Radial Glia and Neuronal-like Ependymal Cells Are Present within the Spinal Cord of the Trunk (Body) in the Leopard Gecko (Eublepharis macularius)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Care
2.2. Tail Autotomy
2.3. 5-Bromo-2′-Deoxyuridine (BrdU) Pulse-Chase Experiments
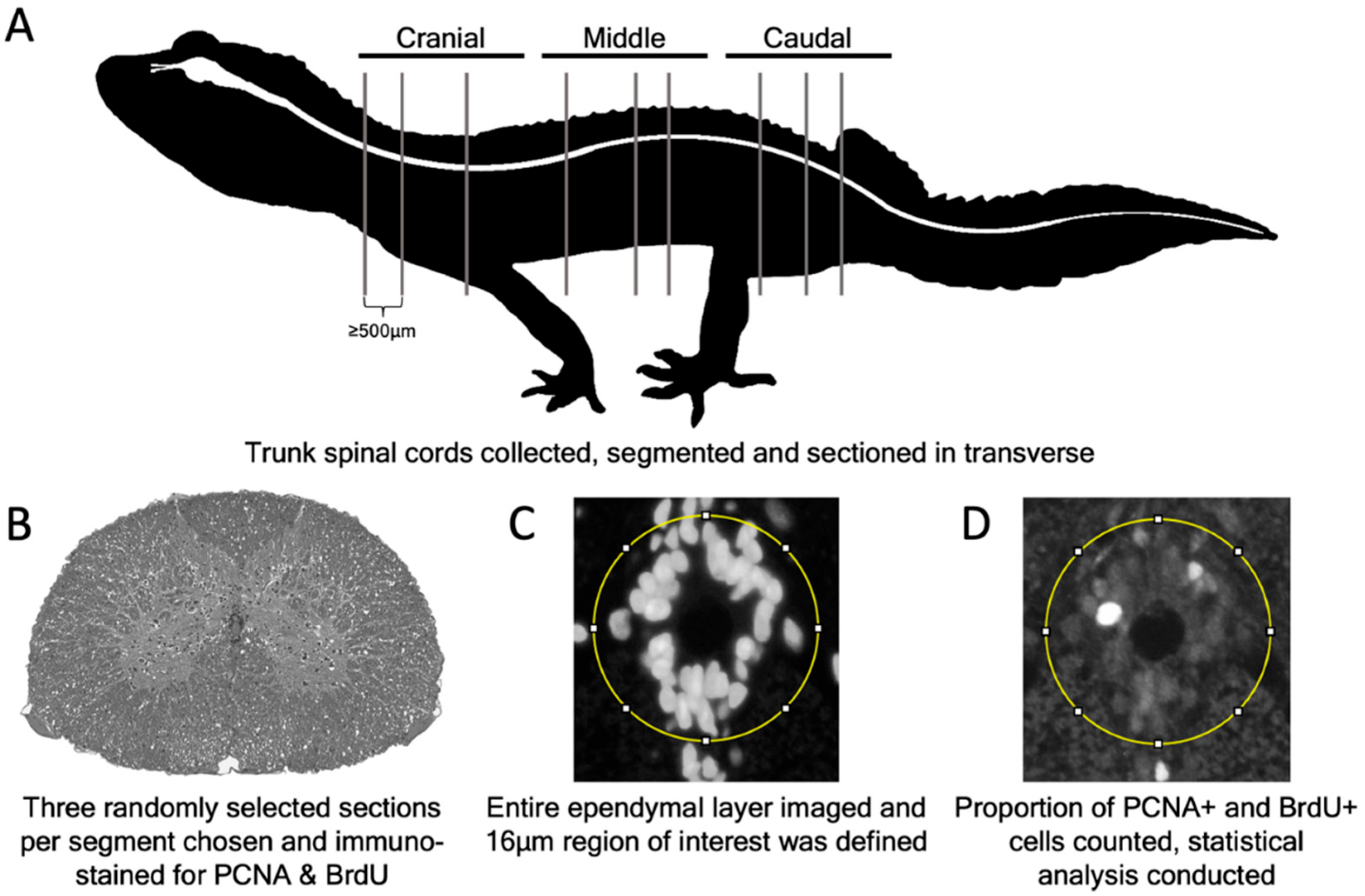
2.4. Tissue Collection and Preparation
2.5. Hematoxylin and Eosin
2.6. Immunofluorescence
2.6.1. Standard Immunofluorescence Protocol
2.6.2. Modified Immunofluorescence Protocol with Tris Retrieval
2.6.3. Modified Immunofluorescence Protocol with Trypsin Retrieval
2.7. Quantification of Slow Cycling and Proliferation Data
2.8. Statistical Analyses
3. Results
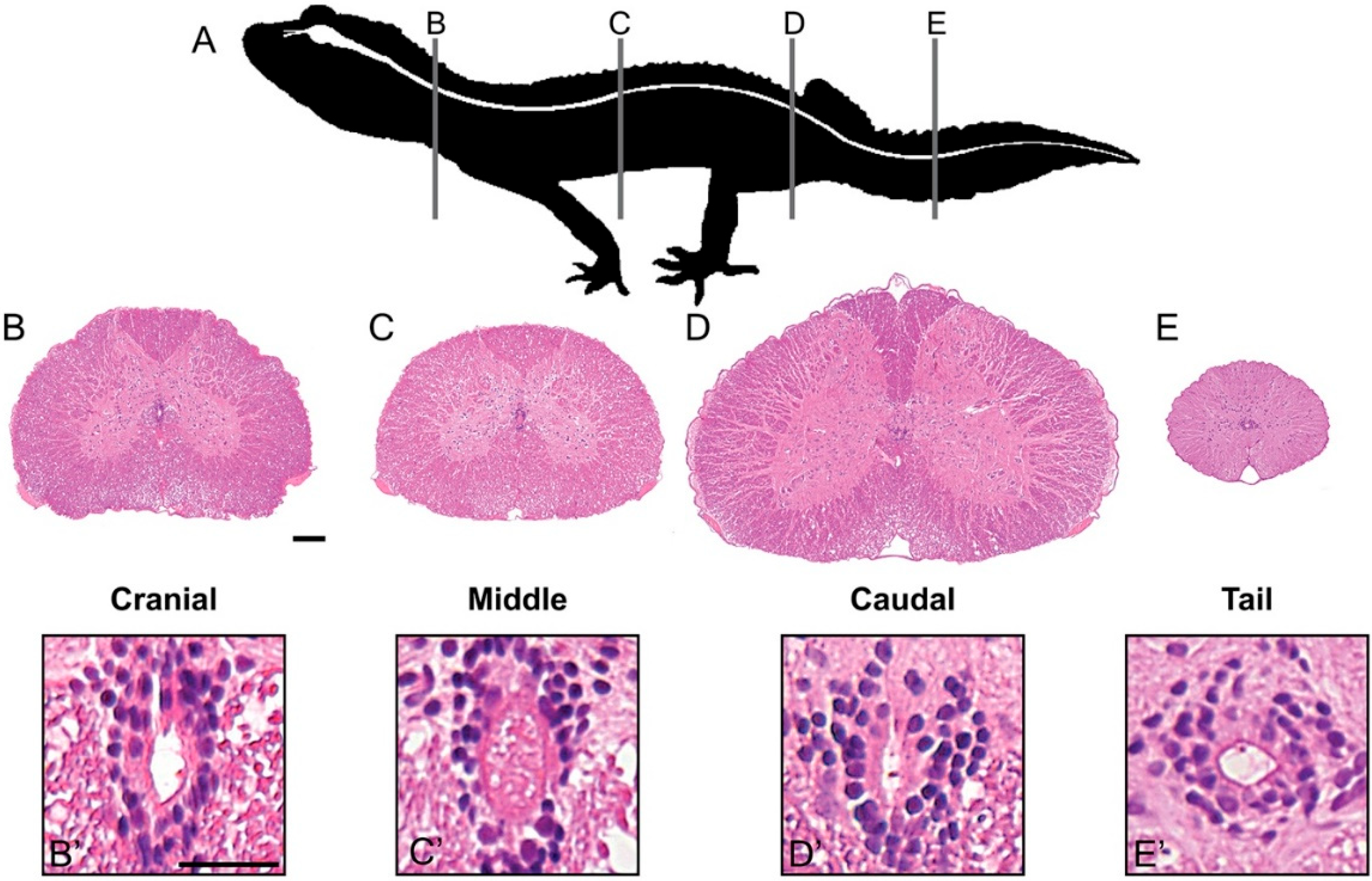
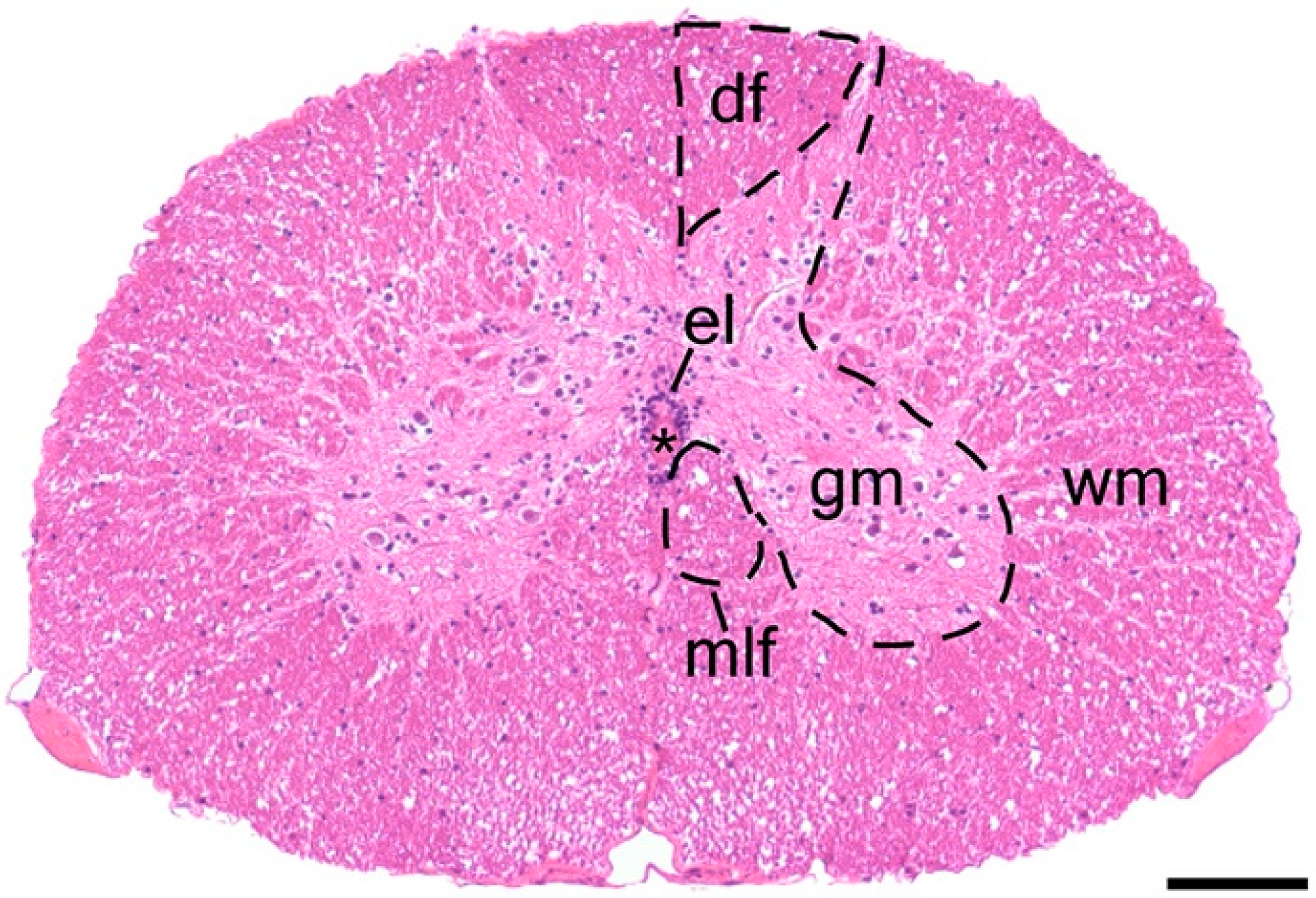
3.1. Spinal Cord Structure and Histology
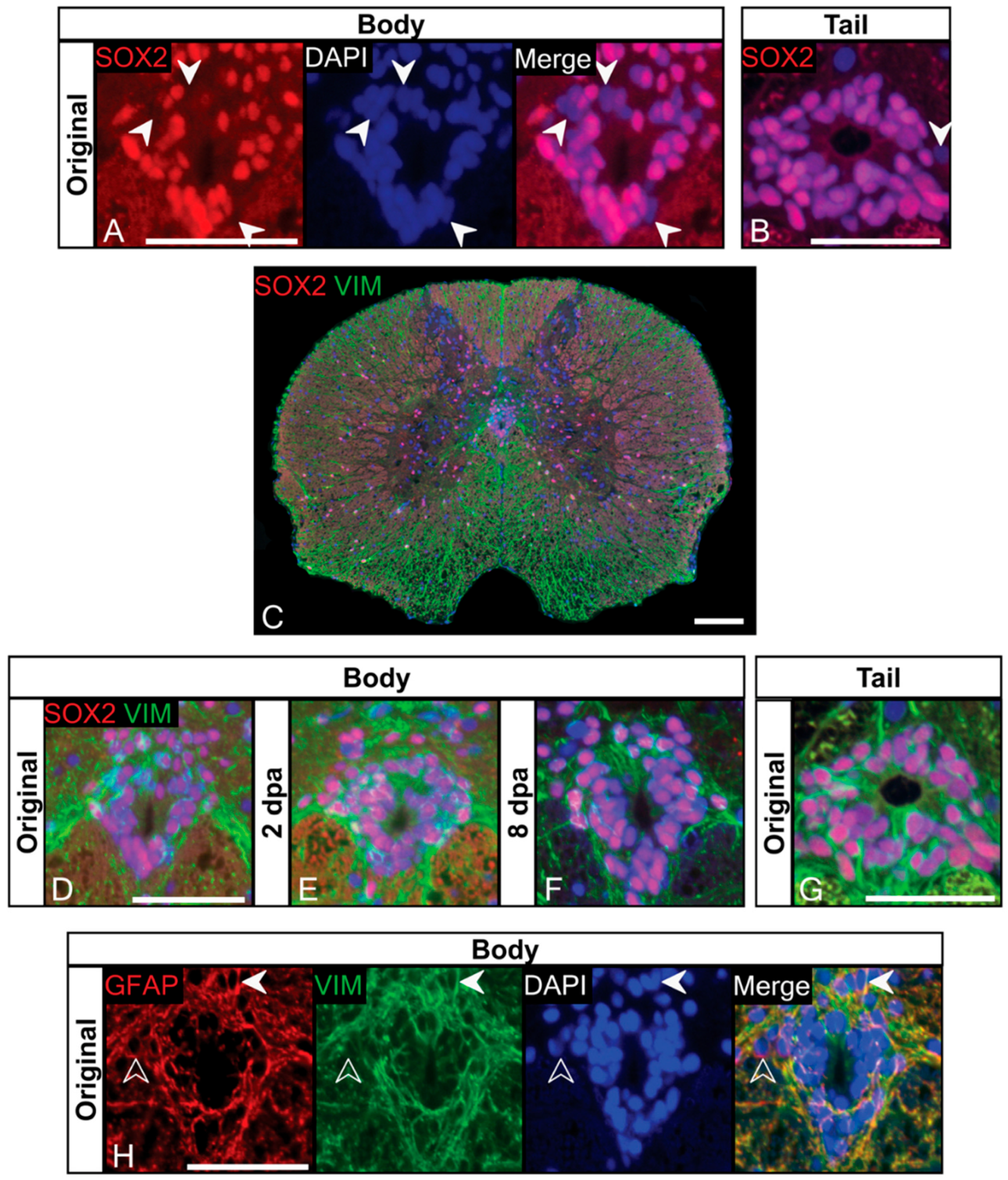
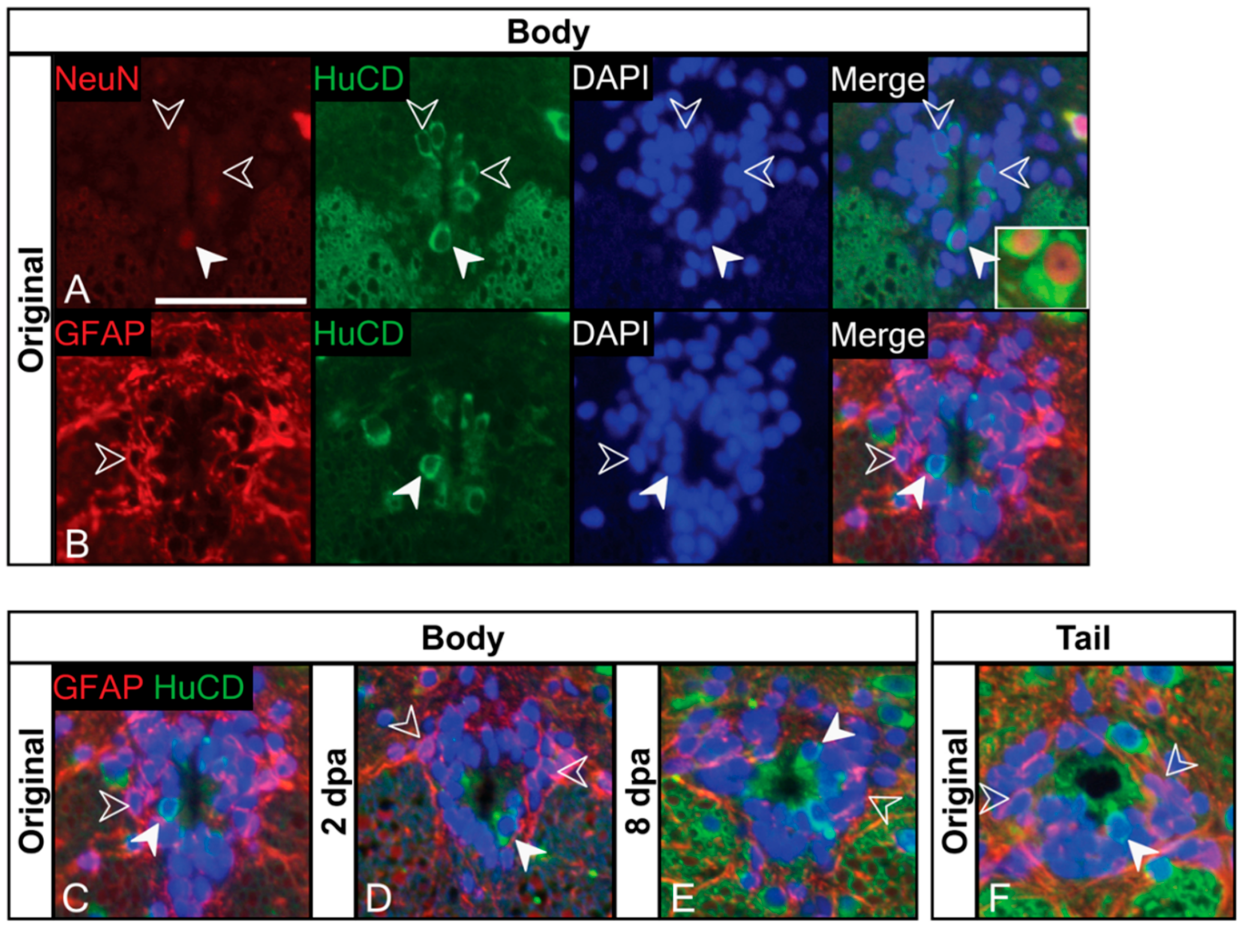
3.2. Ependymal Cells of the Trunk Spinal Cord Are a Heterogeneous Population That Includes NSPCs
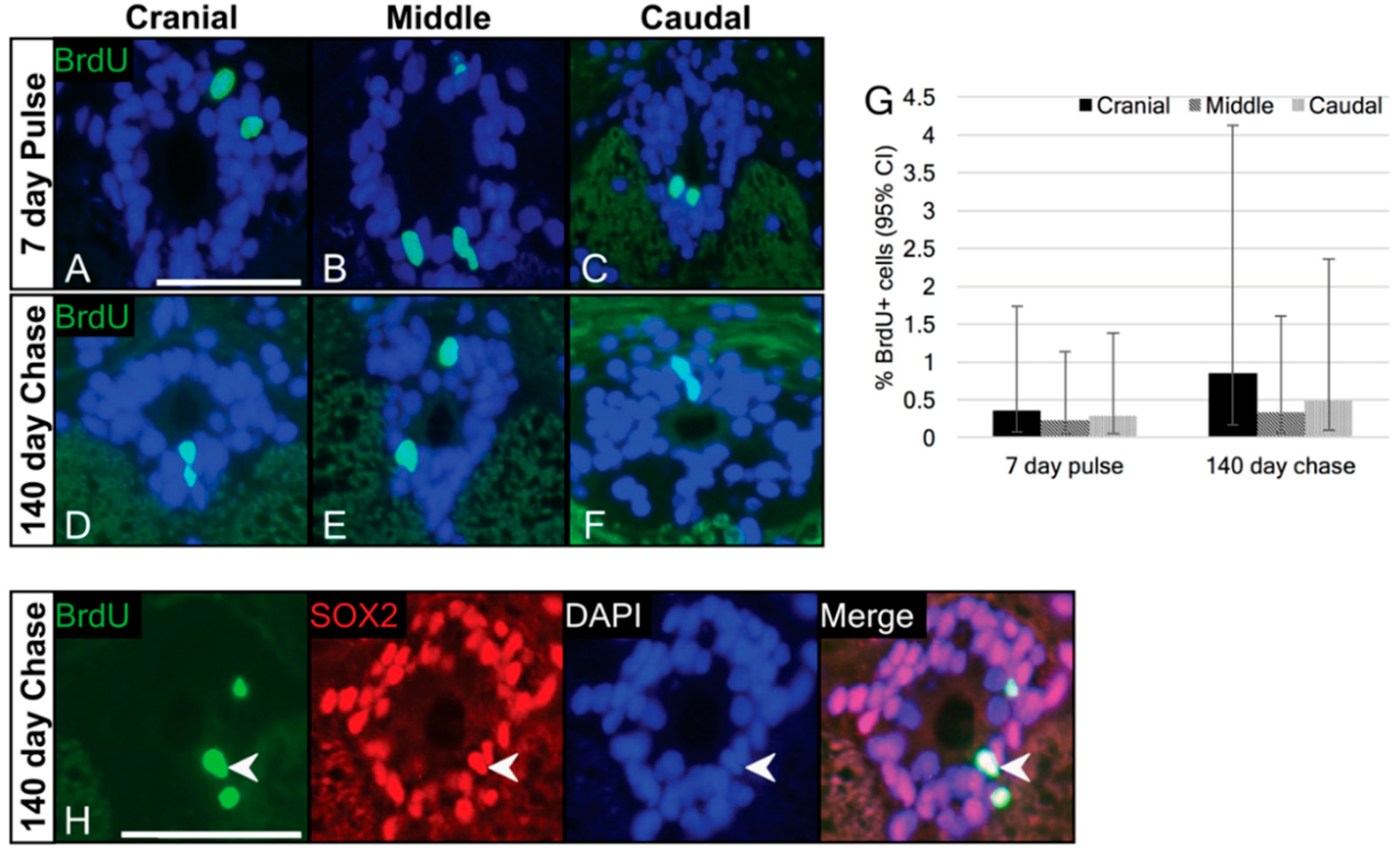
3.3. Label-Retaining Cells Are Present in the Ependymal Layer
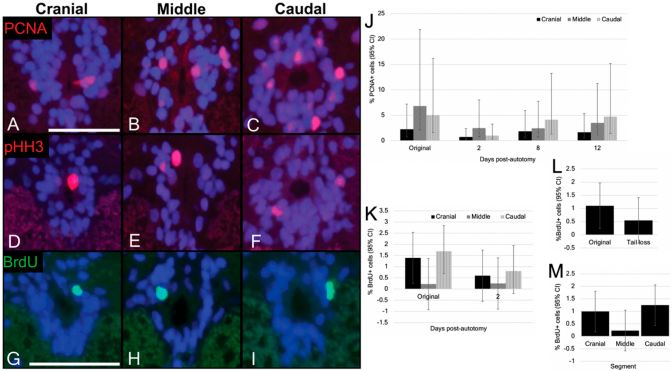
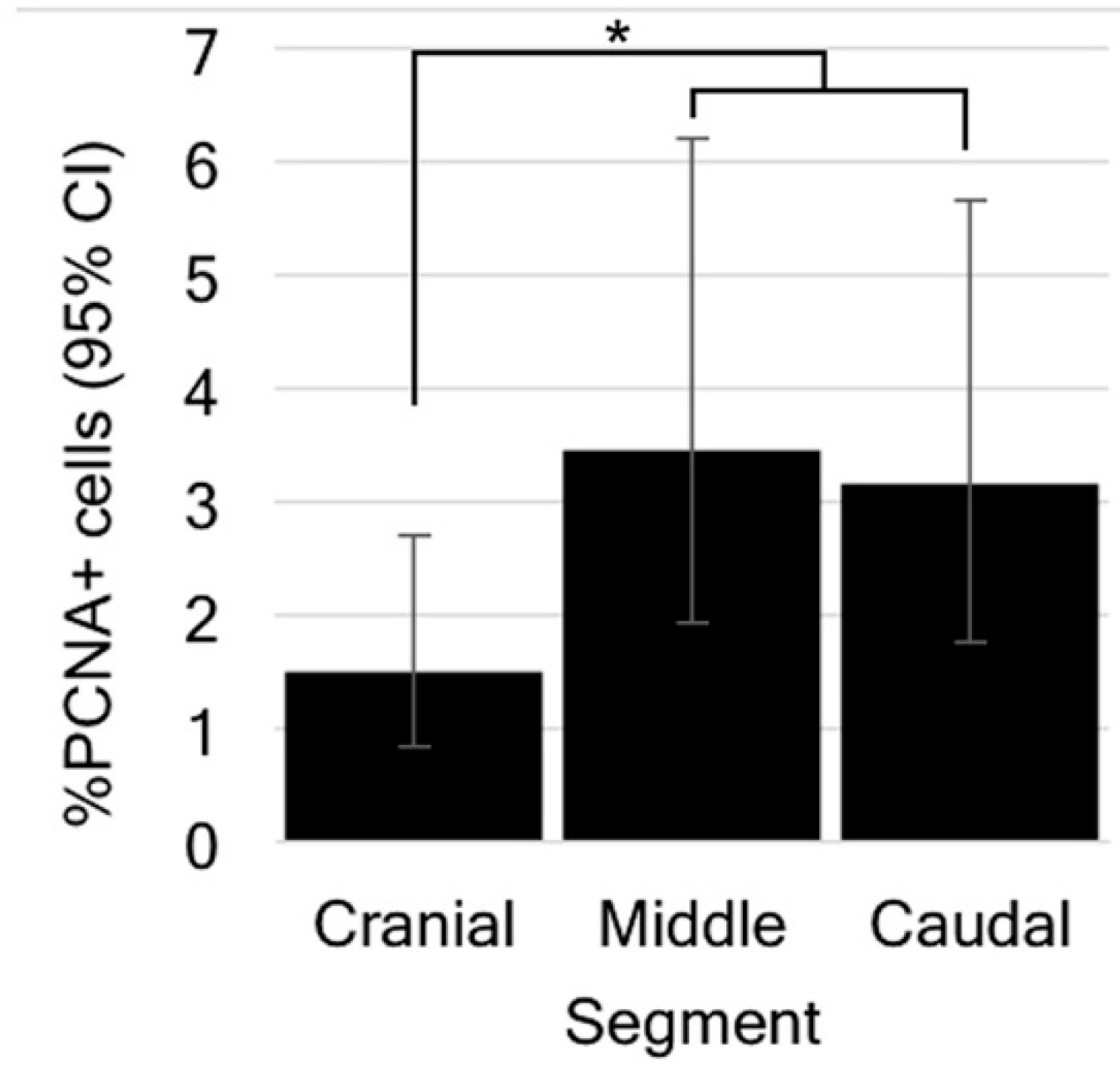
3.4. Cells of the Ependymal Layer Constitutively Proliferate
3.5. Ependymal Cell Proliferation Is Unaltered in Response to Tail Loss
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDonald, J.W.; Sadowsky, C. Spinal-cord injury. Lancet 2002, 359, 417–425. [Google Scholar] [CrossRef]
- Yiu, G.; He, Z. Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 2006, 7, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi-Abdolrezaee, S.; Billakanti, R. Reactive astrogliosis after spinal cord injury—Beneficial and detrimental effects. Mol. Neurobiol. 2012, 46, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Meletis, K.; Barnabé-Heider, F.; Carlén, M.; Evergren, E.; Tomilin, N.; Shupliakov, O.; Frisén, J. Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol. 2008, 6, e182. [Google Scholar] [CrossRef] [PubMed]
- Barnabé-Heider, F.; Göritz, C.; Sabelström, H.; Takebayashi, H.; Pfrieger, F.W.; Meletis, K.; Frisén, J. Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell. 2010, 7, 470–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mothe, A.J.; Tator, C.H. Proliferation, migration, and differentiation of endogenous ependymal region stem/progenitor cells following minimal spinal cord injury in the adult rat. Neuroscience 2005, 131, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Barnabé-Heider, F.; Frisén, J. Stem cells for spinal cord repair. Cell Stem Cell 2008, 3, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Dawley, E.M.; Samson, S.O.; Woodard, K.T.; Matthias, K.A. Spinal cord regeneration in a tail autotomizing urodele. J. Morphol. 2012, 273, 211–225. [Google Scholar] [CrossRef]
- Szarek, D.; Marycz, K.; Lis, A.; Zawada, Z.; Tabakow, P.; Laska, J.; Jarmundowicz, W. Lizard tail spinal cord: A new experimental model of spinal cord injury without limb paralysis. FASEB J. 2015, 30, 1391–1403. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, E.A.B.; Vickaryous, M.K. Neural stem/progenitor cells are activated during tail regeneration in the leopard gecko (Eublepharis macularius). J. Comp. Neurol. 2018, 526, 285–309. [Google Scholar] [CrossRef]
- Sun, A.X.; Londono, R.; Hudall, M.L.; Tuan, R.S.; Lozito, T.P. Differences in neural stem identity and differentiation capacity drive divergent regenerative outcomes in lizards and salamanders. Proc. Natl. Acad. Sci. USA 2018, 115, E8256–E8265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimer, M.M.; Sörensen, I.; Kuscha, V.; Frank, R.E.; Liu, C.; Becker, C.G.; Becker, T. Motor neuron regeneration in adult zebrafish. J. Neurosci. 2008, 28, 8510–8516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monaghan, J.R.; Walker, J.A.; Page, R.B.; Putta, S.; Beachy, C.K.; Voss, S.R. Early gene expression during natural spinal cord regeneration in the salamander Ambystoma mexicanum. J. Neurochem. 2007, 101, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Q.; Li, D.; Zhao, L.; Wang, Y.; Liu, M.; Gu, X.; Liu, Y. Early neurogenesis during caudal spinal cord regeneration in adult Gekko japonicus. J. Mol. Histol. 2013, 44, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Alibardi, L. Observations on lumbar spinal cord recovery after lesion in lizards indicates regeneration of a cellular and fibrous bridge reconnecting the injured cord. J. Dev. Biol. 2014, 2, 210–229. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.W.; Chen, Y.R.; Wang, Y.H.; Hung, K.C.; Lin, S.M. Tail regeneration after autotomy revives survival: A case from a long-term monitored lizard population under avian predation. Proc. R. Soc. B. 2017, 284, 20162538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozito, T.P.; Londono, R.; Sun, A.X.; Hudnall, M.L. Introducing dorsoventral patterning in adult regenerating lizard tails with gene-edited embryonic neural stem cells. Nat. Commun. 2021, 12, 6010. [Google Scholar] [CrossRef]
- Alibardi, L. H3-thymidine labeled cerebrospinal fluid contacting cells in the regenerating caudal spinal cord of the lizard Lampropholis. Ann. Anat. 1994, 176, 347–356. [Google Scholar] [CrossRef]
- Jacyniak, K.; McDonald, R.P.; Vickaryous, M.K. Tail regeneration and other phenomena of wound healing and tissue restoration in lizards. J. Exp. Biol. 2017, 220, 2858–2869. [Google Scholar] [CrossRef] [Green Version]
- Briona, L.K.; Dorsky, R.I. Radial glial progenitors repair the zebrafish spinal cord following transection. Exp. Neurol. 2014, 256, 81–92. [Google Scholar] [CrossRef] [Green Version]
- Than-Trong, E.; Bally-Cuif, L. Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia 2015, 63, 1406–1428. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.; Barragan, J.; Bashiruddin, S.; Smith, C.J.; Tyrrell, C.; Parsons, M.J.; Doris, R.; Kucenas, S.; Downes, G.B.; Velez, C.M.; et al. Gfap-positive radial glial cells are an essential progenitor population for later-born neurons and glia in the zebrafish spinal cord. Glia 2016, 64, 1170–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzari, M.; Franceschini, V. Astroglial cells in the central nervous system of the adult brown anole lizard, Anolis sagrei, revealed by intermediate filament immunohistochemistry. J. Morphol. 2005, 265, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, E.M.; Ferretti, P. Considering the evolution of regeneration in the central nervous system. Nat. Rev. Neurosci. 2009, 10, 713. [Google Scholar] [CrossRef]
- Becker, C.G.; Becker, T. Neuronal regeneration from ependymo-radial glial cells: Cook, little pot, cook! Dev. Cell 2015, 32, 516–527. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Buylla, A.; García-Verdugo, J.M.; Tramontin, A.D. A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2001, 2, 287. [Google Scholar] [CrossRef]
- Doetsch, F. The glial identity of neural stem cells. Nat. Neurosci. 2003, 6, 1127. [Google Scholar] [CrossRef]
- Kriegstein, A.; Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009, 32, 149–184. [Google Scholar] [CrossRef] [Green Version]
- Grandel, H.; Kaslin, J.; Ganz, J.; Wenzel, I.; Brand, M. Neural stem cells and neurogenesis in the adult zebrafish brain: Origin, proliferation dynamics, migration and cell fate. Dev. Biol. 2006, 295, 263–277. [Google Scholar] [CrossRef] [Green Version]
- Chapouton, P.; Skupien, P.; Hesl, B.; Coolen, M.; Moore, J.C.; Madelaine, R.; Kremmer, E.; Faus-Kessler, T.; Blader, P.; Lawson, N.D.; et al. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 2010, 30, 7961–7974. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, E. The tortoise and the hair: Slow-cycling cells in the stem cell race. Cell 2009, 137, 811–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, S.P.; Nag, T.C.; Ghosh, S. Characterization of proliferating neural progenitors after spinal cord injury in adult zebrafish. PLoS ONE 2015, 10, e0143595. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, V.; Khudyakov, J.; Ellis, P.; Pevny, L. SOX2 functions to maintain neural progenitor identity. Neuron 2003, 39, 749–765. [Google Scholar] [CrossRef] [Green Version]
- Oudega, M.; Marani, E. Expression of vimentin and glial fibrillary acidic protein in the developing rat spinal cord: An immunocytochemical study of the spinal cord glial system. J. Anat. 1991, 179, 97. [Google Scholar]
- Dervan, A.G.; Roberts, B.L. Reaction of spinal cord central canal cells to cord transection and their contribution to cord regeneration. J. Comp. Neurol. 2003, 58, 293–306. [Google Scholar] [CrossRef]
- Tapia, C.; Kutzner, H.; Mentzel, T.; Savic, S.; Baumhoer, D.; Glatz, K. Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am. J. Surg. Pathol. 2006, 30, 83–89. [Google Scholar] [CrossRef]
- Halbach, O.V. Immunohistological markers for proliferative events, gliogenesis, and neurogenesis within the adult hippocampus. Cell Tissue Res. 2011, 345, 1–19. [Google Scholar] [CrossRef]
- Roberts, B.L.; Maslam, S.; Scholten, G.; Smit, W. Dopaminergic and GABAergic cerebrospinal fluid-contacting neurons along the central canal of the spinal cord of the eel and trout. J. Comp. Neurol. 1995, 354, 423–437. [Google Scholar] [CrossRef]
- Marichal, N.; García, G.; Radmilovich, M.; Trujillo-Cenóz, O.; Russo, R.E. Enigmatic central canal contacting cells: Immature neurons in “standby mode”? J. Neurosci. 2009, 29, 10010–10024. [Google Scholar] [CrossRef] [Green Version]
- Djenoune, L.; Khabou, H.; Joubert, F.; Quan, F.B.; Nunes Figueiredo, S.; Bodineau, L.; Del Bene, F.; Burcklé, C.; Tostivint, H.; Wyart, C. Investigation of spinal cerebrospinal fluid-contacting neurons expressing PKD2L1: Evidence for a conserved system from fish to primates. Front. Neuroanat. 2014, 8, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petracca, Y.L.; Sartoretti, M.M.; Di Bella, D.J.; Marin-Burgin, A.; Carcagno, A.L.; Schinder, A.F.; Lanuza, G.M. The late and dual origin of cerebrospinal fluid-contacting neurons in the mouse spinal cord. Development 2016, 143, 880–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaro-Cervello, C.; Soriano-Navarro, M.; Mirzadeh, Z.; Alvarez-Buylla, A.; Garcia-Verdugo, J.M. Biciliated ependymal cell proliferation contributes to spinal cord growth. J. Comp. Neurol. 2012, 520, 3528–3552. [Google Scholar] [CrossRef] [Green Version]
- Russo, R.E.; Fernández, A.; Reali, C.; Radmilovich, M.; Trujillo-Cenóz, O. Functional and molecular clues reveal precursor-like cells and immature neurones in the turtle spinal cord. J. Physiol. 2004, 560, 831–838. [Google Scholar] [CrossRef] [Green Version]
- Cruce, W.L.R. Spinal Cord in Lizards. In Biology of Reptilia; Gans, C., Northcutt, R.G., Ulinkski, P., Eds.; Academic Press Inc.: New York, NY, USA, 1979; Volume 10, pp. 111–131. [Google Scholar]
- Ogai, K.; Nakatani, K.; Hisano, S.; Sugitani, K.; Koriyama, Y.; Kato, S. Function of Sox2 in ependymal cells of lesioned spinal cords in adult zebrafish. Neurosci. Res. 2014, 88, 84–87. [Google Scholar] [CrossRef] [Green Version]
- Felix, M.S.; Popa, N.; Djelloul, M.; Boucraut, J.; Gauthier, P.; Bauer, S.; Matarazzo, V.A. Alteration of forebrain neurogenesis after cervical spinal cord injury in the adult rat. Front. Neurosci. 2012, 6, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaete, M.; Muñoz, R.; Sánchez, N.; Tampe, R.; Moreno, M.; Contreras, E.G.; Lee-Liu, D.; Larraín, J. Spinal cord regeneration in Xenopus tadpoles proceeds through activation of Sox2-positive cells. Neural Dev. 2012, 7, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, S.P.; Sengupta, D.; Lee, S.G.; Sen, T.; Kundu, S.; Mathavan, S.; Ghosh, S. Genome wide expression profiling during spinal cord regeneration identifies comprehensive cellular responses in zebrafish. PLoS ONE 2014, 9, e84212. [Google Scholar] [CrossRef]
- Brazel, C.Y.; Limke, T.L.; Osborne, J.K.; Miura, T.; Cai, J.; Pevny, L.; Rao, M.S. Sox2 expression defines a heterogeneous population of neurosphere-forming cells in the adult murine brain. Aging Cell 2005, 4, 197–207. [Google Scholar] [CrossRef]
- Lee, H.J.; Wu, J.; Chung, J.; Wrathall, J.R. SOX2 expression is upregulated in adult spinal cord after contusion injury in both oligodendrocyte lineage and ependymal cells. J. Neurosci. Res. 2013, 91, 196–210. [Google Scholar] [CrossRef]
- Bani-Yaghoub, M.; Tremblay, R.G.; Lei, J.X.; Zhang, D.; Zurakowski, B.; Sandhu, J.K.; Smith, B.; Ribecco-Lutkiewicz, M.; Kennedy, J.; Walker, P.R.; et al. Role of Sox2 in the development of the mouse neocortex. Dev. Biol. 2006, 295, 52–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutton, S.R.; Pevny, L.H. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev. Biol. 2011, 352, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavallaro, M.; Mariani, J.; Lancini, C.; Latorre, E.; Caccia, R.; Gullo, F.; Valotta, M.; DeBiasi, S.; Spinardi, L.; Ronchi, A.; et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 2008, 135, 541–557. [Google Scholar] [CrossRef] [Green Version]
- Ferri, A.L.; Cavallaro, M.; Braida, D.; Di Cristofano, A.; Canta, A.; Vezzani, A.; Ottolenghi, S.; Pandolfi, P.P.; Sala, M.; DeBiasi, S.; et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 2004, 131, 3805–3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taranova, O.V.; Magness, S.T.; Fagan, B.M.; Wu, Y.; Surzenko, N.; Hutton, S.R.; Pevny, L.H. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006, 20, 1187–1202. [Google Scholar] [CrossRef] [Green Version]
- Schnitzer, J.; Franke, W.W.; Schachner, M. Immunocytochemical demonstration of vimentin in astrocytes and ependymal cells of developing and adult mouse nervous system. J. Cell Biol. 1981, 90, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Tapscott, S.J.; Bennett, G.S.; Toyama, Y.; Kleinbart, F.; Holtzer, H. Intermediate filament proteins in the developing chick spinal cord. Dev. Biol. 1981, 86, 40–54. [Google Scholar] [CrossRef]
- Dent, J.A.; Polson, A.G.; Klymkowsky, M.W. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development 1989, 105, 61–74. [Google Scholar] [CrossRef]
- O’Hara, C.M.; Egar, M.W.; Chernoff, E.A. Reorganization of the ependyma during axolotl spinal cord regeneration: Changes in intermediate filament and fibronectin expression. Dev. Dyn. 1992, 193, 103–115. [Google Scholar] [CrossRef]
- Zamora, A.J.; Mutin, M. Vimentin and glial fibrillary acidic protein filaments in radial glia of the adult urodele spinal cord. Neuroscience 1988, 27, 279–288. [Google Scholar] [CrossRef]
- Lazzari, M.; Franceschini, V. Glial fibrillary acidic protein and vimentin immunoreactivity of astroglial cells in the central nervous system of adult Podarcis sicula (Squamata, Lacertidae). J. Anat. 2001, 198, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.J.; Potten, C.S. Highly persistent label-retaining cells in the hair follicles of mice and their fate following induction of anagen. J. Invest. Dermatol. 1999, 112, 470–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeshima, A.; Yamashita, S.; Nojima, Y. Identification of renal progenitor-like tubular cells that participate in the regeneration processes of the kidney. J. Am. Soc. Nephrol. 2003, 14, 3138–3146. [Google Scholar] [CrossRef] [Green Version]
- Tumbar, T.; Guasch, G.; Greco, V.; Blanpain, C.; Lowry, W.E.; Rendl, M.; Fuchs, E. Defining the epithelial stem cell niche in skin. Science 2004, 303, 359–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuwahara, R.; Kofman, A.V.; Landis, C.S.; Swenson, E.S.; Barendswaard, E.; Theise, N.D. The hepatic stem cell niche: Identification by label-retaining cell assay. Hepatology 2008, 47, 1994–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morshead, C.M.; Reynolds, B.A.; Craig, C.G.; McBurney, M.W.; Staines, W.A.; Morassutti, D.; Weiss, S.; van der Kooy, D. Neural stem cells in the adult mammalian forebrain: A relatively quiescent subpopulation of subependymal cells. Neuron 1994, 13, 1071–1082. [Google Scholar] [CrossRef]
- Codega, P.; Silva-Vargas, V.; Paul, A.; Maldonado-Soto, A.R.; DeLeo, A.M.; Pastrana, E.; Doetsch, F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 2014, 82, 545–559. [Google Scholar] [CrossRef] [Green Version]
- März, M.; Chapouton, P.; Diotel, N.; Vaillant, C.; Hesl, B.; Takamiya, M.; Lam, C.S.; Kah, O.; Bally-Cuif, L.; Strähle, U. Heterogeneity in progenitor cell subtypes in the ventricular zone of the zebrafish adult telencephalon. Glia 2010, 58, 870–888. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; Van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef]
- Taupin, P. BrdU immunohistochemistry for studying adult neurogenesis: Paradigms, pitfalls, limitations, and validation. Brain Res. Rev. 2007, 53, 198–214. [Google Scholar] [CrossRef]
- Zacchetti, A.; Van Garderen, E.; Teske, E.; Nederbragt, H.; Dierendonck, J.H.; Rutteman, G.R. Validation of the use of proliferation markers in canine neoplastic and non-neoplastic tissues: Comparison of KI-67 and proliferating cell nuclear antigen (PCNA) expression versus in vivo bromodeoxyuridine labelling by immunohistochemistry. APMIS 2003, 111, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Cooper-Kuhn, C.M.; Kuhn, H.G. Is it all DNA repair?: Methodological considerations for detecting neurogenesis in the adult brain. Dev. Brain Res. 2002, 134, 13–21. [Google Scholar] [CrossRef]
- Bauer, S.; Patterson, P.H. The cell cycle–apoptosis connection revisited in the adult brain. J. Cell Biol. 2005, 171, 641–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, A.R.; Smith, G.T. Spinal transection induces widespread proliferation of cells along the length of the spinal cord in a weakly electric fish. Brain Behav. Evol. 2012, 80, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Blasko, J.; Martoncikova, M.; Lievajova, K.; Saganova, K.; Korimova, A.; Racekova, E. Regional differences of proliferation activity in the spinal cord ependyma of adult rats. Cent. Eur. J. Biol. 2012, 7, 397–403. [Google Scholar] [CrossRef]
- Lacroix, S.; Hamilton, L.K.; Vaugeois, A.; Beaudoin, S.; Breault-Dugas, C.; Pineau, I.; Lévesque, S.A.; Grégoire, C.-A.; Fernandes, K.J.L. Central canal ependymal cells proliferate extensively in response to traumatic spinal cord injury but not demyelinating lesions. PLoS ONE 2014, 9, e85916. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.; Bateman, J.; DiTommaso, T.; Wong, A.Y.; Whited, J.L. Systemic cell cycle activation is induced following complex tissue injury in axolotl. Dev. Biol. 2018, 433, 461–472. [Google Scholar] [CrossRef]
- Orts-Del’Immagine, A.; Wyart, C. Cerebrospinal-fluid-contacting neurons. Curr. Biol. 2017, 27, R1198–R1200. [Google Scholar] [CrossRef] [Green Version]
- Zamora, A.J. The ependymal and glial configuration in the spinal cord of urodeles. Anat. Embryol. 1978, 154, 67–82. [Google Scholar] [CrossRef]
- Kútna, V.; Ševc, J.; Gombalová, Z.; Matiašová, A.; Daxnerová, Z. Enigmatic cerebrospinal fluid-contacting neurons arise even after the termination of neurogenesis in the rat spinal cord during embryonic development and retain their immature-like characteristics until adulthood. Acta Histochem. 2014, 116, 278–285. [Google Scholar] [CrossRef]
- Jalalvand, E.; Robertson, B.; Wallén, P.; Grillner, S. Ciliated neurons lining the central canal sense both fluid movement and pH through ASIC3. Nat. Commun. 2016, 7, 10002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alibardi, L. Electron microscopic observations on the myelination of the long-term regenerated caudal spinal cord in lizards and Sphenodon. Biol. Struct. Morphol. 1990, 3, 147–158. [Google Scholar]
- Simpson, S.B.; Duffy, M. The lizard spinal cord: A model system for the study of spinal cord injury and repair. Prog. Brain Res. 1994, 103, 229–241. [Google Scholar] [PubMed]
- Alibardi, L.; Wibel, R.; Simpson, S.B., Jr. Scanning electron microscopic observations on the central canal of the regenerating tail spinal cord in lizards. Boll. Zool. 1993, 60, 245–252. [Google Scholar] [CrossRef]
- Alibardi, L.; Gibbons, J.; Simpson, S.B., Jr. 3H-GABA administration during tail regeneration of lizards and autoradiographycal localization. J. Hirnforsch. 1993, 34, 67–77. [Google Scholar]
- Alibardi, L. Cerebrospinal fluid-contacting neurons in the regenerating spinal cord of lizards and amphibians are likely mechanoreceptors. J. Morphol. 2019, 280, 1292–1308. [Google Scholar] [CrossRef]
| Antigen | Retrieval | Block | Primary | Secondary |
|---|---|---|---|---|
| BrdU (Anti-5-bromo-2′-deoxyuridine) | Citrate buffer 12 min at 95 °C, 20 min at room temperature in solution 0.1% trypsin in PBS 20 min at 37 °C | 5% NGS in diluent 30 min at 37 °C | 1:100 in diluent overnight at 4 °C (DSHB, mouse monoclonal, G3G4, RRID: AB_2618097) | 1:200 in sterile 1XPBS for 1 h at room temperature (Goat anti-Mouse AlexaFluor 488, Life Technologies, A11001, RRID: AB_2534069) |
| GFAP (Anti-glial fibrillary acidic protein) | Citrate buffer 12 min at 95 °C, 20 min at room temperature in solution 0.1% trypsin in PBS 20 min at 37 °C | 5% NGS in diluent 30 min at 37 °C | 1:400 in diluent overnight at 4 °C (DAKO, rabbit polyclonal, Z0334, RRID: AB_10013382) | 1:1000 in 1XPBS for 1 h at room temperature (Cy3-conjugated Goat anti-Rabbit IgG, 111-165-144, RRID: AB_2338006) |
| HuCD (Anti-human neuronal protein HuC/HuD) | Tris buffer 30 min at 95 °C, 30 min at room temperature in solution | 10% NGS in 0.3% Triton-X-100 in 1XPBS 30 min at room temperature | 1:10 in 1%BSA in 1XPBS overnight at 4 °C (Molecular Probes, mouse monoclonal, 16A11, RRID: AB_221448) | 1:500 in sterile 1XPBS for 1 h at room temperature (Goat anti-Mouse AlexaFluor 488, Life Technologies, A11001, RRID: AB_2534069) |
| NeuN (Anti-neuronal nuclei) | Tris buffer 30 min at 95 °C, 30 min at room temperature in solution | 10% NGS in 0.3% Triton-X-100 in 1XPBS 30 min at room temperature | 1:500 in 1%BSA in 1XPBS overnight at 4 °C (Abcam, rabbit polyclonal, ab104225, RRID: AB_10711153) | 1:1000 in 1XPBS for 1 h at room temperature (Cy3-conjugated Goat anti-Rabbit IgG, 111-165-144, RRID: AB_2338006) |
| PCNA (Anti-proliferating cell nuclear antigen) | none | 3% NGS in 1XPBS 1 h at room temperature | 1:100 in1XPBS overnight at 4 °C (Santa Cruz Biotech., rabbit polyclonal, sc-7907, RRID: AB_2160375) | 1:200 in 1XPBS for 1 h at room temperature (Cy3-conjugated Goat anti-Rabbit IgG, 111-165-144, RRID: AB_2338006) |
| pHH3 (Anti-phospho-histone H3 (Ser10)) | Citrate buffer 12 min at 95 °C, 20 min at room temperature in solution | 3% NGS in 1XPBS 1 h at room temperature | 1:100 in1XPBS overnight at 4 °C (Cell Signaling, rabbit polyclonal, 3377S, RRID: AB_1549592) | 1:250 in 1XPBS for 1 h at room temperature (Cy3-conjugated Goat anti-Rabbit IgG, 111-165-144, RRID: AB_2338006) |
| SOX2 (Anti-SRY (sex-determining box region Y) box2) | Citrate buffer 12 min at 95 °C, 20 min at room temperature in solution 0.1% trypsin in PBS 20 min at 37 °C | 5% NGS in diluent 30 min at 37 °C | 1:50 in diluent overnight at 4 °C (Cell Signaling, rabbit polyclonal, 2748S, RRID: AB_823640) | 1:200 in 1XPBS for 1 h at room temperature (Cy3-conjugated Goat anti-Rabbit IgG, 111-165-144, RRID: AB_2338006) |
| Vimentin (Anti-Vimentin) | Citrate buffer 12 min at 95 °C, 20 min at room temperature in solution 0.1% trypsin in PBS 20 min at 37 °C | 5% NGS in diluent 30 min at 37 °C | 1:50 in diluent overnight at 4 °C (DSHB, mouse monoclonal, H5, RRID: AB_528506) | 1:200 in sterile 1XPBS for 1 h at room temperature (Goat anti-Mouse AlexaFluor 488, Life Technologies, A11001, RRID: AB_2534069) |
| Chase (Days) | Segment | Mean (%) | 95% Confidence Interval | |
|---|---|---|---|---|
| Lower | Upper | |||
| 0 | cranial | 0.356 | 0.073 | 1.733 |
| middle | 0.233 | 0.048 | 1.132 | |
| caudal | 0.285 | 0.058 | 1.323 | |
| 140 | cranial | 0.848 | 0.174 | 4.126 |
| middle | 0.330 | 0.068 | 1.605 | |
| caudal | 0.485 | 0.100 | 2.360 | |
| Group | Segment | Mean (%) | 95% Confidence Interval | |
|---|---|---|---|---|
| Lower | Upper | |||
| Original | cranial | 2.242 | 0.697 | 7.210 |
| middle | 6.809 | 2.118 | 21.894 | |
| caudal | 5.033 | 1.055 | 16.183 | |
| 2 dpa | cranial | 0.740 | 0.230 | 2.380 |
| middle | 2.496 | 0.776 | 8.024 | |
| caudal | 1.013 | 0.315 | 3.256 | |
| 8 dpa | cranial | 1.838 | 0.277 | 5.911 |
| middle | 2.415 | 0.751 | 7.765 | |
| caudal | 4.125 | 0.572 | 13.262 | |
| 12 dpa | cranial | 1.669 | 0.519 | 5.368 |
| middle | 3.493 | 1.086 | 11.232 | |
| caudal | 4.723 | 1.469 | 15.185 | |
| Group | Segment | Mean (%) | 95% Confidence Interval | |
|---|---|---|---|---|
| Lower | Upper | |||
| Original | cranial | 1.391 | 0.244 | 2.538 |
| middle | 0.218 | −0.929 | 1.365 | |
| caudal | 1.691 | 0.544 | 2.838 | |
| 2 dpa | cranial | 0.595 | −0.552 | 1.742 |
| middle | 0.247 | −0.900 | 1.394 | |
| caudal | 0.800 | −0.347 | 1.947 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donato, S.V.; Vickaryous, M.K. Radial Glia and Neuronal-like Ependymal Cells Are Present within the Spinal Cord of the Trunk (Body) in the Leopard Gecko (Eublepharis macularius). J. Dev. Biol. 2022, 10, 21. https://doi.org/10.3390/jdb10020021
Donato SV, Vickaryous MK. Radial Glia and Neuronal-like Ependymal Cells Are Present within the Spinal Cord of the Trunk (Body) in the Leopard Gecko (Eublepharis macularius). Journal of Developmental Biology. 2022; 10(2):21. https://doi.org/10.3390/jdb10020021
Chicago/Turabian StyleDonato, Sarah V., and Matthew K. Vickaryous. 2022. "Radial Glia and Neuronal-like Ependymal Cells Are Present within the Spinal Cord of the Trunk (Body) in the Leopard Gecko (Eublepharis macularius)" Journal of Developmental Biology 10, no. 2: 21. https://doi.org/10.3390/jdb10020021
APA StyleDonato, S. V., & Vickaryous, M. K. (2022). Radial Glia and Neuronal-like Ependymal Cells Are Present within the Spinal Cord of the Trunk (Body) in the Leopard Gecko (Eublepharis macularius). Journal of Developmental Biology, 10(2), 21. https://doi.org/10.3390/jdb10020021





