Krumlauf, R.; Sham, M.H. Regulatory analysis of the mouse hoxb3 gene: Multiple elements work in concert to direct temporal and spatial patterns of expression. Dev. Biol. 2001, 232, 176–190. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Montavon, T.; Duboule, D. Chromatin organization and global regulation of hox gene clusters. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120367. [Google Scholar]
- Duboule, D. The rise and fall of hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [PubMed]
- Pace, R.M.; Grbic, M.; Nagy, L.M. Composition and genomic organization of arthropod hox clusters. EvoDevo 2016, 7, 11. [Google Scholar] [PubMed]
- Sun, D.A.; Patel, N.H. The amphipod crustacean parhyale hawaiensis: An emerging comparative model of arthropod development, evolution, and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e355. [Google Scholar]
- Dearden, P.K.; Wilson, M.J.; Sablan, L.; Osborne, P.W.; Havler, M.; McNaughton, E.; Kimura, K.; Milshina, N.V.; Hasselmann, M.; Gempe, T.; et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006, 16, 1376–1384. [Google Scholar] [CrossRef]
- Shippy, T.D.; Ronshaugen, M.; Cande, J.; He, J.; Beeman, R.W.; Levine, M.; Brown, S.J.; Denell, R.E. Analysis of the tribolium homeotic complex: Insights into mechanisms constraining insect hox clusters. Dev. Genes Evol. 2008, 218, 127–139. [Google Scholar] [CrossRef]
- Freeman, R.; Ikuta, T.; Wu, M.; Koyanagi, R.; Kawashima, T.; Tagawa, K.; Humphreys, T.; Fang, G.C.; Fujiyama, A.; Saiga, H.; et al. Identical genomic organization of two hemichordate hox clusters. Curr. Biol. CB 2012, 22, 2053–2058. [Google Scholar] [CrossRef]
- Arenas-Mena, C.; Cameron, A.R.; Davidson, E.H. Spatial expression of hox cluster genes in the ontogeny of a sea urchin. Development 2000, 127, 4631–4643. [Google Scholar]
- Jorgensen, E.M.; Garber, R.L. Function and misfunction of the two promoters of the drosophila antennapedia gene. Genes Dev. 1987, 1, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Schneuwly, S.; Kuroiwa, A.; Gehring, W.J. Molecular analysis of the dominant homeotic antennapedia phenotype. EMBO J. 1987, 6, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Darbellay, F.; Bochaton, C.; Lopez-Delisle, L.; Mascrez, B.; Tschopp, P.; Delpretti, S.; Zakany, J.; Duboule, D. The constrained architecture of mammalian hox gene clusters. Proc. Natl. Acad. Sci. USA 2019, 116, 13424–13433. [Google Scholar] [CrossRef] [PubMed]
- Duboule, D. Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994, 1994, 135–142. [Google Scholar] [CrossRef]
- Ferrier, D.E.; Holland, P.W. Ciona intestinalis parahox genes: Evolution of hox/parahox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenet. Evol. 2002, 24, 412–417. [Google Scholar]
- Gold, D.A.; Runnegar, B.; Gehling, J.G.; Jacobs, D.K. Ancestral state reconstruction of ontogeny supports a bilaterian affinity for dickinsonia. Evol. Dev. 2015, 17, 315–324. [Google Scholar] [CrossRef]
- Rekaik, H.; Lopez-Delisle, L.; Hintermann, A.; Mascrez, B.; Bochaton, C.; Duboule, D. Sequential and directional insulation by conserved ctcf sites underlies the hox timer in pseudo-embryos. bioRkiv 2022. [Google Scholar] [CrossRef]
- Amandio, A.R.; Beccari, L.; Lopez-Delisle, L.; Mascrez, B.; Zakany, J.; Gitto, S.; Duboule, D. Sequential in cis mutagenesis in vivo reveals various functions for ctcf sites at the mouse hoxd cluster. Genes Dev. 2021, 35, 1490–1509. [Google Scholar]
- Duboule, D. The (unusual) heuristic value of hox gene clusters; a matter of time? Dev. Biol. 2022, 484, 75–87. [Google Scholar] [CrossRef]
- Wei, M.; Qin, Z.; Kong, D.; Liu, D.; Zheng, Q.S.; Bai, S.; Zhang, Z.; Ma, Y. Echiuran hox genes provide new insights into the correspondence between hox subcluster organization and collinearity pattern. Proc. R. Soc. B 2022, 289, 20220705. [Google Scholar] [CrossRef]
- Aronowicz, J.; Lowe, C.J. Hox gene expression in the hemichordate saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Integr. Comp. Biol. 2006, 46, 890–901. [Google Scholar] [PubMed]
- Martin, A.; Serano, J.M.; Jarvis, E.; Bruce, H.S.; Wang, J.; Ray, S.; Barker, C.A.; O’Connell, L.C.; Patel, N.H. Crispr/cas9 mutagenesis reveals versatile roles of hox genes in crustacean limb specification and evolution. Curr. Biol. CB 2016, 26, 14–26. [Google Scholar] [CrossRef]
- Hautier, L.; Charles, C.; Asher, R.J.; Gaunt, S.J. Ossification sequence and genetic patterning in the mouse axial skeleton. J. Exp. Zool. Part B Mol. Dev. Evol. 2014, 322, 631–642. [Google Scholar]
- Rijli, F.M.; Mark, M.; Lakkaraju, S.; Dierich, A.; Dolle, P.; Chambon, P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of hoxa-2, which acts as a selector gene. Cell 1993, 75, 1333–1349. [Google Scholar]
- Gaunt, S.J. Expression patterns of mouse hox genes: Clues to an understanding of developmental and evolutionary strategies. BioEssays News Rev. Mol. Cell. Dev. Biol. 1991, 13, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, S.J.; Dean, W.; Sang, H.; Burton, R.D. Evidence that hoxa expression domains are evolutionarily transposed in spinal ganglia, and are established by forward spreading in paraxial mesoderm. Mech. Dev. 1999, 82, 109–118. [Google Scholar] [CrossRef]
- Duboule, D. Patterning in the vertebrate limb. Curr. Opin. Genet. Dev. 1991, 1, 211–216. [Google Scholar]
- Duboule, D.; Morata, G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. TIG 1994, 10, 358–364. [Google Scholar] [CrossRef]
- Mallo, M.; Alonso, C.R. The regulation of hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef]
- Paro, R. Imprinting a determined state into the chromatin of drosophila. Trends Genet. TIG 1990, 6, 416–421. [Google Scholar]
- Gonzalez-Reyes, A.; Morata, G. The developmental effect of overexpressing a ubx product in drosophila embryos is dependent on its interactions with other homeotic products. Cell 1990, 61, 515–522. [Google Scholar]
- Carapuco, M.; Novoa, A.; Bobola, N.; Mallo, M. Hox genes specify vertebral types in the presomitic mesoderm. Genes Dev. 2005, 19, 2116–2121. [Google Scholar] [CrossRef] [PubMed]
- Shiga, Y.; Sagawa, K.; Takai, R.; Sakaguchi, H.; Yamagata, H.; Hayashi, S. Transcriptional readthrough of hox genes ubx and antp and their divergent post-transcriptional control during crustacean evolution. Evol. Dev. 2006, 8, 407–414. [Google Scholar] [PubMed]
- Simeone, A.; Pannese, M.; Acampora, D.; D’Esposito, M.; Boncinelli, E. At least three human homeoboxes on chromosome 12 belong to the same transcription unit. Nucleic Acids Res. 1988, 16, 5379–5390. [Google Scholar] [CrossRef]
- Mainguy, G.; Koster, J.; Woltering, J.; Jansen, H.; Durston, A. Extensive polycistronism and antisense transcription in the mammalian hox clusters. PLoS ONE 2007, 2, e356. [Google Scholar] [CrossRef]
- Pascual-Anaya, J.; D’Aniello, S.; Kuratani, S.; Garcia-Fernandez, J. Evolution of hox gene clusters in deuterostomes. BMC Dev. Biol. 2013, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Mehta, T.K.; Ravi, V.; Yamasaki, S.; Lee, A.P.; Lian, M.M.; Tay, B.H.; Tohari, S.; Yanai, S.; Tay, A.; Brenner, S.; et al. Evidence for at least six hox clusters in the japanese lamprey (lethenteron japonicum). Proc. Natl. Acad. Sci. USA 2013, 110, 16044–16049. [Google Scholar] [CrossRef]
- Gaunt, S.J.; Krumlauf, R.; Duboule, D. Mouse homeo-genes within a subfamily, hox-1.4, -2.6 and -5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development 1989, 107, 131–141. [Google Scholar]
- Gaunt, S.J.; Sharpe, P.T.; Duboule, D. Spatially restricted domains of homeo-gene transcripts in mouse embryos—Relation to a segmented body plan. Dev. Suppl. 1988, 104, 169–179. [Google Scholar]
- Gaunt, S.J.; Coletta, P.L.; Pravtcheva, D.; Sharpe, P.T. Mouse hox-3.4: Homeobox sequence and embryonic expression patterns compared with other members of the hox gene network. Development 1990, 109, 329–339. [Google Scholar] [CrossRef]
- Geada, A.M.; Gaunt, S.J.; Azzawi, M.; Shimeld, S.M.; Pearce, J.; Sharpe, P.T. Sequence and embryonic expression of the murine hox-3.5 gene. Development 1992, 116, 497–506. [Google Scholar] [PubMed]
- Izpisua-Belmonte, J.C.; Dolle, P.; Renucci, A.; Zappavigna, V.; Falkenstein, H.; Duboule, D. Primary structure and embryonic expression pattern of the mouse hox-4.3 homeobox gene. Development 1990, 110, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Soshnikova, N.; Dewaele, R.; Janvier, P.; Krumlauf, R.; Duboule, D. Duplications of hox gene clusters and the emergence of vertebrates. Dev. Biol. 2013, 378, 194–199. [Google Scholar]
- Di-Poi, N.; Zakany, J.; Duboule, D. Distinct roles and regulations for hoxd genes in metanephric kidney development. PLoS Genet. 2007, 3, e232. [Google Scholar]
- Montavon, T.; Soshnikova, N.; Mascrez, B.; Joye, E.; Thevenet, L.; Splinter, E.; de Laat, W.; Spitz, F.; Duboule, D. A regulatory archipelago controls hox genes transcription in digits. Cell 2011, 147, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Amemiya, C.; Ruddle, F. Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl. Acad. Sci. USA 2003, 100, 14603–14606. [Google Scholar] [CrossRef]
- Smith, F.W.; Boothby, T.C.; Giovannini, I.; Rebecchi, L.; Jockusch, E.L.; Goldstein, B. The compact body plan of tardigrades evolved by the loss of a large body region. Curr. Biol. CB 2016, 26, 224–229. [Google Scholar]
- Moreno, E.; Nadal, M.; Baguna, J.; Martinez, P. Tracking the origins of the bilaterian hox patterning system: Insights from the acoel flatworm symsagittifera roscoffensis. Evol. Dev. 2009, 11, 574–581. [Google Scholar] [CrossRef]
- Hejnol, A.; Martindale, M.Q. Coordinated spatial and temporal expression of hox genes during embryogenesis in the acoel convolutriloba longifissura. BMC Biol. 2009, 7, 65. [Google Scholar]
- Philippe, H.; Brinkmann, H.; Copley, R.R.; Moroz, L.L.; Nakano, H.; Poustka, A.J.; Wallberg, A.; Peterson, K.J.; Telford, M.J. Acoelomorph flatworms are deuterostomes related to xenoturbella. Nature 2011, 470, 255–258. [Google Scholar]
- Philippe, H.; Poustka, A.J.; Chiodin, M.; Hoff, K.J.; Dessimoz, C.; Tomiczek, B.; Schiffer, P.H.; Muller, S.; Domman, D.; Horn, M.; et al. Mitigating anticipated effects of systematic errors supports sister-group relationship between xenacoelomorpha and ambulacraria. Curr. Biol. CB 2019, 29, 1818–1826.e6. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.T.; Vellutini, B.C.; Smith, J., 3rd; Ronquist, F.; Jondelius, U.; Hejnol, A. Xenacoelomorpha is the sister group to nephrozoa. Nature 2016, 530, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Castelli-Gair, J.E.; Capdevila, M.P.; Micol, J.L.; Garcia-Bellido, A. Positive and negative cis-regulatory elements in the bithoraxoid region of the drosophila ultrabithorax gene. Mol. Gen. Genet. MGG 1992, 234, 177–184. [Google Scholar] [CrossRef][Green Version]
- Maeda, R.K.; Karch, F. The abc of the bx-c: The bithorax complex explained. Development 2006, 133, 1413–1422. [Google Scholar]
- Horan, G.S.; Ramirez-Solis, R.; Featherstone, M.S.; Wolgemuth, D.J.; Bradley, A.; Behringer, R.R. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995, 9, 1667–1677. [Google Scholar] [PubMed]
- McIntyre, D.C.; Rakshit, S.; Yallowitz, A.R.; Loken, L.; Jeannotte, L.; Capecchi, M.R.; Wellik, D.M. Hox patterning of the vertebrate rib cage. Development 2007, 134, 2981–2989. [Google Scholar] [CrossRef] [PubMed]
- Wellik, D.M.; Capecchi, M.R. Hox10 and hox11 genes are required to globally pattern the mammalian skeleton. Science 2003, 301, 363–367. [Google Scholar] [PubMed]
- Sanchez-Herrero, E.; Crosby, M.A. The abdominal-b gene of drosophila melanogaster: Overlapping transcripts exhibit two different spatial distributions. EMBO J. 1988, 7, 2163–2173. [Google Scholar] [CrossRef]
- Averof, M.; Akam, M. Hox genes and the diversification of insect and crustacean body plans. Nature 1995, 376, 420–423. [Google Scholar]
- McCarthy-Taylor, J.B.; Kelly, S.R.; VanHook, A.M.; Marques-Souza, H.; Serano, J.M.; Patel, N.H. Expression of abdominal-b in the brine shrimp, artemia franciscana, expands our evolutionary understanding of the crustacean abdomen. Dev. Biol. 2022, 489, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Gebelein, B.; Culi, J.; Ryoo, H.D.; Zhang, W.; Mann, R.S. Specificity of distalless repression and limb primordia development by abdominal hox proteins. Dev. Cell 2002, 3, 487–498. [Google Scholar] [CrossRef]
- Abzhanov, A.; Kaufman, T.C. Crustacean (malacostracan) hox genes and the evolution of the arthropod trunk. Development 2000, 127, 2239–2249. [Google Scholar] [CrossRef] [PubMed]
- Averof, M.; Patel, N.H. Crustacean appendage evolution associated with changes in hox gene expression. Nature 1997, 388, 682–686. [Google Scholar] [CrossRef]
- Burke, A.C.; Nelson, C.E.; Morgan, B.A.; Tabin, C. Hox genes and the evolution of vertebrate axial morphology. Development 1995, 121, 333–346. [Google Scholar] [PubMed]
- Gaunt, S.J. Conservation in the hox code during morphological evolution. Int. J. Dev. Biol. 1994, 38, 549–552. [Google Scholar]
- Vinagre, T.; Moncaut, N.; Carapuco, M.; Novoa, A.; Bom, J.; Mallo, M. Evidence for a myotomal hox/myf cascade governing nonautonomous control of rib specification within global vertebral domains. Dev. Cell 2010, 18, 655–661. [Google Scholar]
- Guerreiro, I.; Nunes, A.; Woltering, J.M.; Casaca, A.; Novoa, A.; Vinagre, T.; Hunter, M.E.; Duboule, D.; Mallo, M. Role of a polymorphism in a hox/pax-responsive enhancer in the evolution of the vertebrate spine. Proc. Natl. Acad. Sci. USA 2013, 110, 10682–10686. [Google Scholar] [CrossRef]
- Chiori, R.; Jager, M.; Denker, E.; Wincker, P.; Da Silva, C.; Le Guyader, H.; Manuel, M.; Queinnec, E. Are hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan clytia hemisphaerica (cnidaria). PLoS ONE 2009, 4, e4231. [Google Scholar] [CrossRef]
- DuBuc, T.Q.; Ryan, J.F.; Shinzato, C.; Satoh, N.; Martindale, M.Q. Coral comparative genomics reveal expanded hox cluster in the cnidarian-bilaterian ancestor. Integr. Comp. Biol. 2012, 52, 835–841. [Google Scholar]
- DuBuc, T.Q.; Stephenson, T.B.; Rock, A.Q.; Martindale, M.Q. Hox and wnt pattern the primary body axis of an anthozoan cnidarian before gastrulation. Nat. Commun. 2018, 9, 2007. [Google Scholar] [CrossRef]
- Kamm, K.; Schierwater, B.; Jakob, W.; Dellaporta, S.L.; Miller, D.J. Axial patterning and diversification in the cnidaria predate the hox system. Curr. Biol. CB 2006, 16, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.C.; Unni, M.K.; Gungi, A.; Agarwal, P.; Galande, S. Evolution of hox-like genes in cnidaria: Study of hydra hox repertoire reveals tailor-made hox-code for cnidarians. Mech. Dev. 2015, 138, 87–96. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaunt, S.J. Seeking Sense in the Hox Gene Cluster. J. Dev. Biol. 2022, 10, 48. https://doi.org/10.3390/jdb10040048
Gaunt SJ. Seeking Sense in the Hox Gene Cluster. Journal of Developmental Biology. 2022; 10(4):48. https://doi.org/10.3390/jdb10040048
Chicago/Turabian StyleGaunt, Stephen J. 2022. "Seeking Sense in the Hox Gene Cluster" Journal of Developmental Biology 10, no. 4: 48. https://doi.org/10.3390/jdb10040048
APA StyleGaunt, S. J. (2022). Seeking Sense in the Hox Gene Cluster. Journal of Developmental Biology, 10(4), 48. https://doi.org/10.3390/jdb10040048






