Abstract
Diverse cell types in the central nervous system (CNS) are generated by a relatively small pool of neural stem cells during early development. Spatial and temporal regulation of stem cell behavior relies on precise coordination of gene expression. Well-studied mechanisms include hormone signaling, transcription factor activity, and chromatin remodeling processes. Much less is known about downstream RNA-dependent mechanisms including posttranscriptional regulation, nuclear export, alternative splicing, and transcript stability. These important functions are carried out by RNA-binding proteins (RBPs). Recent work has begun to explore how RBPs contribute to stem cell function and homeostasis, including their role in metabolism, transport, epigenetic regulation, and turnover of target transcripts. Additional layers of complexity are provided by the different target recognition mechanisms of each RBP as well as the posttranslational modifications of the RBPs themselves that alter function. Altogether, these functions allow RBPs to influence various aspects of RNA metabolism to regulate numerous cellular processes. Here we compile advances in RNA biology that have added to our still limited understanding of the role of RBPs in neurodevelopment.
1. Introduction
The complex question of how a nervous system is built is a fundamental topic in developmental biology. Neurogenesis is one of the earliest events in development that occurs during embryogenesis. During this process, existing cells give rise to new cells that acquire specific identities and move to different locations to begin building precursory tissues within the nervous system (i.e., brain, spinal cord, nerves). This is conducted in a remarkably organized manner to ensure that resulting tissues are of the correct size and have the structure necessary for function. Each cell type that makes up the diverse brain regions has unique transcript profiles, which influences cell identity, proliferation, apoptosis, and differentiation [1,2,3,4]. This helps create the correct number and size of cells with the desired properties and functionalities. The mechanisms by which these unique profiles are established are not clearly understood. In recent years, elegant studies have identified new components and mechanisms that regulate formation of the central nervous system (CNS) [5,6]. Still, several questions surrounding both normal and aberrant development of the CNS remain. One reason for these persistent unknowns is that there are various ways in which a cell can regulate its genetic profile, including transcriptional, hormonal, and metabolic regulation. This raises an important question: if cells in a living organism contain the same genetic material, then how does a cell know when and how to express or repress certain genes? Mechanisms of doing so differ across cell types and each cell must make efficient use of available energy to drive certain pathways. Among them, RNA-binding proteins (RBPs) provide an efficient and timely method to regulate gene expression profiles at the posttranscriptional level. This is performed through direct RNA binding, along with various interactions between RBPs and other gene expression regulators. RBP–RNA interactions can be transient, they can bind early and remain bound until degradation, or they can bind to influence downstream processing such as splicing or transport [7,8,9]. Together with the large number of RBPs estimated in the genome (~1500, [10]), these molecular properties yield an impressive degree of regulation on cell function. Not surprisingly, RBPs have emerged as critical regulators of development.
Here we describe recent advances in developmental neurobiology that contribute to our understanding of the multifaceted processes involved in brain development, focusing on the roles played by RBPs in neural stem cell function and maintenance. This family of proteins is involved in a plethora of cellular processes including localization, splicing, and translation of target RNAs. The wide array of RBP targets, including mRNA, miRNA, and lncRNA, allows amplified regulation of many cellular processes by RBPs, especially in stem cells [11,12]. Additionally, they allow for compartmentalized and temporal regulation of target metabolism. Learning about these processes will aid in understanding how the complex regions of the brain are formed from a relatively small pool of progenitor cells and how they function. To aid in this task, developmental biologists have turned to simple and easy-to-genetically-manipulate organisms such as Drosophila and C. elegans. Studies in these organisms can easily be reiterated in mammalian systems due to the highly conserved nature of many genes and pathways.
2. Neural Stem Cell Development
Early neurogenesis includes the formation of neural cells from ectodermal cells that, in turn, give rise to structures that become the brain, spinal cord, and peripheral nerves. The brain develops from a small pool of stem cells that are programmed to self-renew or differentiate to form an integrated and functional nervous system. Despite their relative low abundance, a high proliferative capacity allows these neural stem cells to continuously self-renew while also generating differentiated progeny throughout development. Should these properties go awry, proper development of an organism can be interrupted. For example, mutations in certain stem cell regulatory pathways have been shown to promote hyperplastic growth, whereas others lead to a decrease in the stem cell pool [13,14,15]. These events may lead to problems later in development and could contribute to several diseases including cancer. The stem cell populations in the brain have become of particular interest because of their potential to treat nervous system disorders as well. This, however, is hindered by the limited knowledge of how neural stem cells are regulated throughout development. In subsequent paragraphs we give a brief overview of neural stem cell development followed by specific studies highlighting the role of RBPs in CNS form and function.
Neural stem cells arise during gastrulation of early embryonic development. These cells will go on to produce all the specialized cell types found in the brain and nervous system. In vertebrates, migratory stem cells, called neural crest cells (NCC), are involved in formation of cephalic structures including ganglia and skeletal structures [16,17]. This process requires accurate and intricate signaling between tissue environments, cells, and proteins [18]. The neural tube is the first structure generated by a series of complex molecular processes and gives rise to the brain and spinal cord [19]. Additional segments and specialized regions arise from the neural tube later in development, and these also rely on various molecular mechanisms. RBPs are important during development of these embryonic and fetal neural structures due to the rapid and simultaneous changes. Cells must express the correct genetic profile and molecular machinery to produce the necessary factors to differentiate or self-renew. These processes, amongst others highlighted in this review, are regulated by RBPs and discussed in detail in subsequent sections.
Drosophila melanogaster (D. melanogaster), the common fruit fly, is a frequently used and established genetic model organism in developmental biology. The various stem cell populations in the fly can be studied across developmental time, and many developmental processes in humans can be easily recapitulated in flies. This has made the fruit fly an attractive model to study stem cells in disease and dysfunction. More specifically, neural stem cell (NSC) biology is often studied in Drosophila because the fruit fly brain contains a fixed population of neural stem cells (neuroblasts) that can be tracked throughout development. Moreover, development of some neuroblasts closely resembles that of neural stem cells, called radial glial cells (RGCs), in the ventricular zone (VZ) and subventricular zone (SVZ) of the mammalian brain where they generate neurons, oligodendrocytes, and astrocytes [20,21,22,23,24,25,26]. Like the neural stem cells in the mammalian brain, Drosophila neuroblasts undergo ACD to produce molecularly and physically distinct progeny [22,27,28]. By this mechanism, a small pool of neuroblasts can give rise to a large population of distinct, specialized cell types that comprise the complex adult brain while maintaining their own population through self-renewal. Neuroblast development relies on temporal and spatial expression of factors assigned at birth that dictate production of a specific subset of neurons/glia [29]. In the subsequent sections we will discuss these factors and how RBPs can regulate their activity.
3. Regulation of Spatial Gene Expression
Compartmentalization makes cellular processes and tissue functions more efficient. Throughout development, individual cells and tissues require spatial regulation of specific processes, notably gene expression, RNA processing, and translation, for example. Spatial regulation of gene expression is particularly important during tissue development. As cell identity is controlled by differential gene expression, stem cells rely on spatial regulation to correctly balance self-renewal and differentiation of their mitotic progeny. An important mode of regulation extends from localization of cognate mRNA to discrete areas within the cell that allow for differential inheritance during cell division. The precise mechanisms of RNA transport are still being elucidated, but several interesting details have been revealed [30,31,32], and we will only mention some commonalities here. Transport signals can be found at the 3′ UTR of mRNA [33], or they can be cis-acting motifs called zipcodes [34] that facilitate formation of a transport complex consisting of molecular motors and RBPs [35,36]. Binding of RBPs to the 3′ UTR is a common regulatory mechanism observed in development. For example, CUGBP ELAV-like family member-2 (CELF2) is an RBP that transports target mRNA between the cytoplasm and the nucleus. This ultimately serves as a mechanism to promote neural progenitor cell self-renewal or differentiation in the murine cerebral cortex [37]. Nuclear-cytoplasmic translocation can occur through formation of complexes that contain molecular motors. The zipcode-binding protein (ZBP1) interacts with a kinesin-I motor complex (Figure 1) in neurons to transport mRNA during murine postnatal development [38]. ZBP1 is closely related to insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in mammals (Imp; Drosophila) (Table 1). IGF2BP1/Imp is involved in RNA localization and translation [39,40]. In Drosophila Imp, the prion-like domains (PLD) regulate formation and homeostasis of neuronal ribonucleoprotein (RNP) granules in axons [41]. Furthermore, Imp regulates Drosophila egg chamber development by regulating Notch activation. More specifically, Imp directs localization of the metalloprotease Kuzbanian to the apical domain to cleave Notch, likely through 3′ UTR-binding [42].
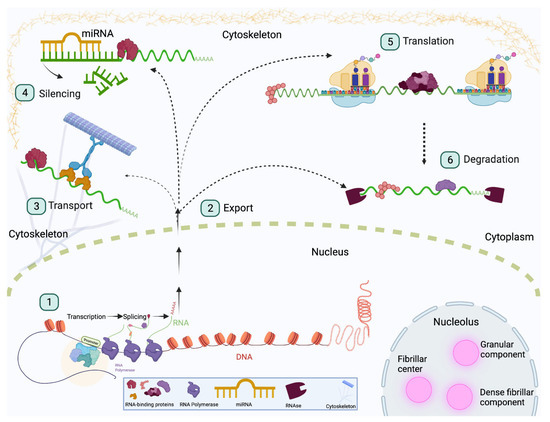
Figure 1.
RNA-binding proteins regulate RNA fate. RBPs are involved in all stages of RNA processing including transcription, translation, splicing, transport, degradation, and silencing. These processes are depicted in the above diagram. RNA targets are bound by RBPs prior to leaving the nucleus and remain associated with RBPs until they are degraded. (1) RBPs such as Rbfox1 and Qki serve as alternative splicing factors to ensure cells have the correct genetic profile. (2) Shuttling of RNA between the nucleus and cytoplasm is also mediated by RBPs, as observed with CELF2. (3) RNA can also be transported to subcellular locations, and this is often conducted through a complex consisting of molecular motors and RBPs, as observed with ZBP1. (4) RBPs also interact with silencing factors such as miRNA to degrade certain targets. The RBP Pumilio facilitates miRNA binding by exposing binding sites that would otherwise be inaccessible. (5) RBPs also promote post-transcriptional processes. For example, Imp binds targets to stabilize mRNA and promote translation. (6) Some RBPs, such as Stau1, promote degradation of certain RNA targets to promote stem cell identity.

Table 1.
Conserved RNA-binding proteins. Table showing RBPs conserved in Drosophila melanogaster (Dmel), humans (Hs), Caenorhabditis elegans (Ce), and Dario rerio (ZF). Resident tissue, function, binding domains, and associated references for each RBP are also listed.
Regulation of gene expression in NCCs is another important process that relies on spatial regulation by RBPs. Gene function during neural tube formation and differentiation of NCCs is particularly important as defects in this process can affect overall tissue architecture and organism development [126,127]. The Wnt/β-catenin signaling pathway is a well-known regulator of cranial neural crest development, particularly epithelial-mesenchymal transition (EMT). NCCs undergo EMT during early development and errors in this process result in various abnormalities. Efforts to identify important components involved in NCC function revealed that EMT causes elevated levels of posttranscriptional regulators and ribosome biogenesis factors in chicken cranial neural crest cells [128]. Both are well known roles of RBPs, pointing to their importance in neural crest cell development and function. An example of this is the RBP Vera (Vg1) that is involved in RNA localization and migration of cells needed to form the neural tube and migration of NCCs in Xenopus oocytes [129]. Embryos lacking Vg1 still generate the appropriate cells, but they fail to migrate to their final tissue [129]. Similarly, the Hu/ELAV family of RBPs regulates migration of NCCs and timely expression of genes involved in neural crest specification in avian embryos [130]. Growth and NCC expansion are further regulated by the RBP cellular nucleic-acid binding protein (CNBP) [131]. Xenopus laevis embryos lacking CNBP had decreased levels of foxD3 and c-myc, two important regulators of NCC growth and specification. Zebrafish forebrain development also employs a similar mechanism involving CNBP. In zebrafish embryos, CNBP promotes survival and regulates proliferation of NCCs [132]. Overall, the ability to transport and localize mRNA at a subcellular level plays an important role in gene expression during brain development.
Brain development further relies on spatial gene expression within various neurogenic niches. These regions are dedicated to maintaining neural stem cell populations, gliogenesis, or repair following injury [133,134]. Along with their specific functions, these niches also express a specific subset of proteins [135]. For example, in the murine CNS, the RBP Quaking (mammalian: Qki, C. elegans: GLD-1, D. melanogaster: HOW, ZF: zqk) [136] regulates cell differentiation through regulation of alternative splicing, translation, and mRNA stability [137]. Qki binds the 3′ UTR of astrocytic mRNA [136] in the murine brain or it binds targets in a sequence-specific manner (CUAAC) in mammalian cells [138]. Its various isoforms are expressed in different areas of the CNS and in different cell types [139]. Within these tissues and cell types, Qki can localize to the cytoplasm or to the nucleus, depending on the isoform. This restricted expression is beneficial for the generation of astrocytes and oligodendrocytes from NSCs [140]. Qki expression can also be restricted to cells that co-express other factors. For instance, Qki5 and -6 are specific to neural stem cells that co-express the neural stem cell transcription factor paired type homeobox 6 (Pax6) [141]. In this way, Qki expression is limited to neural stem cells during early neurogenesis. Qki has additional functions that make it an important regulator of cell differentiation. Those additional functions are discussed in the Regulation of Temporal Gene Expression section below. Spatial regulation by RBPs is a highly conserved mechanism used in various tissues, and the function of Qki and related RBPs highlight an important role that these proteins play specifically in spatially distinct niches controlling neural development.
RBPs can also participate in spatial regulation of gene expression by regulating the location of scaffolding proteins and their cognate mRNA. Drosophila neuroblast development involves precise localization of cellular machinery during mitosis to produce progeny with the correct fate. This is accomplished through establishment of cell polarity and asymmetric cell division. Both processes have been well studied in Drosophila neuroblasts, although some important details of underlying mechanisms remain unknown [142,143]. One well known model in Drosophila is localization of the scaffolding protein Miranda (Mir) to the basal domain of the neuroblast. This event signals for recruitment of important components required for differentiation. For example, targeting of the RBP Staufen (Stau) to the basal domain is accomplished by the scaffolding protein Miranda (Mir) [77]. Basal targeting of Stau and its bound RNAs [144] facilitates their asymmetric segregation during cell division, and thus, differential expression of cell-fate determining genes [145]. Stau functions early in Drosophila development to promote localization of bicoid and oskar RNA in the oocyte [146,147]. Further, Stau is required for basal localization of Prospero (Pros) RNA in mitotic neuroblasts—an event that is associated with production of differentiated cells [148,149]. Loss of Stau leads to improper localization of its target mRNAs, and thus, improper cell fate [145]. Overall, generation of diverse cell types in the embryonic and larval CNS in the fly is highly dependent on spatial gene expression that can be aided through assembly of protein scaffolds directed by intrinsic polarity cues.
Targeting of RBPs and RNA to different subcellular regions establishes the apical and basal domains necessary to guide subsequent divisions. This mechanism is also observed in development of neural precursor cells and neural stem cells in the mammalian cerebral cortex. There are two Staufen genes in humans, Staufen1 and Staufen2 (Stau1/2). Stau1 is more ubiquitously expressed, whereas Stau2 is mostly expressed in the brain. Stau1 is recruited by the zinc-finger protein Kruppel-like factor 4 (Klf4) to the 3′ UTR of mRNAs involved in cortical neurogenesis. Once bound, Stau1 promotes degradation of these targets to maintain neural precursor cell (NPC) identity [150]. Apart from this, Staufen1 also functions in a complex with Barentsz (Btz) to localize mRNA, a function that is conserved in mammalian hippocampal neurons and Drosophila oocytes [151]. Unlike dStaufen, Stau1 is expendable during NPC development and self-renewal [152]. This is likely due to redundant roles between Stau1 and Stau2. During asymmetric cell division, Stau2 is enriched in the intermediate progenitor cell (IPC) where it regulates targets involved in mitosis, transport, and centrosome assembly [153]. Thus, conserved RBPs appear to play similar roles in neurodevelopment.
Spatial expression of RBPs is an efficient way to regulate the gene expression profile of cells and specialized tissues. This method allows for efficient reprogramming to establish cell identity—often observed during the progenitor to differentiated cell transition. These processes are important for tissue establishment during early development. Beyond this, maintenance of cellular identity and tissue homeostasis relies on proper functioning of these RBPs. For some RBPs, this means expression throughout several developmental stages, whereas other RBPs are expressed only during specific developmental windows.
4. Regulation of Temporal Gene Expression
RBPs can influence target metabolism through regulation of RNA splicing, translation, turnover, transcription, and modification. These functions can impact temporal gene expression, which refers to the expression of genes or genetic profiles during specific developmental windows. This property is conserved in many organisms including flies, worms, and zebrafish and assists in specification of cell identity. Temporal gene expression patterns in Drosophila govern the timing of transcriptional reprogramming and this mechanism is also conserved in Caenorhabditis elegans (C. elegans) and Danio rerio (zebrafish) [154,155]. Temporal gene expression in C. elegans regulates memory processing, including storage and retrieval [156]. Transient expression of certain transcription factors in C. elegans regulates gene activation in preparation for neuronal specification [157]. This mechanism is further conserved in mammalian neural stem cells in which the differentiation program is temporally regulated by lncRNAs and other components [158,159]. Translocated in liposarcoma (TLS) is an RBP that inhibits transcription in response to ncRNA signals. Cyclin D1 (CCD1) is a gene involved in cell cycle progression and neural stem cell proliferation [160]. In human cell lines, ncRNAs bind the 5′ region of CCD1 to promote TLS-mediated repression [161]. Similar mechanisms are also in play in the mammalian spinal cord where temporal regulation aids in establishment of neuronal lineages [162]. Not only is the mechanism of temporal gene expression conserved across different species, but certain families of RBPs are also conserved. Mechanisms of temporal gene expression can occur by temporal target regulation by the RBP or via temporal expression of the RBP itself, and notable examples of such regulation are spotlighted in the following paragraphs.
The RBP family of ELAV/Hu embryonic lethal abnormal vision (ELAV) proteins is a widely studied example of temporal gene expression. ELAV/Hu proteins have roles in synaptic plasticity, mRNA stability, differentiation, and nuclear functions [163]. These highly conserved proteins (Humans: HuB, HuC, HuD, HuR, Flies: ELAV, fne, RBP9, C.elegans: exc-7, ZF: ELAVL1a) are necessary for development of neurons that will populate the CNS (Table 1). As such, ELAV is expressed during neuron formation [164]. They have important roles in pre-mRNA splicing of nervous-system-specific isoforms, establishment of brain vasculature during early development, and gene expression in cholinergic motor neurons [63,69,165]. Regulation by ELAV/Hu occurs primarily through modification of target localization and expression levels [166]. For instance, HuD binds targets associated with plasticity and interacts with the survival motor neuron (SMN) protein to transport mRNA within mouse axons [167]. This is accompanied by changes in chromatin architecture to establish a gene expression profile consistent with a differentiated cell [168], which are particularly important during neural stem cell to neuron transition. Many of the specific roles of the ELAV family of proteins have been described [169,170,171,172], so we will not discuss them further here.
Expression of certain RBPs with opposing functions is another common mode of temporal gene expression. An example of this is the expression pattern of IGF-II mRNA-binding (Imp/IGF2BP2) and Syncrip (Syp) in Drosophila where they regulate neuroblast growth and termination [5]. Imp/IGF2BP2 and Syp have opposing expression levels throughout development. Both RBPs regulate target localization, stability, and translation [55,173]. Specifically, developmentally timed expression of Imp and Syp in Drosophila NSCs regulates cell fate and promotes neuronal diversity [5,174,175]. Imp is highly expressed during early development and suppresses expression of Syp. Imp regulates the mushroom body (MB) lineage in Drosophila by regulating expression of Chinmo mRNA. Chinmo is responsible for limiting neuroblast self-renewal [176], and thus, is an important regulator of cell specification. Imp is also a key regulator of neuroblast size and growth rate through interaction with Myc mRNA [175]. The Imp–Myc mRNA interaction stabilizes the transcript to increase Myc protein levels and promote neuroblast growth [175]. Apart from being important in early development of the brain, Imp and Syp are also important in halting brain growth by preventing future neuroblast divisions through initiation of timely decommissioning and differentiation. In contrast to Imp, Syp exhibits minimal expression during early development. Levels of Syp gradually increase and reach significant expression levels by later larval stages. This shift in Syp expression serves to inhibit Imp expression during later developmental stages [5]. This event marks the end of neurogenesis, and thus, is an important step in forming tissues of the proper size (see RBPs in disease and dysfunction, below). Additionally, Syp regulates late developmental stages through interaction with miRNAs. Recently, it was shown that Syp also interacts with the miRNA pri-let-7a to regulate the larvae–adult transition in flies and worms and to suppress tumors in mammalian cells (let-7) [177,178]. Syp is also involved in neuroblast differentiation through regulation of Pros mRNA with a long 3′ UTR. Binding of Syp to the extended 3′ UTR of Pros mRNA stabilizes the transcript to promote increased Pros protein production [179]. This isoform of Pros is not expressed earlier in development [180] but is necessary for production of larval neurons [179]. Similar mechanisms of mRNA regulation are observed in mammalian neurons where Syncrip regulates plasticity, neuronal development, and differentiation through 3′ UTR-mediated repression or stabilization of target mRNAs [181]. Thus, gene regulatory networks involving multiple RBPs and their respective targets can participate in temporal gene expression patterns that facilitate proper developmental transitions in neural stem cells.
Finally, alternative splicing offers yet another form of RBP-mediated temporal gene expression. RBPs are important mediators of alternative splicing in neural tissue during early and late development [182,183] (Figure 1). Protein variants and transcripts found in different neural cell types are often generated through alternative splicing [184]. For example, RNA-binding Fox-1 homolog 1 (Rbfox1) is itself alternatively spliced, and transcript isoforms containing specific exons are expressed in specific regions of the brain [185]. During the neural progenitor cell-to-neuron transition, Rbfox1 in turn ensures correct inclusion of neuronal exons during alternative splicing in the cerebral cortex [186]. Another example is the previously mentioned RBP Qki. Its roles in alternative splicing are important for NPC differentiation and, like Rbfox1, Qki isoforms are present in specific tissues and developmental stages [187]. Remarkably, Qki in oligodendrocytes autoregulates its own splicing events [188]. Qki5 has been shown to regulate targets in early embryonic neural stem cells. Axon development and microtubule dynamics were shown to be two processes dependent on Qki5 function [189]. Isoform specificity further extends to differentiated cell types such as neurons and oligodendrocytes [188]. For example, Qki promotes a GABAergic neuron profile, and loss of Qki alters the gene expression to promote a glutamatergic neuron profile [189]. A comprehensive review of the role of RBPs in neuronal differentiation can be found in [190]. In oligodendrocytes, mRNAs associated with myelination are significantly downregulated [188]. Interestingly, expression and function of the RBPs themselves are also regulated, with many RBPs being essential in certain tissues and stages of development. For instance, the RBP Hu antigen R (HuR) is dispensable during embryonic development but essential during adult neurogenesis [191]. Further, HuR regulates a group of lncRNAs involved in differentiation. More directly, HuR expression is necessary to maintain stemness and prevent aberrant increases in cells expressing neuronal markers [192]. Together, these examples point to the importance of regulating gene expression profiles to obtain cells with the desired properties. Still, not all temporal regulation occurs at the transcriptional level. Many mechanisms are in effect following transcription and these events are described in the next sections.
5. Posttranscriptional Regulation by RNA-Binding Proteins
Posttranscriptional regulation is an emerging means of maintaining stem cell homeostasis. This mechanism aids in the response to cellular and environmental cues throughout development. During early development, stem cells must undergo transcriptional remodeling to become differentiated cell types. This includes expression of the important protein factors, such as differentiated cell markers, and repression of stemness genes including growth promoting factors. The expansive repertoire of RBPs can aid in regulating this process through control of protein translation, a process particularly important in stem cells [193,194,195]. Apart from directly influencing target metabolism (splicing, localization, degradation, etc.), RBPs can also regulate targets indirectly. One such mechanism is regulation of microRNA function and biogenesis. This mechanism is common in brain development and recent work has highlighted the important role of miRNAs in regulation of growth and proliferation [196,197]. Moreover, the complexity of cell types in the brain has been attributed to wide expression of miRNAs [198]. Until recently, few RBPs were thought to regulate miRNA structure and function, with only a handful of RBPs being able to bind miRNAs. Further studies have showed that RBPs facilitate the miRNA–mRNA interaction. They do so by exposing binding sites that would otherwise be inaccessible by the miRNA (Figure 1). Pumilio does this by binding the 3′ UTR of CDKN1B to allow binding by its regulatory miRNA [199]. Conversely, RBPs can also inhibit binding site access, including the mammalian RNA-binding motif protein 38 (RBM38) and insulin-like growth factor 2 mRNA-binding protein 2 (Imp2) [200,201]. Apart from influencing the interaction between miRNA and its targets, RBPs can regulate miRNA processing.
RBPs can also regulate targets by interacting with miRNAs or with targets that resemble the miRNA structure. One example of this is the RBP DiGeorge syndrome critical region 8 (DGCR8). This RBP is part of an important miRNA biogenesis component known as the microprocessor complex. DGCR8 and accompanying factors function to produce primary miRNAs (pri-miRNA), including some involved in neural stem cell growth [202,203,204]. For instance, this complex is responsible for processing of the pri-miRNA-bantam. This miRNA is part of a feedback loop with Numb and Notch to impart neural stem cell growth in the Drosophila brain [205]. The Drosha-DGCR8 complex is a well-known mechanism of miRNA processing. Details of this mechanism have been reviewed elsewhere [206]; thus, we will highlight the role of RBPs in miRNA processing and some of the miRNAs involved. RBPs have been shown to regulate gene expression through processing of miRNAs that act in a tissue-specific manner [202]. In addition to their roles in miRNA processing, DGCR8 and Drosha function to regulate expression of key transcription factors involved in murine embryonic neurogenesis. One of these factors is the transcription factor Neurogenin 2 (Neurog2). The Drosha–DGCR8 complex promotes degradation of Neurog2 mRNA to prevent differentiation [207]. Interestingly, this inhibition is independent of the canonical Microprocessor functions. Rather, it is facilitated by the structure of Neurog2 mRNA that resembles pri-miRNA [207]. Further, DGCR8 also functions in neural progenitor stem cell maintenance and cortical development [208]. Several of the miRNA targets regulate differentiation potential and proliferation of neural stem cells [209]. For instance, the 3′ UTR of p53 undergoes regulation by the miRNA miR-302. Consistent with this, cells depleted of miR-302 exhibit increased p53 activation that prevents cell differentiation [210]. Similar defects are observed when the RBP components of the miRNA processing machinery are misexpressed. Overexpression of DGCR8 in the murine cortex promotes an expansion of the neural progenitor cell pool, whereas loss of DGCR8 leads to apoptosis and defective corticogenesis [208,211]. Similarly, the RBP LIN28A/B, which is highly expressed in undifferentiated cells, prevents miRNA-dependent differentiation, and influences growth of neuroblasts (neuroblast: in fruit flies, these are the neural stem cells that produce glia and neurons) and neural progenitor cells (neural progenitor cell: cells with limited differentiation potential that produce specific neuron and glial subtypes) in mice [212]. As described, miRNAs have important roles in cell growth and differentiation. They provide additional means to maintain tissue homeostasis through their various functions across different tissues. RBPs regulate these various functions through miRNA processing and binding of miRNA to their targets. Thus, RBPs ensure that miRNA functions are carried out when it is developmentally relevant.
Target metabolism is also regulated by RBPs through interference with translational machinery. For example, the fragile X mental retardation protein (FMRP) regulates protein translation through ribosome stalling [213]. In this way, translation of certain mRNAs is reduced, and this is central to the cellular response to signals or to adapting to a changing environment. Similarly, cells rely on ribosome stalling during cellular differentiation, namely, neuronal differentiation, where transcripts are regulated posttranscriptionally through 3′ UTRs rather than through mRNA levels [214,215]. Similar mechanisms are used during early murine forebrain development where the protein synthesis machinery is downregulated at later stages of forebrain establishment [216]. Translation can also be inhibited before the ribosomes are loaded onto the mRNA by acting directly on the target. Binding of the RBP to the transcript prevents ribosome loading, and thus, protein synthesis. Conversely, RBPs may enhance protein synthesis by promoting recruitment of the necessary machinery or by creating a favorable RNA structure, such as with LIN28-mediated effects on neural stem cell maintenance [217].
The RBP cold-shock domain-containing E1 (CSDE1) regulates targets at the posttranscriptional level to temper their stability and translation. Interestingly, CSDE1 can function at different levels of translation and can tailor its target metabolism according to the tissue or cell type [218,219]. More specifically, CSDE1 promotes a stem cell profile in human embryonic stem cells (hESC) to prevent neural differentiation. This is performed through negative regulation of targets involved in radial glial cell formation (RGC). Consistent with this, loss of CSDE1 results in precocious neurogenesis, whereas overexpression leads to impaired neurogenesis [220]. Similarly, the RBP CUGBP ELAV1 family member 1 (CELF1) has an important role in murine neocortical development by regulating production of glutamatergic neurons. This is accomplished through translational repression of ELAV4 through a mechanism involving the 5′ UTR-binding. Unlike previously discussed RBPs, CELF1 is involved in temporal regulation of different isoforms of target RNAs, namely, ELAVl4. That is, isoforms are differentially regulated in early and late corticogenesis, and this is determined by the 5′ UTR of the target mRNA [221]. The RBP Pumilio2 (Pum2) also acts posttranscriptionally to regulate mammalian neuronal differentiation. Pum2 does this by initiating translation of elF4E to ultimately promote neuronal growth [222]. This mechanism adds an additional level of complexity to RBP-mediated regulation of gene expression.
Posttranscriptional regulation is an efficient mechanism that can be used for rapid changes in gene expression. Transitions from stem cells to more differentiated cell types must be accurate and rapid to keep up with a developing organism. More importantly, posttranscriptional regulation is a mechanism often observed during early development. For example, the Drosophila embryo relies exclusively on maternally inherited mRNA and proteins for the first three hours following fertilization. During this time, posttranscriptional regulation of these maternal genes and proteins guide developmental stages until transcription is turned on later in development [223,224,225]. Maternal-zygotic transition in Xenopus also depends on posttranscriptional regulation to degrade maternal transcripts during cell-fate determination [226]. Together with temporal and spatial gene expression, posttranscriptional mechanisms provide multiple layers of regulation. The benefits and malfunctions of these complexities will be discussed in the following sections.
6. RBPs and Phase Separation
Cells contain microenvironments that facilitate biological processes including transcription, degradation, and translation. To separate the various processes happening concurrently and ensure fidelity, cells contain canonical membrane-bound organelles along with more recently appreciated membraneless structures. Formation of membrane-bound organelles ensures that components can be restricted to discrete locales, whereas membraneless condensates allow for more dynamic movement of relevant molecules, as well as asymmetric subcellular localization in some cases. Still, these membraneless structures must also be regulated to maintain cellular homeostasis. Membraneless condensates are increasingly understood to be formed and regulated through liquid–liquid phase separation (LLPS) [227]. This process results in compartments that resemble an emulsion of oil droplets and water [228]. The transient and less restricted nature of the resulting membraneless structures, termed biomolecular condensates, makes them useful for rapid changes, such as cell specialization. Biomolecular condensates provide an efficient environment to carry out biochemical reactions and compartmentalize certain activities. These dynamic structures can be made up of different proteins and molecules, depending on the cell type, including a growing list of RBPs and bound RNA. Further, they can be formed in different parts of the cell, including the nucleus, and their formation can result from various conditions including stress and cell growth [229]. Structural specifications, such as intrinsic disorder, drive LLPS. More information about protein structure and the propensity to undergo LLPS can be found in excellent reviews [9,230,231,232,233].
Subcellular compartments such as P-granules (C. elegans), stress granules, and ribonucleoprotein (RNP) granules play important roles in posttranscriptional regulation. As such, these granules contain RBPs that are responsible for recruiting and regulating targets [234], as well as many of the other functions mentioned thus far. Further, structural properties such as intrinsically disordered regions (IDRs) (Figure 2) of many RBPs favor LLPS, and thus, creation of these subcellular structures (Figure 2) [235]. RBPs in biomolecular condensates have recently been shown to regulate stem cell homeostasis. The RBP fused in sarcoma (FUS) is involved in RNA localization, transcription, splicing, and DNA repair [236]. It has a role in maintenance of neural stem progenitor cells (NSPC), specifically through cell cycle regulation [237] and progression (S, G2/M) [238,239]. FUS acts in transcriptional condensates formed via LLPS to influence gene expression through recruitment of transcriptional machinery [240]. Interestingly, downstream functions of FUS are phase separation dependent. Namely, FUS that does not undergo LLPS interacts with RNA, whereas FUS that phase separates is involved in DNA damage repair and chromatin remodeling [241]. Moreover, it is responsible for regulating early differentiation. FUS contains an N-terminal prion-like domain (PLD) that facilitates reversible LLPS. This process is required for FUS functions including DNA repair [242], and this is important during stem cell divisions to ensure genome integrity is preserved. Stem cells are further regulated by RNP granules that promote translation of select targets. These highly conserved structures sequester mRNA and are formed through LLPS. Imp/ZBP1 is one RBP found in RNP granules in the Drosophila brain where it resides inside RNP granules along with translationally repressed mRNAs. Release of Imp and mRNAs from the granule results in translational activation [243]. Conversely, binding of RNA to RBPs can also promote formation of biomolecular condensates. Binding of RNA to hnRNPA1, an RBP enriched in the murine CNS [244], promotes phase separation and incorporation into stress granules [245]. Stress granules house stalled mRNA and RBPs and are frequently observed during stem cell differentiation. Like other biomolecular condensates, stress granules are not static. They disassemble once stressful conditions have been resolved and mRNA translation is reinstated [246]. Together with other resident proteins and nucleic acids, RBPs found in biomolecular condensates regulate cell fate spatially and temporally. Although much work remains to fully unravel the diverse roles that LLPS likely plays in neurodevelopment, it is clear this process represents yet another dynamic mode of RBP-dependent regulation to neural stem cell function.
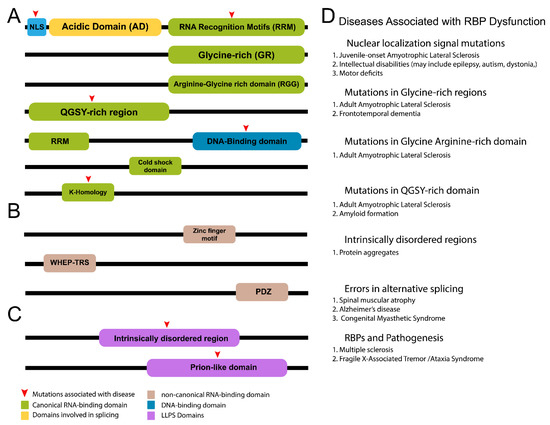
Figure 2.
Structural Properties of RNA-binding proteins. Illustration depicts common RBP domains. (A) Most RBPs contain common RNA-binding domains, such as RRMs, GRs, and QGSY regions. Nuclear localization signals and acidic domains [247] (splicing factors) are also found in various RBPs. (B) Some RBPs contain noncanonical domains that can bind RNA/DNA [184], such as Zn-finger motifs, WHEP-TRS domains [248], and PDZ domains. (C) Other RBPs contain important regions involved in liquid–liquid phase separation that promote formation of biological condensates important for RBP function, such as stress granule formation and transcription. (D) List of RBP domains associated with disease and dysfunction. Common diseases and abnormalities are included [249,250,251,252,253,254].
7. RBPs Aid in Maintaining Tissue Homeostasis
The previous sections highlighted the importance of regulating cell growth and proliferation to ensure proper tissue establishment. However, RBPs also play an important regulatory role to maintain tissue function and prevent disease. For instance, an important feature of stem cells that must be controlled throughout development is their ability to self-renew and differentiate. This fundamental feature helps determine the number and size of cells that comprise a tissue, with aberrant regulation of these often leading to several developmental diseases. For example, the YAP/TAZ (Drosophila; Hippo signaling) pathway has been implicated in regulation of tissue size and cell number through functioning of its effector YAP (Yorkie in Drosophila). In the Drosophila brain, neuroblast proliferation, quiescence, and overall brain size is modulated by the Hippo pathway [255,256]. In mammalian systems, YAP/TAZ/Yorkie dysfunction has been linked to aberrant tissue growth in both stem cells and epithelial tissue [256,257,258,259]. Thus, the Hippo pathway is an important regulator of tissue homeostasis, although the mechanisms that regulate the pathway itself remain unclear. Recently, regulation of the Hippo pathway has been attributed to posttranscriptional regulation and RBPs. Recent work showed that the ribonucleoprotein Hrb27C modulates the phosphorylation state of the effector proteins YAP/TAZ/Yorkie to promote growth [260]. Consistent with known roles of the Hippo pathway in growth and cell differentiation, loss of Hrb27C also results in decreased proliferation and aberrant structure of differentiated cells [260]. Loss of Hrb27C influences Yorkie-target gene expression; however, a molecular mechanism remains undefined. Regulation of Hippo also occurs through posttranscriptional control of Yki RNA. The RBP Rox8 prevents anomalous Yki signaling by promoting degradation of Yki RNA via two mechanisms. Rox8 binds the 3′ UTR of Yki mRNA (see [261,262]) to promote Yki mRNA decay. Rox8 also interacts with the microRNA miR-8 to recruit RISC to degrade Yki mRNA. A similar mechanism is observed when Drosophila cells are transfected with mammalian Rox8, TIAR [105]. In mouse embryonic stem cells, loss of TIAR promotes self-renewal [263]. Collectively, these studies illustrate the important role of RBPs in the regulation of an essential and evolutionarily-conserved cell growth pathway that mediates homeostatic control of tissue and organ size.
8. RNA-Binding Proteins in Dysfunction and Disease
The widespread role of RBPs in development also makes their dysfunction a common source of disease. Notably, RBP dysfunction in the CNS often presents as neurodegeneration or morphological defects [264,265]. Mutations resulting in altered function and RBP localization have been shown to lead to disease (Figure 2). Many neurodegenerative diseases are caused by protein aggression or mutations in genes that regulate autophagy. Protein aggregates result from errors in protein folding, denaturation, stress conditions, or due to age [266,267]. They contain a mixture of components including elevated amounts of RNA and are observed in numerous motor neuron disorders [268,269]. The RBPs FUS, TDP-43, hnRNPA1, and MATR3 have all been shown to aggregate and cause disease [270]. Recent work on the RBP Musashi (Msi) highlighted its role in Tau protein aggregation, commonly seen in Alzheimer’s disease. Interaction between Msi and Tau results in formation of aggregates and a similar response is observed when Tau interacts with TIA1 [271]. TIA1 promotes Tau phase separation and generation of toxic, oligomerized Tau [272]. Decreased TIA1 levels in an aberrant Tau background results in increased neuroinflammation [273]. Similarly, disease can also arise from dysfunction in regulatory pathways. Returning to the example of the Hippo, this pathway is also regulated by RBPs in several diseases including brain, liver, and pancreatic cancer pathogenesis [274,275,276]. In glioblastoma, levels of the RBP cytotoxic granule-associated RNA-binding protein (TIA1) (Drosophila: Rox8) (Table 1) increase following YAP knockdown to prevent cell invasion in U87 glioblastoma cells [276].
As another important example, Staufen, the RBP essential for neural stem cell development both in the fruit fly and in mammals, has been associated with diseased states including amyotrophic lateral sclerosis (ALS) and certain types of dementia. In one study, spinal cord samples from patients with ALS showed significantly elevated levels of Stau1 along with increased mTOR signaling [277]. Stau1 has been shown to inhibit autophagy through activation of mTOR [278]. In ALS, mTOR activation leads to stimulation of certain astrocytes that results in motor neuron toxicity and death. Many homeostatic mechanisms rely on autophagy to clear the cell of debris and toxic compounds. Thus, a disruption in the autophagy process contributes to neurodegeneration. Other sources of disease include incomplete development of tissue or tissue of the wrong size. Microcephaly and neural tube defects are two conditions caused by improper regulation of cell growth and proliferation. Loss of the RBP LIN28A leads to microcephaly in mice and a double deletion of LIN28A/B leads to neural tube defects. The underlying cause for these phenotypes is reduced protein synthesis resulting from defective ribosome biogenesis and translation [217]. Protein synthesis is an important process necessary to maintain cellular functions. As such, deviations from translational homeostasis lead to cognitive defects. Fragile X mental retardation protein (FMRP) is an RBP that is expressed in the brain where it regulates translation. Not surprisingly, loss of FMRP in adult neural stem cells results in decreased production of neurons and defective hippocampal processes [279,280]. Diseases resulting from defective FMRP signaling or absence include autism, intellectual disabilities, and a role in addiction [281,282,283]. Much of the disease is caused by over proliferation of adult neural stem cells at the expense of neurons [284].
9. Conclusions
Stem cells have the potential to generate diverse progeny from a small population of cells. Diversity of the progeny is attributed to several mechanisms including hormone signaling, transcription factor activity, and changes in cellular metabolism [285,286,287]. It is important to note that some metabolic enzymes have RNA-binding activity, and some metabolic processes can result in posttranslational modifications that in turn influence enzyme interactions with RNA [288]. This is an interesting avenue that will require further investigation. Complexity among distinct cell progeny is further increased by the varying proteome of each cell type. Stem cell regulation at the transcriptional level has been widely studied; however, posttranscriptional and translational regulation are more contemporary fields that warrant further studying. Notable among the outstanding research topics are the functions of RBPs in both normal and disease processes in brain development. Some RBP functions are associated with healthy tissues, whereas others, such as LLPS, have primarily been associated with disease. As discussed in this review, LLPS of TDP-43 causes disease, whereas that of FUS is critical for DNA repair. Characteristics governing disease-associated LLPS have been elucidated for a limited number of RBPs; however, details remain unknown for several others. For instance, LLPS of TDP-43 results in disease; however, aggresome formation also leads to disease [289]. Additional pathways that lead to generation of aggresomes have not been determined for other proteins known to be involved in brain diseases, including hnRNPA1 and FUS [290]. Furthermore, molecular characteristics such as disordered regions, phosphorylation, and amino acid composition and their role in spatial and temporal regulation of aggresome formation and LLPS remain to be explored [291,292]. From a pharmacological standpoint, these details could further efforts to identify drug targets beyond the conventional single protein strategies [293,294].
More well-known, but equally important, contributors to disease are errors in RNA processing. Brain diseases can also arise from errors in RNA splicing, localization, and translation. These include errors in processing of target RNA or expression/processing of the RBP transcript itself, as observed with FMRP in fragile X syndrome (FXS) [295]. The initial need here is to identify potential regulators of RBP expression. These may be tissue specific or ubiquitous, depending on the RBP. That is, expression of RBPs that are tissue-specific must be induced by upstream activators and these may be different, depending on the tissue. Large-scale identification and analysis of RBP targets will also aid in understanding the role of RBPs in disease. Recent advances in technology have made this possible [296]; however, these studies would be well complemented by follow up studies in tissues of interest.
Stem cells and their progeny have different molecular components and express varying transcriptional and translational regulatory proteins, including RBPs [297,298,299]. These genetic profiles differ according to cell function and resident tissue. Thus, it is important that cells have the appropriate gene expression pattern to ensure correct development. The previous sections outlined the important roles of RBPs in establishing cell identity and maintaining tissue homeostasis. The diverse roles of RBPs allow them to be involved in many cellular processes including translation, miRNA processing, transport, splicing, and others mentioned here. Additionally, numerous RBPs function in a complex to exert different effects on targets (i.e., localization). Further, RBPs can alter when and where targets are expressed. Spatial and temporal regulation of targets can be particularly important during early and late developmental stages (see the Regulation of Temporal Gene Expression section). These various functions, coupled with the numerous RBPs identified to date, make them important regulators of neurodevelopment. This review discussed some of the important mechanisms in neurodevelopment that involve RBPs, which have only recently been identified as important regulators of cell fate. Many questions remain open for future exploration, including identification of additional RBPs and their key RNA targets across model organisms. Moreover, unraveling additional gene regulatory networks that rely on RBPs for proper regulation of gene expression is needed.
Author Contributions
Conceptualization, A.S.P. and C.A.J.; methodology, A.S.P. and C.A.J.; investigation, A.S.P. and C.A.J.; writing—original draft, A.S.P. and C.A.J.; writing—review and editing, A.S.P. and C.A.J.; funding acquisition, A.S.P. and C.A.J.; resources, A.S.P. and C.A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a Faculty Development Award (A.S.P.), and by a grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number R01 GM108756 (C.A.J.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was conducted as part of the University of New Mexico Women in STEM Faculty Development Fund (A.S.P.). We apologize to those authors whose excellent work has not been included in this review due to space limitations.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Yuan, F.; Pan, X.; Zeng, T.; Zhang, Y.H.; Chen, L.; Gan, Z.; Huang, T.; Cai, Y.D. Identifying Cell-Type Specific Genes and Expression Rules Based on Single-Cell Transcriptomic Atlas Data. Front. Bioeng. Biotechnol. 2020, 8, 350. [Google Scholar] [CrossRef] [PubMed]
- Cervenka, J.; Tyleckova, J.; Kupcova Skalnikova, H.; Vodickova Kepkova, K.; Poliakh, I.; Valekova, I.; Pfeiferova, L.; Kolar, M.; Vaskovicova, M.; Pankova, T.; et al. Proteomic Characterization of Human Neural Stem Cells and Their Secretome During in vitro Differentiation. Front. Cell Neurosci. 2020, 14, 612560. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Hu, J.; Ralls, S.; Kitamura, T.; Loh, Y.P.; Yang, Y.; Mukouyama, Y.S.; Ahn, S. The molecular profiles of neural stem cell niche in the adult subventricular zone. PLoS ONE 2012, 7, e50501. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Chen, L.; Zheng, S. Global Reprogramming of Apoptosis-Related Genes during Brain Development. Cells 2021, 10, 2901. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, C.P.; Sugino, K.; Fu, C.C.; Liu, L.Y.; Yao, X.; Lee, L.P.; Lee, T. Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science 2015, 350, 317–320. [Google Scholar] [CrossRef]
- Ren, Q.; Yang, C.P.; Liu, Z.; Sugino, K.; Mok, K.; He, Y.; Ito, M.; Nern, A.; Otsuna, H.; Lee, T. Stem Cell-Intrinsic, Seven-up-Triggered Temporal Factor Gradients Diversify Intermediate Neural Progenitors. Curr. Biol. 2017, 27, 1303–1313. [Google Scholar] [CrossRef]
- Quattrone, A.; Dassi, E. The Architecture of the Human RNA-Binding Protein Regulatory Network. iScience 2019, 21, 706–719. [Google Scholar] [CrossRef]
- Dassi, E. Handshakes and Fights: The Regulatory Interplay of RNA-Binding Proteins. Front. Mol. Biosci. 2017, 4, 67. [Google Scholar] [CrossRef]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, H.; Zhang, Q.; Zhang, Y. The functions of long non-coding RNAs in neural stem cell proliferation and differentiation. Cell Biosci. 2020, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.D.; Andersen, R.E.; Liu, S.J.; Nowakowski, T.J.; Hong, S.J.; Gertz, C.; Salinas, R.D.; Zarabi, H.; Kriegstein, A.R.; Lim, D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015, 16, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Robinson, K.J.; Doe, C.Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 2006, 439, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Penning, A.; Tosoni, G.; Abiega, O.; Bielefeld, P.; Gasperini, C.; De Pietri Tonelli, D.; Fitzsimons, C.P.; Salta, E. Adult Neural Stem Cell Regulation by Small Non-coding RNAs: Physiological Significance and Pathological Implications. Front. Cell Neurosci. 2021, 15, 781434. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Chen, W.; Orr, B.A.; Spitsbergen, J.M.; Jia, S.; Eden, C.J.; Henson, H.E.; Taylor, M.R. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol. Cancer 2015, 14, 18. [Google Scholar] [CrossRef]
- Bronner, M.E. Formation and migration of neural crest cells in the vertebrate embryo. Histochem. Cell Biol. 2012, 138, 179–186. [Google Scholar] [CrossRef]
- Mendez-Maldonado, K.; Vega-Lopez, G.A.; Aybar, M.J.; Velasco, I. Neurogenesis From Neural Crest Cells: Molecular Mechanisms in the Formation of Cranial Nerves and Ganglia. Front. Cell Dev. Biol. 2020, 8, 635. [Google Scholar] [CrossRef]
- Stiles, J.; Jernigan, T.L. The basics of brain development. Neuropsychol. Rev. 2010, 20, 327–348. [Google Scholar] [CrossRef]
- Copp, A.J.; Greene, N.D. Genetics and development of neural tube defects. J. Pathol. 2010, 220, 217–230. [Google Scholar] [CrossRef]
- Gotz, M.; Barde, Y.A. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron 2005, 46, 369–372. [Google Scholar] [CrossRef]
- Taverna, E.; Gotz, M.; Huttner, W.B. The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014, 30, 465–502. [Google Scholar] [CrossRef] [PubMed]
- Dewey, E.B.; Taylor, D.T.; Johnston, C.A. Cell Fate Decision Making through Oriented Cell Division. J. Dev. Biol. 2015, 3, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Merkle, F.T.; Tramontin, A.D.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. USA 2004, 101, 17528–17532. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Kawaguchi, A.; Saito, K.; Kawano, M.; Muto, T.; Ogawa, M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 2004, 131, 3133–3145. [Google Scholar] [CrossRef] [PubMed]
- Ming, G.L.; Song, H. Adult neurogenesis in the mammalian central nervous system. Annu. Rev. Neurosci. 2005, 28, 223–250. [Google Scholar] [CrossRef] [PubMed]
- Shitamukai, A.; Matsuzaki, F. Control of asymmetric cell division of mammalian neural progenitors. Dev. Growth Differ. 2012, 54, 277–286. [Google Scholar] [CrossRef] [PubMed]
- LaMonica, B.E.; Lui, J.H.; Hansen, D.V.; Kriegstein, A.R. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat. Commun. 2013, 4, 1665. [Google Scholar] [CrossRef]
- Noctor, S.C.; Martinez-Cerdeno, V.; Ivic, L.; Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004, 7, 136–144. [Google Scholar] [CrossRef]
- Urbach, R.; Technau, G.M. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 2003, 130, 3621–3637. [Google Scholar] [CrossRef]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review). Int. J. Mol. Med. 2014, 33, 747–762. [Google Scholar] [CrossRef]
- Chin, A.; Lecuyer, E. RNA localization: Making its way to the center stage. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2956–2970. [Google Scholar] [CrossRef] [PubMed]
- Suter, B. RNA localization and transport. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 938–951. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C. What Are 3 UTRs Doing? Csh Perspect. Biol. 2019, 11, a034728. [Google Scholar] [CrossRef]
- Kislauskis, E.H.; Singer, R.H. Determinants of mRNA localization. Curr. Opin. Cell Biol. 1992, 4, 975–978. [Google Scholar] [CrossRef]
- Dimitrova-Paternoga, L.; Jagtap, P.K.A.; Cyrklaff, A.; Vaishali; Lapouge, K.; Sehr, P.; Perez, K.; Heber, S.; Löw, C.; Hennig, J.; et al. Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex. Gene Dev. 2021, 35, 976. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Pazyra-Murphy, M.F.; Silagi, E.S.; Tasdemir-Yilmaz, O.E.; Li, Y.H.; Rose, L.; Yeoh, Z.C.; Vangos, N.E.; Geffken, E.A.; Seo, H.S.; et al. Binding and transport of SFPQ-RNA granules by KIF5A/KLC1 motors promotes axon survival. J. Cell Biol. 2021, 220, e202005051. [Google Scholar] [CrossRef]
- Wang, R.; Wang, R.; Liu, M.; Yuan, W.; Zhao, Z.; Liu, X.; Peng, Y.; Yang, X.; Sun, Y.; Tang, W. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2101838118. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, J.; Zhu, T.; Cohen, I.; Dictenberg, J. A kinesin adapter directly mediates dendritic mRNA localization during neural development in mice. J. Biol. Chem. 2020, 295, 6605–6628. [Google Scholar] [CrossRef]
- Biswas, J.; Nunez, L.; Das, S.; Yoon, Y.J.; Eliscovich, C.; Singer, R.H. Zipcode Binding Protein 1 (ZBP1; IGF2BP1): A Model for Sequence-Specific RNA Regulation. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 1–10. [Google Scholar] [CrossRef]
- Glass, M.; Misiak, D.; Bley, N.; Muller, S.; Hagemann, S.; Busch, B.; Rausch, A.; Huttelmaier, S. IGF2BP1, a Conserved Regulator of RNA Turnover in Cancer. Front. Mol. Biosci. 2021, 8, 632219. [Google Scholar] [CrossRef]
- Vijayakumar, J.; Perrois, C.; Heim, M.; Bousset, L.; Alberti, S.; Besse, F. The prion-like domain of Drosophila Imp promotes axonal transport of RNP granules in vivo. Nat. Commun. 2019, 10, 2593. [Google Scholar] [CrossRef] [PubMed]
- Fic, W.; Faria, C.; St Johnston, D. IMP regulates Kuzbanian to control the timing of Notch signalling in Drosophila follicle cells. Development 2019, 146, dev168963. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Christiansen, J.; Lykke-Andersen, J.; Johnsen, A.H.; Wewer, U.M.; Nielsen, F.C. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell Biol. 1999, 19, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Pillasch, F.; Lacher, U.; Wallrapp, C.; Micha, A.; Zimmerhackl, F.; Hameister, H.; Varga, G.; Friess, H.; Buchler, M.; Beger, H.G.; et al. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene 1997, 14, 2729–2733. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, L.; Qiang, P. The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int. 2021, 21, 99. [Google Scholar] [CrossRef] [PubMed]
- Degrauwe, N.; Suva, M.L.; Janiszewska, M.; Riggi, N.; Stamenkovic, I. IMPs: An RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016, 30, 2459–2474. [Google Scholar] [CrossRef] [PubMed]
- Toledano, H.; D’Alterio, C.; Czech, B.; Levine, E.; Jones, D.L. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 2012, 485, 605–610. [Google Scholar] [CrossRef]
- Chao, J.A.; Patskovsky, Y.; Patel, V.; Levy, M.; Almo, S.C.; Singer, R.H. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010, 24, 148–158. [Google Scholar] [CrossRef]
- Pan, F.; Huttelmaier, S.; Singer, R.H.; Gu, W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell Biol. 2007, 27, 8340–8351. [Google Scholar] [CrossRef]
- Ren, F.; Lin, Q.; Gong, G.; Du, X.; Dan, H.; Qin, W.; Miao, R.; Xiong, Y.; Xiao, R.; Li, X.; et al. Igf2bp3 maintains maternal RNA stability and ensuRes. early embryo development in zebrafish. Commun. Biol. 2020, 3, 94. [Google Scholar] [CrossRef]
- Vong, Y.H.; Sivashanmugam, L.; Leech, R.; Zaucker, A.; Jones, A.; Sampath, K. The RNA-binding protein Igf2bp3 is critical for embryonic and germline development in zebrafish. PLoS Genet. 2021, 17, e1009667. [Google Scholar] [CrossRef] [PubMed]
- McDermott, S.M.; Yang, L.; Halstead, J.M.; Hamilton, R.S.; Meignin, C.; Davis, I. Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction. RNA 2014, 20, 1593–1606. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.P.; Samuels, T.J.; Huang, Y.; Yang, L.; Ish-Horowicz, D.; Davis, I.; Lee, T. Imp and Syp RNA-binding proteins govern decommissioning of Drosophila neural stem cells. Development 2017, 144, 3454–3464. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Hobor, F.; Dallmann, A.; Ball, N.J.; Cicchini, C.; Battistelli, C.; Ogrodowicz, R.W.; Christodoulou, E.; Martin, S.R.; Castello, A.; Tripodi, M.; et al. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 2018, 9, 831. [Google Scholar] [CrossRef]
- Rizzo, F.; Nizzardo, M.; Vashisht, S.; Molteni, E.; Melzi, V.; Taiana, M.; Salani, S.; Santonicola, P.; Di Schiavi, E.; Bucchia, M.; et al. Key role of SMN/SYNCRIP and RNA-Motif 7 in spinal muscular atrophy: RNA-Seq and motif analysis of human motor neurons. Brain 2019, 142, 276–294. [Google Scholar] [CrossRef] [PubMed]
- Kinnaird, J.H.; Maitland, K.; Walker, G.A.; Wheatley, I.; Thompson, F.J.; Devaney, E. HRP-2, a heterogeneous nuclear ribonucleoprotein, is essential for embryogenesis and oogenesis in Caenorhabditis elegans. Exp. Cell Res. 2004, 298, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Kabat, J.L.; Barberan-Soler, S.; Zahler, A.M. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J. Biol. Chem. 2009, 284, 28490–28497. [Google Scholar] [CrossRef] [PubMed]
- UniProt. Heterogeneous Nuclear RibonucleoProtein (HnRNP) R Homolog. 2021. Available online: https://www.uniprot.org/uniprot/Q9NLD1 (accessed on 19 May 2022).
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- Bayes, A.; Collins, M.O.; Reig-Viader, R.; Gou, G.; Goulding, D.; Izquierdo, A.; Choudhary, J.S.; Emes, R.D.; Grant, S.G. Evolution of complexity in the zebrafish synapse proteome. Nat. Commun. 2017, 8, 14613. [Google Scholar] [CrossRef]
- Toni, M.; Angiulli, E.; Miccoli, G.; Cioni, C.; Alleva, E.; Frabetti, F.; Pizzetti, F.; Grassi Scalvini, F.; Nonnis, S.; Negri, A.; et al. Environmental temperature variation affects brain protein expression and cognitive abilities in adult zebrafish (Danio rerio): A proteomic and behavioural study. J. Proteomics 2019, 204, 103396. [Google Scholar] [CrossRef] [PubMed]
- Koushika, S.P.; Lisbin, M.J.; White, K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr. Biol. 1996, 6, 1634–1641. [Google Scholar] [CrossRef]
- Lisbin, M.J.; Gordon, M.; Yannoni, Y.M.; White, K. Function of RRM domains of Drosophila melanogaster ELAV: Rnp1 mutations and rrm domain replacements with ELAV family proteins and SXL. Genetics 2000, 155, 1789–1798. [Google Scholar] [CrossRef] [PubMed]
- Hilgers, V.; Lemke, S.B.; Levine, M. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 2012, 26, 2259–2264. [Google Scholar] [CrossRef]
- Fukao, A.; Sasano, Y.; Imataka, H.; Inoue, K.; Sakamoto, H.; Sonenberg, N.; Thoma, C.; Fujiwara, T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol. Cell 2009, 36, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Bronicki, L.M.; Jasmin, B.J. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA 2013, 19, 1019–1037. [Google Scholar] [CrossRef]
- Fujita, M.; Hawkinson, D.; King, K.V.; Hall, D.H.; Sakamoto, H.; Buechner, M. The role of the ELAV homologue EXC-7 in the development of the Caenorhabditis elegans excretory canals. Dev. Biol. 2003, 256, 290–301. [Google Scholar] [CrossRef]
- Loria, P.M.; Duke, A.; Rand, J.B.; Hobert, O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr. Biol. 2003, 13, 1317–1323. [Google Scholar] [CrossRef]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 1998, 282, 2012–2018. [CrossRef]
- Tallafuss, A.; Kelly, M.; Gay, L.; Gibson, D.; Batzel, P.; Karfilis, K.V.; Eisen, J.; Stankunas, K.; Postlethwait, J.H.; Washbourne, P. Transcriptomes of post-mitotic neurons identify the usage of alternative pathways during adult and embryonic neuronal differentiation. BMC Genom. 2015, 16, 1100. [Google Scholar] [CrossRef]
- Hoang, T.; Wang, J.; Boyd, P.; Wang, F.; Santiago, C.; Jiang, L.; Yoo, S.; Lahne, M.; Todd, L.J.; Jia, M.; et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science 2020, 370, eabb8598. [Google Scholar] [CrossRef] [PubMed]
- UniProtKb. ELAV-Like Protein. 2004. Available online: https://www.uniprot.org/uniprot/Q6P105 (accessed on 19 May 2022).
- Park, E.; Maquat, L.E. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 2013, 4, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Gleghorn, M.L.; Maquat, L.E. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. USA 2013, 110, 405–412. [Google Scholar] [CrossRef]
- St Johnston, D.; Beuchle, D.; Nusslein-Volhard, C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 1991, 66, 51–63. [Google Scholar] [CrossRef]
- Jia, M.; Shan, Z.; Yang, Y.; Liu, C.; Li, J.; Luo, Z.G.; Zhang, M.; Cai, Y.; Wen, W.; Wang, W. The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division. Nat. Commun. 2015, 6, 8381. [Google Scholar] [CrossRef]
- Ferrandon, D.; Elphick, L.; Nusslein-Volhard, C.; St Johnston, D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 1994, 79, 1221–1232. [Google Scholar] [CrossRef]
- St Johnston, D.; Brown, N.H.; Gall, J.G.; Jantsch, M. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. USA 1992, 89, 10979–10983. [Google Scholar] [CrossRef]
- Marion, R.M.; Fortes, P.; Beloso, A.; Dotti, C.; Ortin, J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell Biol. 1999, 19, 2212–2219. [Google Scholar] [CrossRef]
- Lebeau, G.; Maher-Laporte, M.; Topolnik, L.; Laurent, C.E.; Sossin, W.; Desgroseillers, L.; Lacaille, J.C. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol. Cell Biol. 2008, 28, 2896–2907. [Google Scholar] [CrossRef]
- Gowravaram, M.; Schwarz, J.; Khilji, S.K.; Urlaub, H.; Chakrabarti, S. Insights into the assembly and architecture of a Staufen-mediated mRNA decay (SMD)-competent mRNP. Nat. Commun. 2019, 10, 5054. [Google Scholar] [CrossRef]
- Kohrmann, M.; Luo, M.; Kaether, C.; DesGroseillers, L.; Dotti, C.G.; Kiebler, M.A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 1999, 10, 2945–2953. [Google Scholar] [CrossRef] [PubMed]
- Ricci, E.P.; Kucukural, A.; Cenik, C.; Mercier, B.C.; Singh, G.; Heyer, E.E.; Ashar-Patel, A.; Peng, L.; Moore, M.J. Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol. 2014, 21, 26–35. [Google Scholar] [CrossRef] [PubMed]
- LeGendre, J.B.; Campbell, Z.T.; Kroll-Conner, P.; Anderson, P.; Kimble, J.; Wickens, M. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J. Biol. Chem. 2013, 288, 2532–2545. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Veksler-Lublinsky, I.; Morrissey, D.; Ambros, V. Staufen Negatively Modulates MicroRNA Activity in Caenorhabditis elegans. G3 (Bethesda) 2016, 6, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Bateman, M.J.; Cornell, R.; d’Alencon, C.; Sandra, A. Expression of the zebrafish Staufen gene in the embryo and adult. Gene Expr. Patterns 2004, 5, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Wang, H.; Quach, H.N.; Sampath, K. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev. Biol. 2006, 292, 393–406. [Google Scholar] [CrossRef]
- Nakamura, M.; Okano, H.; Blendy, J.A.; Montell, C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 1994, 13, 67–81. [Google Scholar] [CrossRef]
- Forouzanfar, M.; Lachinani, L.; Dormiani, K.; Nasr-Esfahani, M.H.; Gure, A.O.; Ghaedi, K. Intracellular functions of RNA-binding protein, Musashi1, in stem and cancer cells. Stem Cell Res. Ther. 2020, 11, 193. [Google Scholar] [CrossRef]
- Kaneko, Y.; Sakakibara, S.; Imai, T.; Suzuki, A.; Nakamura, Y.; Sawamoto, K.; Ogawa, Y.; Toyama, Y.; Miyata, T.; Okano, H. Musashi1: An evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev. Neurosci. 2000, 22, 139–153. [Google Scholar] [CrossRef]
- Siddall, N.A.; McLaughlin, E.A.; Marriner, N.L.; Hime, G.R. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc. Natl. Acad. Sci. USA 2006, 103, 8402–8407. [Google Scholar] [CrossRef]
- Bertolin, A.P.; Katz, M.J.; Yano, M.; Pozzi, B.; Acevedo, J.M.; Blanco-Obregon, D.; Gandara, L.; Sorianello, E.; Kanda, H.; Okano, H.; et al. Musashi mediates translational repression of the Drosophila hypoxia inducible factor. Nucleic Acids Res. 2016, 44, 7555–7567. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Vandivier, L.; Parada, K.; Woo, D.H.; Wang, S.; Naqvi, A.S.; Rao, S.; et al. The Msi Family of RNA-Binding Proteins Function Redundantly as Intestinal Oncoproteins. Cell Rep. 2015, 13, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Parada, K.; Rao, S.; Minuesa, G.; Katz, Y.; Gregory, B.D.; et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat. Commun. 2015, 6, 6517. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Shan, Y.T.; Ma, H.L.; Elshoura, I.; Nafees, M.; Yang, K.Y.; Yin, W. Identification of a novel splice variant of the human musashi-1 gene. Oncol. Lett. 2018, 16, 5441–5448. [Google Scholar] [CrossRef] [PubMed]
- Good, P.; Yoda, A.; Sakakibara, S.; Yamamoto, A.; Imai, T.; Sawa, H.; Ikeuchi, T.; Tsuji, S.; Satoh, H.; Okano, H. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics 1998, 52, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Mastrandreas, P.; Vukojevic, V.; Boglari, C.; Peter, F.; de Quervain, D.; Papassotiropoulos, A.; Stetak, A. The Role of Musashi Rna Binding Proteins in Associative Learning and Memory. Eur. Neuropsychopharm. 2019, 29, S855. [Google Scholar] [CrossRef]
- Hadziselimovic, N.; Vukojevic, V.; Peter, F.; Milnik, A.; Fastenrath, M.; Fenyves, B.G.; Hieber, P.; Demougin, P.; Vogler, C.; de Quervain, D.J.; et al. Forgetting is regulated via Musashi-mediated translational control of the Arp2/3 complex. Cell 2014, 156, 1153–1166. [Google Scholar] [CrossRef]
- Yoda, A.; Sawa, H.; Okano, H. MSI-1, a neural RNA-binding protein, is involved in male mating behaviour in Caenorhabditis elegans. Genes Cells 2000, 5, 885–895. [Google Scholar] [CrossRef]
- Zearfoss, N.R.; Deveau, L.M.; Clingman, C.C.; Schmidt, E.; Johnson, E.S.; Massi, F.; Ryder, S.P. A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J. Biol. Chem. 2014, 289, 35530–35541. [Google Scholar] [CrossRef]
- Hochgreb-Hagele, T.; Koo, D.E.; Das, N.M.; Bronner, M.E. Zebrafish stem/progenitor factor msi2b exhibits two phases of activity mediated by different splice variants. Stem Cells 2014, 32, 558–571. [Google Scholar] [CrossRef][Green Version]
- Shibata, S.; Umei, M.; Kawahara, H.; Yano, M.; Makino, S.; Okano, H. Characterization of the RNA-binding protein Musashi1 in zebrafish. Brain Res. 2012, 1462, 162–173. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deng, X.; Meller, V.H. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 2006, 174, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Sun, Y.; Azad, T.; Janse van Rensburg, H.J.; Luo, J.; Yang, S.; Liu, P.; Lv, Z.; Zhan, M.; Lu, L.; et al. Rox8 promotes microRNA-dependent yki messenger RNA decay. Proc. Natl. Acad. Sci. USA 2020, 117, 30520–30530. [Google Scholar] [CrossRef] [PubMed]
- Brand, S.; Bourbon, H.M. The developmentally-regulated Drosophila gene rox8 encodes an RRM-type RNA binding protein structurally related to human TIA-1-type nucleolysins. Nucleic Acids Res. 1993, 21, 3699–3704. [Google Scholar] [CrossRef][Green Version]
- Park, J.W.; Parisky, K.; Celotto, A.M.; Reenan, R.A.; Graveley, B.R. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 15974–15979. [Google Scholar] [CrossRef]
- Mazan-Mamczarz, K.; Lal, A.; Martindale, J.L.; Kawai, T.; Gorospe, M. Translational repression by RNA-binding protein TIAR. Mol. Cell Biol. 2006, 26, 2716–2727. [Google Scholar] [CrossRef]
- Kawakami, A.; Tian, Q.; Duan, X.; Streuli, M.; Schlossman, S.F.; Anderson, P. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 1992, 89, 8681–8685. [Google Scholar] [CrossRef]
- Dember, L.M.; Kim, N.D.; Liu, K.Q.; Anderson, P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996, 271, 2783–2788. [Google Scholar] [CrossRef]
- Beck, A.R.; Miller, I.J.; Anderson, P.; Streuli, M. RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl. Acad. Sci. USA 1998, 95, 2331–2336. [Google Scholar] [CrossRef]
- Garrido-Godino, A.I.; Garcia-Lopez, M.C.; Navarro, F. Correct assembly of RNA polymerase II depends on the foot domain and is required for multiple steps of transcription in Saccharomyces cerevisiae. Mol. Cell Biol. 2013, 33, 3611–3626. [Google Scholar] [CrossRef]
- Huelgas-Morales, G.; Silva-Garcia, C.G.; Salinas, L.S.; Greenstein, D.; Navarro, R.E. The Stress Granule RNA-Binding Protein TIAR-1 Protects Female Germ Cells from Heat Shock in Caenorhabditis elegans. G3 (Bethesda) 2016, 6, 1031–1047. [Google Scholar] [CrossRef] [PubMed]
- Andrusiak, M.G.; Sharifnia, P.; Lyu, X.; Wang, Z.; Dickey, A.M.; Wu, Z.; Chisholm, A.D.; Jin, Y. Inhibition of Axon Regeneration by Liquid-like TIAR-2 Granules. Neuron 2019, 104, 290–304.e8. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, K.; Handa, H.; Kawakami, K. Illuminating ALS Motor Neurons with Optogenetics in Zebrafish. Front. Cell Dev. Biol. 2021, 9, 640414. [Google Scholar] [CrossRef]
- Chen, C.H.; Luhur, A.; Sokol, N. Lin-28 promotes symmetric stem cell division and drives adaptive growth in the adult Drosophila intestine. Development 2015, 142, 3478–3487. [Google Scholar] [CrossRef] [PubMed]
- Stratoulias, V.; Heino, T.I.; Michon, F. Lin-28 regulates oogenesis and muscle formation in Drosophila melanogaster. PLoS ONE 2014, 9, e101141. [Google Scholar] [CrossRef]
- Moss, E.G.; Tang, L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003, 258, 432–442. [Google Scholar] [CrossRef]
- Polesskaya, A.; Cuvellier, S.; Naguibneva, I.; Duquet, A.; Moss, E.G.; Harel-Bellan, A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007, 21, 1125–1138. [Google Scholar] [CrossRef]
- De Luca, L.; Trino, S.; Laurenzana, I.; Tagliaferri, D.; Falco, G.; Grieco, V.; Bianchino, G.; Nozza, F.; Campia, V.; D’Alessio, F.; et al. Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. Cell Death Dis. 2017, 8, e2849. [Google Scholar] [CrossRef]
- Liu, X.R.; Tian, W.H.; Dong, X.Y.; Wu, X.Z.; Lv, J.X.; Wu, X.B. Effects of Lin28a and Lin28b on let-7 family activity. Bing Du Xue Bao 2011, 27, 533–541. [Google Scholar]
- Ambros, V.; Horvitz, H.R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 1984, 226, 409–416. [Google Scholar] [CrossRef]
- Moss, E.G.; Lee, R.C.; Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997, 88, 637–646. [Google Scholar] [CrossRef]
- Mitra, S.; Sharma, P.; Kaur, S.; Khursheed, M.A.; Gupta, S.; Chaudhary, M.; Kurup, A.J.; Ramachandran, R. Dual regulation of lin28a by Myc is necessary during zebrafish retina regeneration. J. Cell Biol. 2019, 218, 489–507. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, Y.; Yamamoto, J.; Iwamoto, T. The heterochronic genes lin-28a and lin-28b play an essential and evolutionarily conserved role in early zebrafish development. PLoS ONE 2014, 9, e88086. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, F.; Piacentini, M.; Fimia, G.M. The involvement of cell death and survival in neural tube defects: A distinct role for apoptosis and autophagy? Cell Death Differ. 2008, 15, 1170–1177. [Google Scholar] [CrossRef]
- Vega-Lopez, G.A.; Cerrizuela, S.; Tribulo, C.; Aybar, M.J. Neurocristopathies: New insights 150 years after the neural crest discovery. Dev. Biol. 2018, 444 (Suppl. S1), S110–S143. [Google Scholar] [CrossRef]
- Hutchins, E.J.; Piacentino, M.L.; Bronner, M.E. Transcriptomic Identification of Draxin-Responsive Targets During Cranial Neural Crest EMT. Front. Physiol. 2021, 12, 624037. [Google Scholar] [CrossRef]
- Yaniv, K.; Fainsod, A.; Kalcheim, C.; Yisraeli, J.K. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development 2003, 130, 5649–5661. [Google Scholar] [CrossRef]
- Wakamatsu, Y.; Weston, J.A. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development 1997, 124, 3449–3460. [Google Scholar] [CrossRef]
- Armas, P.; Aguero, T.H.; Borgognone, M.; Aybar, M.J.; Calcaterra, N.B. Dissecting CNBP, a zinc-finger protein required for neural crest development, in its structural and functional domains. J. Mol. Biol. 2008, 382, 1043–1056. [Google Scholar] [CrossRef]
- Weiner, A.M.; Allende, M.L.; Becker, T.S.; Calcaterra, N.B. CNBP mediates neural crest cell expansion by controlling cell proliferation and cell survival during rostral head development. J. Cell Biochem. 2007, 102, 1553–1570. [Google Scholar] [CrossRef]
- Lim, D.A.; Alvarez-Buylla, A. The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Csh Perspect. Biol. 2016, 8, a018820. [Google Scholar] [CrossRef] [PubMed]
- Frisen, J. Neurogenesis and Gliogenesis in Nervous System Plasticity and Repair. Annu. Rev. Cell Dev. Biol. 2016, 32, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Kjell, J.; Fischer-Sternjak, J.; Thompson, A.J.; Friess, C.; Sticco, M.J.; Salinas, F.; Cox, J.; Martinelli, D.C.; Ninkovic, J.; Franze, K.; et al. Defining the Adult Neural Stem Cell Niche Proteome Identifies Key Regulators of Adult Neurogenesis. Cell Stem Cell 2020, 26, 277. [Google Scholar] [CrossRef] [PubMed]
- Sakers, K.; Liu, Y.; Llaci, L.; Lee, S.M.; Vasek, M.J.; Rieger, M.A.; Brophy, S.; Tycksen, E.; Lewis, R.; Maloney, S.E.; et al. Loss of Quaking RNA binding protein disrupts the expression of genes associated with astrocyte maturation in mouse brain. Nat. Commun. 2021, 12, 1537. [Google Scholar] [CrossRef]
- Neumann, D.P.; Goodall, G.J.; Gregory, P.A. The Quaking RNA-binding proteins as regulators of cell differentiation. Wiley Interdiscip Rev. RNA 2022, e1724. [Google Scholar] [CrossRef]
- Sharma, M.; Sharma, S.; Alawada, A. Understanding the binding specificities of mRNA targets by the mammalian Quaking protein. Nucleic Acids Res. 2019, 47, 10564–10579. [Google Scholar] [CrossRef]
- Hayakawa-Yano, Y.; Suyama, S.; Nogami, M.; Yugami, M.; Koya, I.; Furukawa, T.; Zhou, L.; Abe, M.; Sakimura, K.; Takebayashi, H.; et al. An RNA-binding protein, Qki5, regulates embryonic neural stem cells through pre-mRNA processing in cell adhesion signaling. Genes Dev. 2017, 31, 1910–1925. [Google Scholar] [CrossRef]
- Takeuchi, A.; Takahashi, Y.; Iida, K.; Hosokawa, M.; Irie, K.; Ito, M.; Brown, J.B.; Ohno, K.; Nakashima, K.; Hagiwara, M. Identification of Qk as a Glial Precursor Cell Marker that Governs the Fate Specification of Neural Stem Cells to a Glial Cell Lineage. Stem Cell Rep. 2020, 15, 883–897. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, C.T.; Chen, J.; Pankratz, M.T.; Xi, J.; Li, J.; Yang, Y.; Lavaute, T.M.; Li, X.J.; Ayala, M.; et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 2010, 7, 90–100. [Google Scholar] [CrossRef]
- Siegrist, S.E.; Doe, C.Q. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 2005, 123, 1323–1335. [Google Scholar] [CrossRef]
- Wodarz, A.; Ramrath, A.; Grimm, A.; Knust, E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 2000, 150, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Laver, J.D.; Li, X.; Ancevicius, K.; Westwood, J.T.; Smibert, C.A.; Morris, Q.D.; Lipshitz, H.D. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structuRes. that confer target specificity. Nucleic Acids Res. 2013, 41, 9438–9460. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, X.; Wasser, M.; Cai, Y.; Chia, W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell 1997, 90, 437–447. [Google Scholar] [CrossRef]
- Broadus, J.; Fuerstenberg, S.; Doe, C.Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 1998, 391, 792–795. [Google Scholar] [CrossRef]
- Kim-Ha, J.; Smith, J.L.; Macdonald, P.M. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 1991, 66, 23–35. [Google Scholar] [CrossRef]
- Lai, S.L.; Doe, C.Q. Transient nuclear Prospero induces neural progenitor quiescence. Elife 2014, 3, e03363. [Google Scholar] [CrossRef]
- Matsuzaki, F.; Koizumi, K.; Hama, C.; Yoshioka, T.; Nabeshima, Y. Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem. Biophys. Res. Commun. 1992, 182, 1326–1332. [Google Scholar] [CrossRef]
- Moon, B.S.; Bai, J.; Cai, M.; Liu, C.; Shi, J.; Lu, W. Kruppel-like factor 4-dependent Staufen1-mediated mRNA decay regulates cortical neurogenesis. Nat. Commun. 2018, 9, 401. [Google Scholar] [CrossRef]
- Macchi, P.; Kroening, S.; Palacios, I.M.; Baldassa, S.; Grunewald, B.; Ambrosino, C.; Goetze, B.; Lupas, A.; St Johnston, D.; Kiebler, M. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J. Neurosci. 2003, 23, 5778–5788. [Google Scholar] [CrossRef]
- Kuc, C.A.; Brott, J.T.; Thorpe, H.H.A.; Smart, A.; Vessey, J.P. Staufen 1 is expressed by neural precursor cells in the developing murine cortex but is dispensable for NPC self-renewal and neuronal differentiation in vitro. Brain Res. 2021, 1773, 147700. [Google Scholar] [CrossRef]
- Kusek, G.; Campbell, M.; Doyle, F.; Tenenbaum, S.A.; Kiebler, M.; Temple, S. Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell 2012, 11, 505–516. [Google Scholar] [CrossRef]
- Lin, S.Y.; Johnson, S.M.; Abraham, M.; Vella, M.C.; Pasquinelli, A.; Gamberi, C.; Gottlieb, E.; Slack, F.J. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 2003, 4, 639–650. [Google Scholar] [CrossRef]
- Grosshans, H.; Johnson, T.; Reinert, K.L.; Gerstein, M.; Slack, F.J. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell 2005, 8, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Freytag, V.; Probst, S.; Hadziselimovic, N.; Boglari, C.; Hauser, Y.; Peter, F.; Gabor Fenyves, B.; Milnik, A.; Demougin, P.; Vukojevic, V.; et al. Genome-Wide Temporal Expression Profiling in Caenorhabditis elegans Identifies a Core Gene Set Related to Long-Term Memory. J. Neurosci. 2017, 37, 6661–6672. [Google Scholar] [CrossRef]
- Charest, J.; Daniele, T.; Wang, J.; Bykov, A.; Mandlbauer, A.; Asparuhova, M.; Rohsner, J.; Gutierrez-Perez, P.; Cochella, L. Combinatorial Action of Temporally Segregated Transcription Factors. Dev. Cell 2020, 55, 483–499.e487. [Google Scholar] [CrossRef]
- Li, J.; Sun, L.; Peng, X.L.; Yu, X.M.; Qi, S.J.; Lu, Z.J.; Han, J.J.; Shen, Q. Integrative genomic analysis of early neurogenesis reveals a temporal genetic program for differentiation and specification of preplate and Cajal-Retzius neurons. PLoS Genet. 2021, 17, e1009355. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.J.; Wang, C.; Sintupisut, N.; Niou, Z.X.; Lin, C.H.; Li, K.C.; Yeang, C.H. Analysis of spatial-temporal gene expression patterns reveals dynamics and regionalization in developing mouse brain. Sci. Rep. 2016, 6, 19274. [Google Scholar] [CrossRef] [PubMed]
- Bizen, N.; Inoue, T.; Shimizu, T.; Tabu, K.; Kagawa, T.; Taga, T. A growth-promoting signaling component cyclin D1 in neural stem cells has antiastrogliogenic function to execute self-renewal. Stem Cells 2014, 32, 1602–1615. [Google Scholar] [CrossRef]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef]
- Delile, J.; Rayon, T.; Melchionda, M.; Edwards, A.; Briscoe, J.; Sagner, A. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 2019, 146, dev173807. [Google Scholar] [CrossRef]
- Colombrita, C.; Silani, V.; Ratti, A. ELAV proteins along evolution: Back to the nucleus? Mol. Cell Neurosci. 2013, 56, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Antic, D.; Keene, J.D. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 1997, 61, 273–278. [Google Scholar] [CrossRef]
- Chowdhury, T.A.; Koceja, C.; Eisa-Beygi, S.; Kleinstiver, B.P.; Kumar, S.N.; Lin, C.W.; Li, K.; Prabhudesai, S.; Joung, J.K.; Ramchandran, R. Temporal and Spatial Post-Transcriptional Regulation of Zebrafish tie1 mRNA by Long Noncoding RNA During Brain Vascular Assembly. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1562–1575. [Google Scholar] [CrossRef] [PubMed]
- Zaharieva, E.; Haussmann, I.U.; Brauer, U.; Soller, M. Concentration and Localization of Coexpressed ELAV/Hu Proteins Control Specificity of mRNA Processing. Mol. Cell Biol. 2015, 35, 3104–3115. [Google Scholar] [CrossRef] [PubMed]
- Akten, B.; Kye, M.J.; Hao le, T.; Wertz, M.H.; Singh, S.; Nie, D.; Huang, J.; Merianda, T.T.; Twiss, J.L.; Beattie, C.E.; et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. USA 2011, 108, 10337–10342. [Google Scholar] [CrossRef]
- Ye, Y.; Li, M.; Gu, L.; Chen, X.; Shi, J.; Zhang, X.; Jiang, C. Chromatin remodeling during in vivo neural stem cells differentiating to neurons in early Drosophila embryos. Cell Death Differ. 2017, 24, 409–420. [Google Scholar] [CrossRef]
- Ratti, A.; Fallini, C.; Cova, L.; Fantozzi, R.; Calzarossa, C.; Zennaro, E.; Pascale, A.; Quattrone, A.; Silani, V. A role for the ELAV RNA-binding proteins in neural stem cells: Stabilization of Msi1 mRNA. J. Cell Sci. 2006, 119, 1442–1452. [Google Scholar] [CrossRef]
- Berger, C.; Renner, S.; Luer, K.; Technau, G.M. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev. Dyn. 2007, 236, 3562–3568. [Google Scholar] [CrossRef]
- Wang, F.; Tidei, J.J.; Polich, E.D.; Gao, Y.; Zhao, H.; Perrone-Bizzozero, N.I.; Guo, W.; Zhao, X. Positive feedback between RNA-binding protein HuD and transcription factor SATB1 promotes neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E4995–E5004. [Google Scholar] [CrossRef]
- Kim, D.Y. Post-transcriptional regulation of gene expression in neural stem cells. Cell Biochem. Funct. 2016, 34, 197–208. [Google Scholar] [CrossRef]
- Geng, C.; Macdonald, P.M. Imp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol. Cell Biol. 2006, 26, 9508–9516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Syed, M.H.; Mark, B.; Doe, C.Q. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. eLife 2017, 6, e26287. [Google Scholar] [CrossRef] [PubMed]
- Samuels, T.J.; Jarvelin, A.I.; Ish-Horowicz, D.; Davis, I. Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. Elife 2020, 9, e51529. [Google Scholar] [CrossRef] [PubMed]
- Dillard, C.; Narbonne-Reveau, K.; Foppolo, S.; Lanet, E.; Maurange, C. Two distinct mechanisms silence chinmo in Drosophila neuroblasts and neuroepithelial cells to limit their self-renewal. Development 2018, 145, dev154534. [Google Scholar] [CrossRef]
- Chen, Y.; Chan, J.; Chen, W.; Li, J.; Sun, M.; Kannan, G.S.; Mok, Y.K.; Yuan, Y.A.; Jobichen, C. SYNCRIP, a new player in pri-let-7a processing. RNA 2020, 26, 290–305. [Google Scholar] [CrossRef]
- Bashirullah, A.; Pasquinelli, A.E.; Kiger, A.A.; Perrimon, N.; Ruvkun, G.; Thummel, C.S. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev. Biol. 2003, 259, 1–8. [Google Scholar] [CrossRef]
- Samuels, T.J.; Arava, Y.; Jarvelin, A.I.; Robertson, F.; Lee, J.Y.; Yang, L.; Yang, C.P.; Lee, T.; Ish-Horowicz, D.; Davis, I. Neuronal upregulation of Prospero protein is driven by alternative mRNA polyadenylation and Syncrip-mediated mRNA stabilisation. Biol. Open 2020, 9, bio049684. [Google Scholar] [CrossRef]
- Thurmond, J.; Goodman, J.L.; Strelets, V.B.; Attrill, H.; Gramates, L.S.; Marygold, S.J.; Matthews, B.B.; Millburn, G.; Antonazzo, G.; Trovisco, V.; et al. FlyBase 2.0: The next generation. Nucleic Acids Res. 2019, 47, D759–D765. [Google Scholar] [CrossRef]
- Khudayberdiev, S.; Soutschek, M.; Ammann, I.; Heinze, A.; Rust, M.B.; Baumeister, S.; Schratt, G. The cytoplasmic SYNCRIP mRNA interactome of mammalian neurons. RNA Biol. 2021, 18, 1252–1264. [Google Scholar] [CrossRef]
- Weyn-Vanhentenryck, S.M.; Feng, H.; Ustianenko, D.; Duffie, R.; Yan, Q.; Jacko, M.; Martinez, J.C.; Goodwin, M.; Zhang, X.; Hengst, U.; et al. Precise temporal regulation of alternative splicing during neural development. Nat. Commun. 2018, 9, 2189. [Google Scholar] [CrossRef]
- Tomassoni-Ardori, F.; Fulgenzi, G.; Becker, J.; Barrick, C.; Palko, M.E.; Kuhn, S.; Koparde, V.; Cam, M.; Yanpallewar, S.; Oberdoerffer, S.; et al. Rbfox1 up-regulation impairs BDNF-dependent hippocampal LTP by dysregulating TrkB isoform expression levels. Elife 2019, 8, e49673. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.Y.; Cody, N.A.L.; Dominguez, D.; et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 2020, 583, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Casanovas, S.; Schlichtholz, L.; Muhlbauer, S.; Dewi, S.; Schule, M.; Strand, D.; Strand, S.; Zografidou, L.; Winter, J. Rbfox1 Is Expressed in the Mouse Brain in the Form of Multiple Transcript Variants and Contains Functional E Boxes in Its Alternative Promoters. Front. Mol. Neurosci. 2020, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, M.H.; Wu, X.; Kodani, A.; Fan, J.; Doan, R.; Ozawa, M.; Ma, J.; Yoshida, N.; Reiter, J.F.; et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 2016, 166, 1147–1162.e1115. [Google Scholar] [CrossRef]
- Pilotte, J.; Larocque, D.; Richard, S. Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev. 2001, 15, 845–858. [Google Scholar] [CrossRef]
- Darbelli, L.; Choquet, K.; Richard, S.; Kleinman, C.L. Transcriptome profiling of mouse brains with qkI-deficient oligodendrocytes reveals major alternative splicing defects including self-splicing. Sci. Rep. 2017, 7, 7554. [Google Scholar] [CrossRef]
- Hayakawa-Yano, Y.; Yano, M. An RNA Switch of a Large Exon of Ninein Is Regulated by the Neural Stem Cell Specific-RNA Binding Protein, Qki5. Int. J. Mol. Sci. 2019, 20, 1010. [Google Scholar] [CrossRef]
- Agnes, F.; Perron, M. RNA-binding proteins and neural development: A matter of targets and complexes. Neuroreport 2004, 15, 2567–2570. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Guo, Y.; Tang, C.; Han, X.; Xu, M.; Sun, J.; Zhao, Y.; Zhang, Y.; Wang, M.; Cao, X.; et al. Developmental Cytoplasmic-to-Nuclear Translocation of RNA-Binding Protein HuR Is Required for Adult Neurogenesis. Cell Rep. 2019, 29, 3101–3117.e3107. [Google Scholar] [CrossRef]
- Carelli, S.; Giallongo, T.; Rey, F.; Latorre, E.; Bordoni, M.; Mazzucchelli, S.; Gorio, M.C.; Pansarasa, O.; Provenzani, A.; Cereda, C.; et al. HuR interacts with lincBRN1a and lincBRN1b during neuronal stem cells differentiation. RNA Biol. 2019, 16, 1471–1485. [Google Scholar] [CrossRef]
- Saba, J.A.; Liakath-Ali, K.; Green, R.; Watt, F.M. Translational control of stem cell function. Nat. Rev. Mol. Cell Biol. 2021, 22, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Gabut, M.; Bourdelais, F.; Durand, S. Ribosome and Translational Control in Stem Cells. Cells 2020, 9, 497. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Yang, F.; Jin, H.; Wang, X. The regulation of protein translation and its implications for cancer. Signal Transduct. Target Ther. 2021, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Weng, R.; Cohen, S.M. Control of Drosophila Type I and Type II central brain neuroblast proliferation by bantam microRNA. Development 2015, 142, 3713–3720. [Google Scholar] [CrossRef]
- Monticelli, S.; Ansel, K.M.; Xiao, C.; Socci, N.D.; Krichevsky, A.M.; Thai, T.H.; Rajewsky, N.; Marks, D.S.; Sander, C.; Rajewsky, K.; et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005, 6, R71. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Rani, N.; Golkaram, M.; Zhou, H.R.; Alvarado, B.; Huch, K.; West, J.A.; Leyrat, A.; Pollen, A.A.; Kriegstein, A.R.; et al. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat. Neurosci. 2018, 21, 1784–1792. [Google Scholar] [CrossRef]
- Kedde, M.; van Kouwenhove, M.; Zwart, W.; Oude Vrielink, J.A.; Elkon, R.; Agami, R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010, 12, 1014–1020. [Google Scholar] [CrossRef]
- Degrauwe, N.; Schlumpf, T.B.; Janiszewska, M.; Martin, P.; Cauderay, A.; Provero, P.; Riggi, N.; Suva, M.L.; Paro, R.; Stamenkovic, I. The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell Rep. 2016, 15, 1634–1647. [Google Scholar] [CrossRef]
- Leveille, N.; Elkon, R.; Davalos, V.; Manoharan, V.; Hollingworth, D.; Oude Vrielink, J.; le Sage, C.; Melo, C.A.; Horlings, H.M.; Wesseling, J.; et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat. Commun. 2011, 2, 513. [Google Scholar] [CrossRef]
- Michlewski, G.; Caceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Lee, K.S.; Song, Y.; Gehrke, S.; Lu, B. The bantam microRNA acts through Numb to exert cell growth control and feedback regulation of Notch in tumor-forming stem cells in the Drosophila brain. PLoS Genet. 2017, 13, e1006785. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Knuckles, P.; Vogt, M.A.; Lugert, S.; Milo, M.; Chong, M.M.; Hautbergue, G.M.; Wilson, S.A.; Littman, D.R.; Taylor, V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012, 15, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Marinaro, F.; Marzi, M.J.; Hoffmann, N.; Amin, H.; Pelizzoli, R.; Niola, F.; Nicassio, F.; De Pietri Tonelli, D. MicroRNA-independent functions of DGCR8 are essential for neocortical development and TBR1 expression. EMBO Rep. 2017, 18, 603–618. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, C.; Khodadadi-Jamayran, A.; Dang, L.; Han, X.; Kim, K.; Li, H.; Zhao, R. Canonical microRNAs Enable Differentiation, Protect Against DNA Damage, and Promote Cholesterol Biosynthesis in Neural Stem Cells. Stem Cells Dev. 2017, 26, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, C.; Skamagki, M.; Khodadadi-Jamayran, A.; Zhang, W.; Kong, D.; Chang, C.W.; Feng, J.; Han, X.; Townes, T.M.; et al. Elevated p53 Activities Restrict Differentiation Potential of MicroRNA-Deficient Pluripotent Stem Cells. Stem Cell Rep. 2017, 9, 1604–1617. [Google Scholar] [CrossRef]
- Hoffmann, N.; Weise, S.C.; Marinaro, F.; Vogel, T.; De Pietri Tonelli, D. DGCR8 Promotes Neural Progenitor Expansion and Represses Neurogenesis in the Mouse Embryonic Neocortex. Front. Neurosci. 2018, 12, 281. [Google Scholar] [CrossRef]
- Romer-Seibert, J.S.; Hartman, N.W.; Moss, E.G. The RNA-binding protein LIN28 controls progenitor and neuronal cell fate during postnatal neurogenesis. FASEB J. 2019, 33, 3291–3303. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.D.; Hockemeyer, D.; Doudna, J.A.; Bateup, H.S.; Floor, S.N. Widespread Translational Remodeling during Human Neuronal Differentiation. Cell Rep. 2017, 21, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, S.; Federspiel, J.D.; Hartenian, E.; Cristea, I.M.; Glaunsinger, B. Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife 2018, 7, e37663. [Google Scholar] [CrossRef] [PubMed]
- Chau, K.F.; Shannon, M.L.; Fame, R.M.; Fonseca, E.; Mullan, H.; Johnson, M.B.; Sendamarai, A.K.; Springel, M.W.; Laurent, B.; Lehtinen, M.K. Downregulation of ribosome biogenesis during early forebrain development. Elife 2018, 7, e36998. [Google Scholar] [CrossRef] [PubMed]
- Herrlinger, S.; Shao, Q.; Yang, M.; Chang, Q.; Liu, Y.; Pan, X.; Yin, H.; Xie, L.W.; Chen, J.F. Lin28-mediated temporal promotion of protein synthesis is crucial for neural progenitor cell maintenance and brain development in mice. Development 2019, 146, dev173765. [Google Scholar] [CrossRef]
- Dinur, M.; Kilav, R.; Sela-Brown, A.; Jacquemin-Sablon, H.; Naveh-Many, T. In vitro evidence that upstream of N-ras participates in the regulation of parathyroid hormone messenger ribonucleic acid stability. Mol. Endocrinol. 2006, 20, 1652–1660. [Google Scholar] [CrossRef]
- Chang, T.C.; Yamashita, A.; Chen, C.Y.; Yamashita, Y.; Zhu, W.; Durdan, S.; Kahvejian, A.; Sonenberg, N.; Shyu, A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004, 18, 2010–2023. [Google Scholar] [CrossRef]
- Ju Lee, H.; Bartsch, D.; Xiao, C.; Guerrero, S.; Ahuja, G.; Schindler, C.; Moresco, J.J.; Yates, J.R., 3rd; Gebauer, F.; Bazzi, H.; et al. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat. Commun. 2017, 8, 1456. [Google Scholar] [CrossRef]
- Popovitchenko, T.; Park, Y.; Page, N.F.; Luo, X.; Krsnik, Z.; Liu, Y.; Salamon, I.; Stephenson, J.D.; Kraushar, M.L.; Volk, N.L.; et al. Translational derepression of Elavl4 isoforms at their alternative 5′ UTRs determines neuronal development. Nat. Commun. 2020, 11, 1674. [Google Scholar] [CrossRef]
- Zahr, S.K.; Yang, G.; Kazan, H.; Borrett, M.J.; Yuzwa, S.A.; Voronova, A.; Kaplan, D.R.; Miller, F.D. A Translational Repression Complex in Developing Mammalian Neural Stem Cells that Regulates Neuronal Specification. Neuron 2018, 97, 520–537.e526. [Google Scholar] [CrossRef]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Gouw, J.W.; Pinkse, M.W.; Vos, H.R.; Moshkin, Y.; Verrijzer, C.P.; Heck, A.J.; Krijgsveld, J. In vivo stable isotope labeling of fruit flies reveals post-transcriptional regulation in the maternal-to-zygotic transition. Mol. Cell Proteom. 2009, 8, 1566–1578. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Bluhm, A.; Casas-Vila, N.; Dinges, N.; Dejung, M.; Sayols, S.; Kreutz, C.; Roignant, J.Y.; Butter, F.; Legewie, S. Quantifying post-transcriptional regulation in the development of Drosophila melanogaster. Nat. Commun. 2018, 9, 4970. [Google Scholar] [CrossRef] [PubMed]
- Bentaya, S.; Ghogomu, S.M.; Vanhomwegen, J.; Van Campenhout, C.; Thelie, A.; Dhainaut, M.; Bellefroid, E.J.; Souopgui, J. The RNA-binding protein XSeb4R regulates maternal Sox3 at the posttranscriptional level during maternal-zygotic transition in Xenopus. Dev. Biol. 2012, 363, 362–372. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Tang, L. Liquid phase separation. Nat. Methods 2019, 16, 18. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal 2016, 14, 1. [Google Scholar] [CrossRef]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef]
- Martin, E.W.; Holehouse, A.S. Intrinsically disordered protein regions and phase separation: Sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 2020, 4, 307–329. [Google Scholar] [CrossRef]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Nozawa, R.S.; Jia, T.Z.; Saio, T.; Mori, E. Biological phase separation: Cell biology meets biophysics. Biophys. Rev. 2020, 12, 519–539. [Google Scholar] [CrossRef] [PubMed]
- Marcelo, A.; Koppenol, R.; de Almeida, L.P.; Matos, C.A.; Nobrega, C. Stress granules, RNA-binding proteins and polyglutamine diseases: Too much aggregation? Cell Death Dis. 2021, 12, 592. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, X.; Wu, X.; Zhang, M. Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications. J. Biol. Chem. 2019, 294, 14823–14835. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.; Rademakers, R.; Neumann, M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010, 9, 995–1007. [Google Scholar] [CrossRef]
- Colombrita, C.; Onesto, E.; Megiorni, F.; Pizzuti, A.; Baralle, F.E.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 2012, 287, 15635–15647. [Google Scholar] [CrossRef]
- Stronati, E.; Biagioni, S.; Fiore, M.; Giorgi, M.; Poiana, G.; Toselli, C.; Cacci, E. Wild-Type and Mutant FUS Expression Reduce Proliferation and Neuronal Differentiation Properties of Neural Stem Progenitor Cells. Int. J. Mol. Sci. 2021, 22, 7566. [Google Scholar] [CrossRef]
- Ward, C.L.; Boggio, K.J.; Johnson, B.N.; Boyd, J.B.; Douthwright, S.; Shaffer, S.A.; Landers, J.E.; Glicksman, M.A.; Bosco, D.A. A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis. 2014, 5, e1572. [Google Scholar] [CrossRef]
- Davis, R.B.; Kaur, T.; Moosa, M.M.; Banerjee, P.R. FUS oncofusion protein condensates recruit mSWI/SNF chromatin remodeler via heterotypic interactions between prion-like domains. Protein Sci. 2021, 30, 1454–1466. [Google Scholar] [CrossRef]
- Reber, S.; Jutzi, D.; Lindsay, H.; Devoy, A.; Mechtersheimer, J.; Levone, B.R.; Domanski, M.; Bentmann, E.; Dormann, D.; Muhlemann, O.; et al. The phase separation-dependent FUS interactome reveals nuclear and cytoplasmic function of liquid-liquid phase separation. Nucleic Acids Res. 2021, 49, 7713–7731. [Google Scholar] [CrossRef]
- Levone, B.R.; Lenzken, S.C.; Antonaci, M.; Maiser, A.; Rapp, A.; Conte, F.; Reber, S.; Mechtersheimer, J.; Ronchi, A.E.; Muhlemann, O.; et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J. Cell Biol. 2021, 220, e202008030. [Google Scholar] [CrossRef]
- Formicola, N.; Heim, M.; Dufourt, J.; Lancelot, A.S.; Nakamura, A.; Lagha, M.; Besse, F. Tyramine induces dynamic RNP granule remodeling and translation activation in the Drosophila brain. Elife 2021, 10, e65742. [Google Scholar] [CrossRef] [PubMed]
- Gagne, M.; Deshaies, J.E.; Sidibe, H.; Benchaar, Y.; Arbour, D.; Dubinski, A.; Litt, G.; Peyrard, S.; Robitaille, R.; Sephton, C.F.; et al. hnRNP A1B, a Splice Variant of HNRNPA1, Is Spatially and Temporally Regulated. Front. Neurosci 2021, 15, 724307. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.G.; Ohn, T.; Jang, C.H.; Vijayakumar, K.; Cho, G.W. The Role of Stress Granules in the Neuronal Differentiation of Stem Cells. Mol. Cells 2020, 43, 848–855. [Google Scholar] [CrossRef]
- Beuck, C.; Williamson, J.R.; Wuthrich, K.; Serrano, P. The acidic domain is a unique structural feature of the splicing factor SYNCRIP. Protein Sci. 2016, 25, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [PubMed]
- Notaro, A.; Messina, A.; La Bella, V. A Deletion of the Nuclear Localization Signal Domain in the Fus Protein Induces Stable Post-stress Cytoplasmic Inclusions in SH-SY5Y Cells. Front. Neurosci. 2021, 15, 759659. [Google Scholar] [CrossRef]
- Shoubridge, C.; Tan, M.H.; Fullston, T.; Cloosterman, D.; Coman, D.; McGillivray, G.; Mancini, G.M.; Kleefstra, T.; Gecz, J. Mutations in the nuclear localization sequence of the Aristaless related homeobox; sequestration of mutant ARX with IPO13 disrupts normal subcellular distribution of the transcription factor and retards cell division. Pathogenetics 2010, 3, 1. [Google Scholar] [CrossRef]
- Pesiridis, G.S.; Lee, V.M.; Trojanowski, J.Q. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009, 18, R156–R162. [Google Scholar] [CrossRef]
- Doron-Mandel, E.; Koppel, I.; Abraham, O.; Rishal, I.; Smith, T.P.; Buchanan, C.N.; Sahoo, P.K.; Kadlec, J.; Oses-Prieto, J.A.; Kawaguchi, R.; et al. The glycine arginine-rich domain of the RNA-binding protein nucleolin regulates its subcellular localization. EMBO J. 2021, 40, e107158. [Google Scholar] [CrossRef]
- Gebauer, F.; Schwarzl, T.; Valcarcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Low, Y.H.; Asi, Y.; Foti, S.C.; Lashley, T. Heterogeneous Nuclear Ribonucleoproteins: Implications in Neurological Diseases. Mol. Neuro Biol. 2021, 58, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Weynans, K.; Bossing, T.; Barros, C.S.; Berger, C. The Hippo signalling pathway maintains quiescence in Drosophila neural stem cells. Nat. Commun. 2016, 7, 10510. [Google Scholar] [CrossRef]
- Poon, C.L.; Mitchell, K.A.; Kondo, S.; Cheng, L.Y.; Harvey, K.F. The Hippo Pathway Regulates Neuroblasts and Brain Size in Drosophila melanogaster. Curr. Biol. 2016, 26, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.S.; Johnston, C.A. Mud Loss Restricts Yki-Dependent Hyperplasia in Drosophila Epithelia. J. Dev. Biol. 2020, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Lee, S.M.; Kwon, M.; Noh, H.; Lee, J.H.; Yoon, Y.; Cho, J.Y.; Yoon, K. YAP Enhances FGF2-Dependent Neural Stem Cell Proliferation by Induction of FGF Receptor Expression. Stem Cells Dev. 2020, 29, 1240–1246. [Google Scholar] [CrossRef]
- Mach, J.; Atkins, M.; Gajewski, K.M.; Mottier-Pavie, V.; Sansores-Garcia, L.; Xie, J.; Mills, R.A.; Kowalczyk, W.; Van Huffel, L.; Mills, G.B.; et al. Modulation of the Hippo pathway and organ growth by RNA processing proteins. Proc. Natl. Acad. Sci. USA 2018, 115, 10684–10689. [Google Scholar] [CrossRef]
- Katz, N.; Cohen, R.; Solomon, O.; Kaufmann, B.; Atar, O.; Yakhini, Z.; Goldberg, S.; Amit, R. Synthetic 5′ UTRs Can Either Up- or Downregulate Expression upon RNA-Binding Protein Binding. Cell Syst. 2019, 9, 93–106.e108. [Google Scholar] [CrossRef]
- Moore, K.S.; von Lindern, M. RNA Binding Proteins and Regulation of mRNA Translation in Erythropoiesis. Front. Physiol. 2018, 9, 910. [Google Scholar] [CrossRef]
- Geng, Z.; Li, P.; Tan, L.; Song, H. Targeted Knockdown of RNA-Binding Protein TIAR for Promoting Self-Renewal and Attenuating Differentiation of Mouse Embryonic Stem Cells. Stem Cells Int. 2015, 2015, 657325. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.M.; Yin, L.; Thambisetty, M.; Troncoso, J.C.; Lah, J.J.; Levey, A.I.; Seyfried, N.T. Deep proteomic network analysis of Alzheimer’s disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Mol. Neurodegener. 2018, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Kapeli, K.; Martinez, F.J.; Yeo, G.W. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum. Genet. 2017, 136, 1193–1214. [Google Scholar] [CrossRef] [PubMed]
- Jamar, N.H.; Kritsiligkou, P.; Grant, C.M. Loss of mRNA surveillance pathways results in widespread protein aggregation. Sci. Rep. 2018, 8, 3894. [Google Scholar] [CrossRef] [PubMed]
- Surguchev, A.; Surguchov, A. Conformational diseases: Looking into the eyes. Brain Res. Bull. 2010, 81, 12–24. [Google Scholar] [CrossRef]
- Shmookler Reis, R.J.; Atluri, R.; Balasubramaniam, M.; Johnson, J.; Ganne, A.; Ayyadevara, S. “Protein aggregates” contain RNA and DNA, entrapped by misfolded proteins but largely rescued by slowing translational elongation. Aging Cell 2021, 20, e13326. [Google Scholar] [CrossRef]
- Maziuk, B.; Ballance, H.I.; Wolozin, B. Dysregulation of RNA Binding Protein Aggregation in Neurodegenerative Disorders. Front. Mol. Neurosci. 2017, 10, 89. [Google Scholar] [CrossRef]
- Xue, Y.C.; Ng, C.S.; Xiang, P.; Liu, H.; Zhang, K.; Mohamud, Y.; Luo, H. Dysregulation of RNA-Binding Proteins in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2020, 13, 78. [Google Scholar] [CrossRef]
- Apicco, D.J.; Ash, P.E.A.; Maziuk, B.; LeBlang, C.; Medalla, M.; Al Abdullatif, A.; Ferragud, A.; Botelho, E.; Ballance, H.I.; Dhawan, U.; et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 2018, 21, 72–80. [Google Scholar] [CrossRef]
- Ash, P.E.A.; Lei, S.; Shattuck, J.; Boudeau, S.; Carlomagno, Y.; Medalla, M.; Mashimo, B.L.; Socorro, G.; Al-Mohanna, L.F.A.; Jiang, L.; et al. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc. Natl. Acad. Sci. USA 2021, 118, e2014188118. [Google Scholar] [CrossRef]
- LeBlang, C.J.; Medalla, M.; Nicoletti, N.W.; Hays, E.C.; Zhao, J.; Shattuck, J.; Cruz, A.L.; Wolozin, B.; Luebke, J.I. Reduction of the RNA Binding Protein TIA1 Exacerbates Neuroinflammation in Tauopathy. Front. Neurosci. 2020, 14, 285. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hu, J.; Chen, J.; Chen, Z.; Jiao, F.; Cui, J.; Quan, M.; Wang, L. RNA-binding protein Musashi2 regulates Hippo signaling via SAV1 and MOB1 in pancreatic cancer. Med. Oncol. 2020, 37, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wei, D.; Li, X.; Wang, F.; Yan, F.; Xing, Z.; Yan, Z.; Lu, H.; Zhai, D.; Ye, Z.; et al. RNA-binding protein QKI regulates contact inhibition via Yes-associate protein in ccRCC. Acta Biochim. Biophys. Sin. 2019, 51, 9–19. [Google Scholar] [CrossRef]
- Ma, X.; Wang, H.; Ji, J.; Xu, W.; Sun, Y.; Li, W.; Zhang, X.; Chen, J.; Xue, L. Hippo signaling promotes JNK-dependent cell migration. Proc. Natl. Acad. Sci. USA 2017, 114, 1934–1939. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Dansithong, W.; Figueroa, K.P.; Gandelman, M.; Scoles, D.R.; Pulst, S.M. Staufen1 in Human Neurodegeneration. Ann. Neurol. 2021, 89, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Allan, A.M.; Zong, R.; Zhang, L.; Johnson, E.B.; Schaller, E.G.; Murthy, A.C.; Goggin, S.L.; Eisch, A.J.; Oostra, B.A.; et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat. Med. 2011, 17, 559–565. [Google Scholar] [CrossRef]
- Luo, Y.; Shan, G.; Guo, W.; Smrt, R.D.; Johnson, E.B.; Li, X.; Pfeiffer, R.L.; Szulwach, K.E.; Duan, R.; Barkho, B.Z.; et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010, 6, e1000898. [Google Scholar] [CrossRef]
- Nelson, D.L.; Orr, H.T.; Warren, S.T. The unstable repeats—Three evolving faces of neurological disease. Neuron 2013, 77, 825–843. [Google Scholar] [CrossRef]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Bryant, C.D.; Yazdani, N. RNA-binding proteins, neural development and the addictions. Genes Brain Behav. 2016, 15, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, Y.; Stackpole, E.E.; Novak, A.; Gao, Y.; Zhao, Y.; Zhao, X.; Richter, J.D. Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. USA 2018, 115, E11397–E11405. [Google Scholar] [CrossRef] [PubMed]
- Homem, C.C.; Repic, M.; Knoblich, J.A. Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 2015, 16, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Brannvall, K.; Korhonen, L.; Lindholm, D. Estrogen-receptor-dependent regulation of neural stem cell proliferation and differentiation. Mol. Cell Neurosci. 2002, 21, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.; Clark, S.G.; McDermott, J.B.; Aamodt, E.; Horvitz, H.R. PAG-3, a Zn-finger transcription factor, determines neuroblast fate in C. elegans. Development 2002, 129, 1763–1774. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Hentze, M.W.; Preiss, T. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol. Metab. 2015, 26, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Inami, H.; Oiwa, K.; Murata, Y.; Sakai, S.; Komine, O.; Sobue, A.; Iguchi, Y.; Katsuno, M.; Yamanaka, K. Aggresome formation and liquid-liquid phase separation independently induce cytoplasmic aggregation of TAR DNA-binding protein 43. Cell Death Dis. 2020, 11, 909. [Google Scholar] [CrossRef]
- Libner, C.D.; Salapa, H.E.; Levin, M.C. The Potential Contribution of Dysfunctional RNA-Binding Proteins to the Pathogenesis of Neurodegeneration in Multiple Sclerosis and Relevant Models. Int. J. Mol. Sci. 2020, 21, 4571. [Google Scholar] [CrossRef]
- Hofweber, M.; Dormann, D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Alberti, S. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J. Biol. Chem. 2019, 294, 7128–7136. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Li, L.; Chin, L.S. Aggresome formation and neurodegenerative diseases: Therapeutic implications. Curr. Med. Chem. 2008, 15, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.J. Therapeutics-how to treat phase separation-associated diseases. Emerg. Top. Life Sci. 2020, 4, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Nussbacher, J.K.; Tabet, R.; Yeo, G.W.; Lagier-Tourenne, C. Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 2019, 102, 294–320. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Lee, D.; Feng, J.J.; Lochovsky, L.; Lou, S.; Rutenberg-Schoenberg, M.; Gerstein, M. RADAR: Annotation and prioritization of variants in the post-transcriptional regulome of RNA-binding proteins. Genome Biol. 2020, 21, 151. [Google Scholar] [CrossRef] [PubMed]
- Robledinos-Anton, N.; Rojo, A.I.; Ferreiro, E.; Nunez, A.; Krause, K.H.; Jaquet, V.; Cuadrado, A. Transcription factor NRF2 controls the fate of neural stem cells in the subgranular zone of the hippocampus. Redox Biol. 2017, 13, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cheng, H.; Li, X.; Lu, W.; Wang, K.; Wen, T. Regulation of neural stem cell differentiation by transcription factors HNF4-1 and MAZ-1. Mol. Neuro Biol. 2013, 47, 228–240. [Google Scholar] [CrossRef]
- Sen, S.Q.; Chanchani, S.; Southall, T.D.; Doe, C.Q. Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. Elife 2019, 8, e44036. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).