Transcriptional Regulation and Implications for Controlling Hox Gene Expression
Abstract
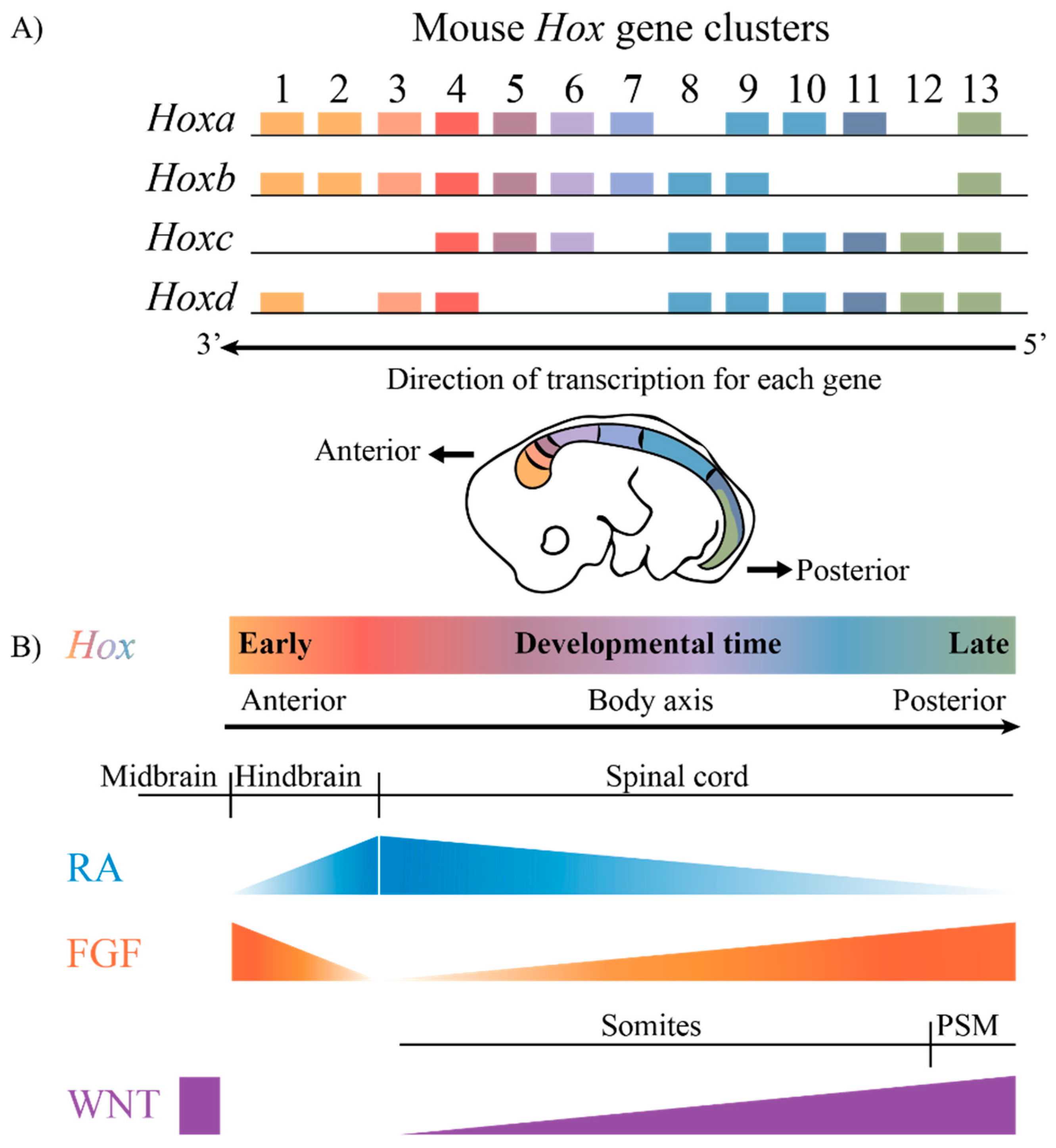
1. Introduction
2. Regulatory Features
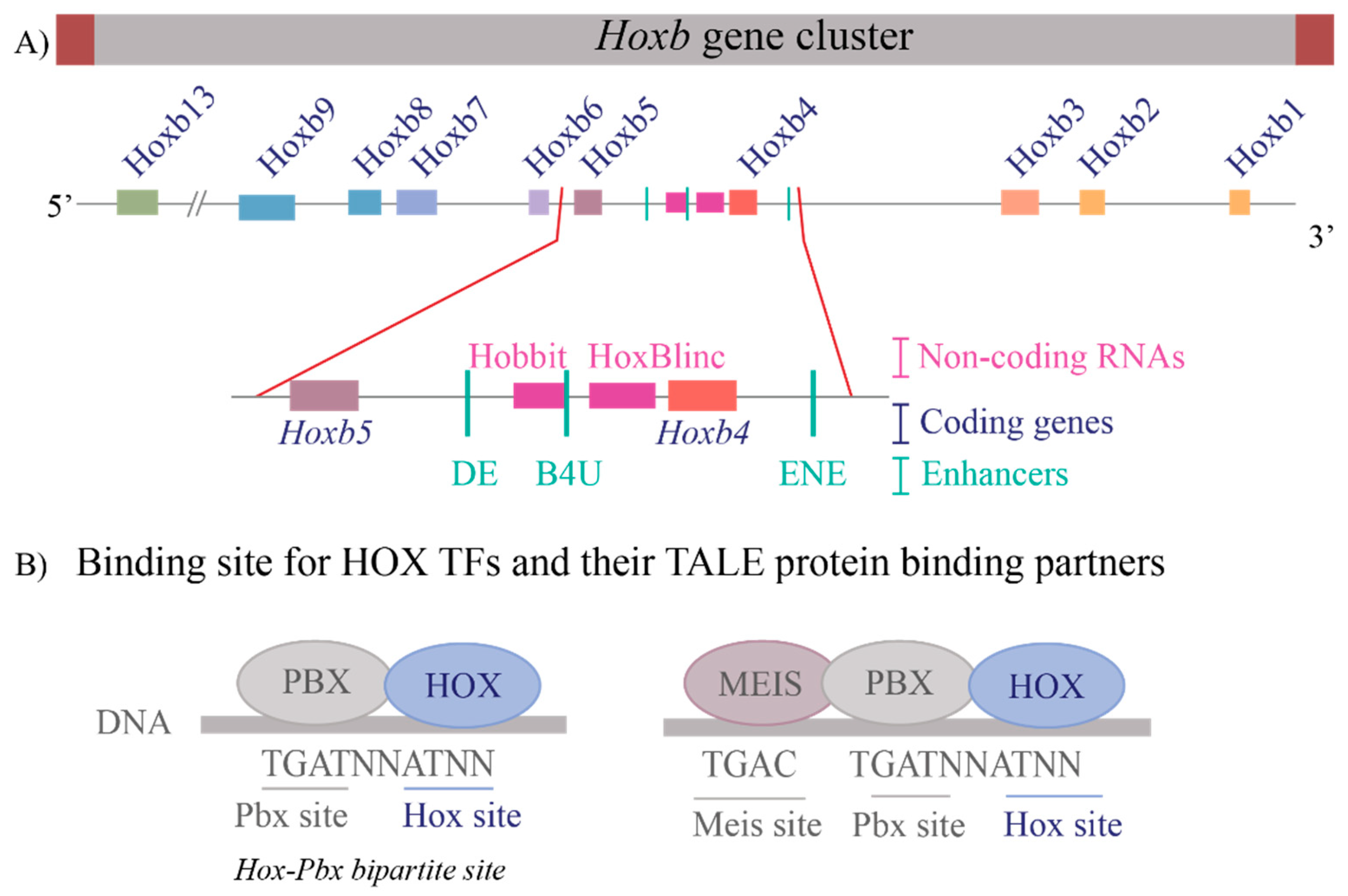
2.1. Enhancers
2.2. Transcription Factors and Their Impact on Enhancer Activity
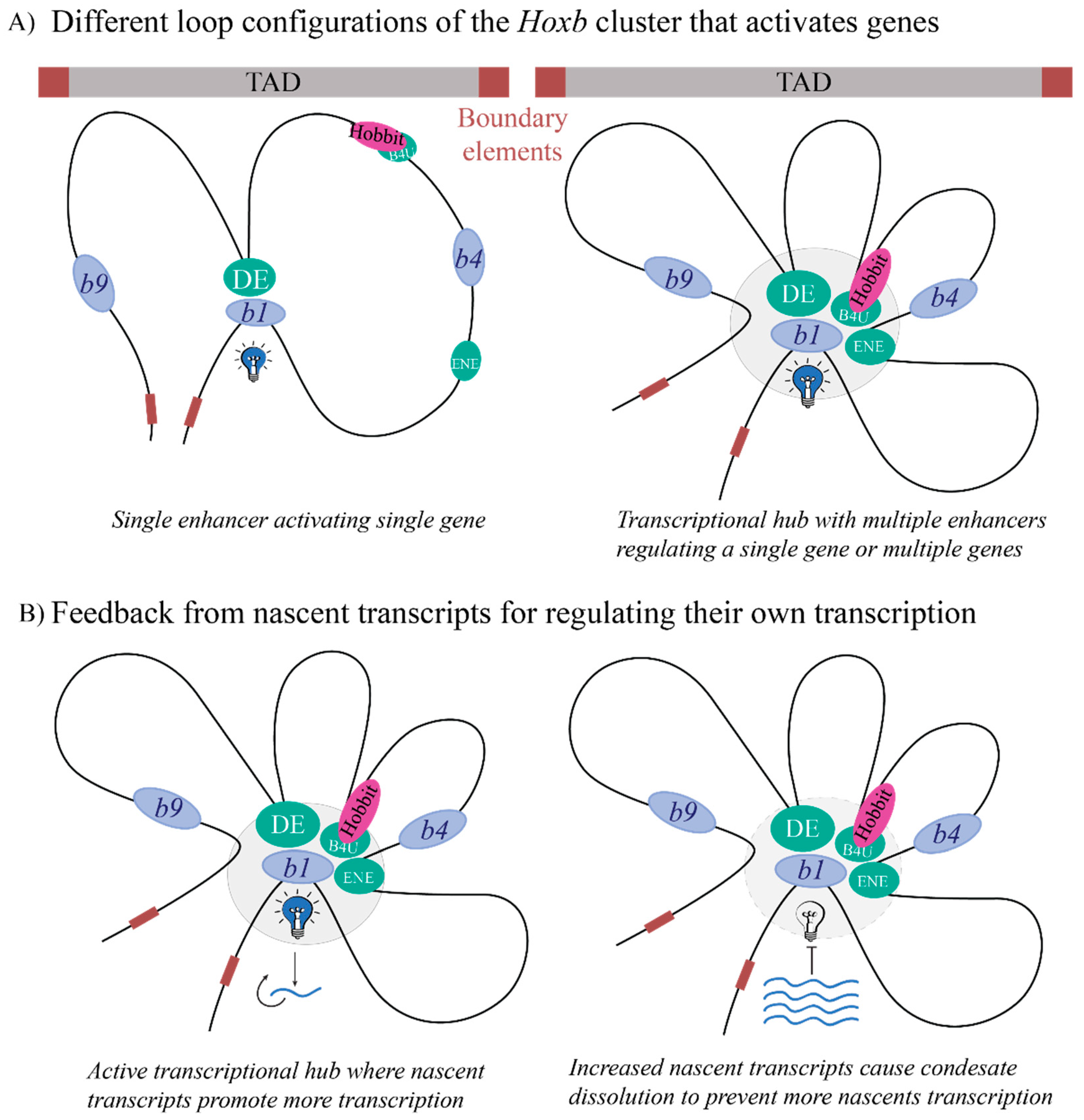
2.3. Genome Organization and Its Impact on Enhancer Activity
2.4. Deciphering Enhancer Activity through Nascent Expression
2.5. Additional Features in Regulation of Transcription of Hox Genes
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swalla, B.J. Building divergent body plans with similar genetic pathways. Heredity 2006, 97, 235–243. [Google Scholar] [CrossRef]
- Lowe, C.J.; Clarke, D.N.; Medeiros, D.M.; Rokhsar, D.S.; Gerhart, J. The deuterostome context of chordate origins. Nature 2015, 520, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Lowe, C.J.; Wu, M.; Salic, A.; Evans, L.; Lander, E.; Stange-Thomann, N.; Gruber, C.E.; Gerhart, J.; Kirschner, M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 2003, 113, 853–865. [Google Scholar] [CrossRef]
- Pani, A.M.; Mullarkey, E.E.; Aronowicz, J.; Assimacopoulos, S.; Grove, E.A.; Lowe, C.J. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 2012, 483, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Darras, S.; Fritzenwanker, J.H.; Uhlinger, K.R.; Farrelly, E.; Pani, A.M.; Hurley, I.A.; Norris, R.P.; Osovitz, M.; Terasaki, M.; Wu, M.; et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018, 16, e2003698. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Minor, P.J.; Zhao, Y.T.; Jeong, Y.; Pani, A.M.; King, A.N.; Symmons, O.; Gan, L.; Cardoso, W.V.; Spitz, F.; et al. Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nat. Genet. 2016, 48, 575–580. [Google Scholar] [CrossRef]
- Carroll, S.B.; Weatherbee, S.D.; Langeland, J.A. Homeotic genes and the regulation and evolution of insect wing number. Nature 1995, 375, 58–61. [Google Scholar] [CrossRef]
- Carroll, S.B. Homeotic genes and the evolution of arthropods and chordates. Nature 1995, 376, 479–485. [Google Scholar] [CrossRef]
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010, 344, 7–15. [Google Scholar] [CrossRef]
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Arendt, D. Hox genes and body segmentation. Science 2018, 361, 1310–1311. [Google Scholar] [CrossRef]
- Duboule, D.; Dolle, P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989, 8, 1497–1505. [Google Scholar] [CrossRef]
- Gaunt, S.J.; Sharpe, P.T.; Duboule, D. Spatially restricted domains of homeo-gene transcripts in mouse embryos: Relation to a segmented body plan. Development 1988, 104, 169–181. [Google Scholar] [CrossRef]
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Dollé, P.; Izpisùa-Belmonte, J.C.; Falkenstein, H.; Renucci, A.; Duboule, D. Co-ordinate expression of the murine Hox-5 complex homeobox-containing genes during limb pattern formation. Nature 1989, 342, 767–772. [Google Scholar] [CrossRef]
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef] [PubMed]
- Nolte, C.; De Kumar, B.; Krumlauf, R. Hox genes: Downstream “effectors” of retinoic acid signaling in vertebrate embryogenesis. Genesis 2019, 57, e23306. [Google Scholar] [CrossRef]
- Parker, H.J.; Krumlauf, R. Segmental arithmetic: Summing up the Hox gene regulatory network for hindbrain development in chordates. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e286. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.; Sela-Donenfeld, D. Hindbrain induction and patterning during early vertebrate development. Cell. Mol. Life Sci. 2019, 76, 941–960. [Google Scholar] [CrossRef]
- Deschamps, J.; van Nes, J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005, 132, 2931–2942. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Diez del Corral, R.; Storey, K.G. Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays 2004, 26, 857–869. [Google Scholar] [CrossRef]
- Wilson, V.; Olivera-Martinez, I.; Storey, K.G. Stem cells, signals and vertebrate body axis extension. Development 2009, 136, 1591–1604. [Google Scholar] [CrossRef]
- Young, T.; Rowland, J.E.; van de Ven, C.; Bialecka, M.; Novoa, A.; Carapuco, M.; van Nes, J.; de Graaff, W.; Duluc, I.; Freund, J.N.; et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 2009, 17, 516–526. [Google Scholar] [CrossRef]
- Simeone, A.; Acampora, D.; Arcioni, L.; Andrews, P.W.; Boncinelli, E.; Mavilio, F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 1990, 346, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Schilling, T.F.; Nie, Q.; Lander, A.D. Dynamics and precision in retinoic acid morphogen gradients. Curr. Opin. Genet. Dev. 2012, 22, 562–569. [Google Scholar] [CrossRef]
- Bedois, A.M.H.; Parker, H.J.; Krumlauf, R. Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification. Diversity 2021, 13, 398. [Google Scholar] [CrossRef]
- Krumlauf, R.; Wilkinson, D.G. Segmentation and patterning of the vertebrate hindbrain. Development 2021, 148, dev186460. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef]
- Rhinn, M.; Dolle, P. Retinoic acid signalling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef]
- Niederreither, K.; Dolle, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553. [Google Scholar] [CrossRef]
- Ahn, Y.; Mullan, H.E.; Krumlauf, R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev. Biol. 2014, 388, 134–144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qian, P.; De Kumar, B.; He, X.C.; Nolte, C.; Gogol, M.; Ahn, Y.; Chen, S.; Li, Z.; Xu, H.; Perry, J.M.; et al. Retinoid-Sensitive Epigenetic Regulation of the Hoxb Cluster Maintains Normal Hematopoiesis and Inhibits Leukemogenesis. Cell Stem Cell 2018, 22, 740–754.e7. [Google Scholar] [CrossRef] [PubMed]
- Nolte, C.; Jinks, T.; Wang, X.; Martinez Pastor, M.T.; Krumlauf, R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev. Biol. 2013, 383, 158–173. [Google Scholar] [CrossRef] [PubMed]
- Oosterveen, T.; Niederreither, K.; Dolle, P.; Chambon, P.; Meijlink, F.; Deschamps, J. Retinoids regulate the anterior expression boundaries of 5’ Hoxb genes in posterior hindbrain. EMBO J. 2003, 22, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, J.; Nonchev, S.; Gould, A.; Whiting, J.; Krumlauf, R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998, 17, 1788–1798. [Google Scholar] [CrossRef]
- Gould, A.; Morrison, A.; Sproat, G.; White, R.A.; Krumlauf, R. Positive cross-regulation and enhancer sharing: Two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997, 11, 900–913. [Google Scholar] [CrossRef]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef]
- Narayanan, A.; Meriin, A.; Andrews, J.O.; Spille, J.H.; Sherman, M.Y.; Cisse, I.I. A first order phase transition mechanism underlies protein aggregation in mammalian cells. eLife 2019, 8, e39695. [Google Scholar] [CrossRef]
- Henninger, J.E.; Oksuz, O.; Shrinivas, K.; Sagi, I.; LeRoy, G.; Zheng, M.M.; Andrews, J.O.; Zamudio, A.V.; Lazaris, C.; Hannett, N.M.; et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 2021, 184, 207–225.e24. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.E.; Manteiga, J.C.; Henninger, J.E.; Sabari, B.R.; Dall’Agnese, A.; Hannett, N.M.; Spille, J.H.; Afeyan, L.K.; Zamudio, A.V.; Shrinivas, K.; et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Zamudio, A.V.; Dall’Agnese, A.; Henninger, J.E.; Manteiga, J.C.; Afeyan, L.K.; Hannett, N.M.; Coffey, E.L.; Li, C.H.; Oksuz, O.; Sabari, B.R.; et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol. Cell 2019, 76, 753–766.e6. [Google Scholar] [CrossRef]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2021, 28, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.; Stadler, M.R.; Ortiz, S.A.; Hannon, C.E.; Harrison, M.M.; Darzacq, X.; Eisen, M.B. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 2018, 7, e40497. [Google Scholar] [CrossRef]
- Berrocal, A.; Lammers, N.C.; Garcia, H.G.; Eisen, M.B. Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene. eLife 2020, 9, e61635. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.; Dugast-Darzacq, C.; Liu, Z.; Dong, P.; Dailey, G.M.; Cattoglio, C.; Heckert, A.; Banala, S.; Lavis, L.; Darzacq, X.; et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 2018, 361, aar2555. [Google Scholar] [CrossRef]
- Liu, Z.; Tjian, R. Visualizing transcription factor dynamics in living cells. J. Cell Biol. 2018, 217, 1181–1191. [Google Scholar] [CrossRef]
- Zhang, Z.; Tjian, R. Measuring dynamics of eukaryotic transcription initiation: Challenges, insights and opportunities. Transcription 2018, 9, 159–165. [Google Scholar] [CrossRef]
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308. [Google Scholar] [CrossRef]
- Shen, Y.; Yue, F.; McCleary, D.F.; Ye, Z.; Edsall, L.; Kuan, S.; Wagner, U.; Dixon, J.; Lee, L.; Lobanenkov, V.V.; et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012, 488, 116–120. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five essential questions. Nat. Rev. Genet. 2013, 14, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Avsec, Z.; Weilert, M.; Shrikumar, A.; Krueger, S.; Alexandari, A.; Dalal, K.; Fropf, R.; McAnany, C.; Gagneur, J.; Kundaje, A.; et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 2021, 53, 354–366. [Google Scholar] [CrossRef]
- Zeitlinger, J. Seven myths of how transcription factors read the cis-regulatory code. Curr. Opin. Syst. Biol. 2020, 23, 22–31. [Google Scholar] [CrossRef]
- He, Q.; Johnston, J.; Zeitlinger, J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 2015, 33, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Girskis, K.M.; Stergachis, A.B.; DeGennaro, E.M.; Doan, R.N.; Qian, X.; Johnson, M.B.; Wang, P.P.; Sejourne, G.M.; Nagy, M.A.; Pollina, E.A.; et al. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 2021, 109, 3239–3251.e7. [Google Scholar] [CrossRef]
- Snetkova, V.; Pennacchio, L.A.; Visel, A.; Dickel, D.E. Perfect and imperfect views of ultraconserved sequences. Nat. Rev. Genet. 2021. [Google Scholar] [CrossRef]
- Snetkova, V.; Ypsilanti, A.R.; Akiyama, J.A.; Mannion, B.J.; Plajzer-Frick, I.; Novak, C.S.; Harrington, A.N.; Pham, Q.T.; Kato, M.; Zhu, Y.; et al. Ultraconserved enhancer function does not require perfect sequence conservation. Nat. Genet. 2021, 53, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, T.; Jordan, J.; van Breugel, M.E.; Halavatyi, A.; Tischer, C.; Polidoro, P.; Abe, N.; Tsai, A.; Mann, R.S.; Stern, D.L.; et al. Dense and pleiotropic regulatory information in a developmental enhancer. Nature 2020, 587, 235–239. [Google Scholar] [CrossRef]
- Marshall, H.; Studer, M.; Popperl, H.; Aparicio, S.; Kuroiwa, A.; Brenner, S.; Krumlauf, R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 1994, 370, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.J.; De Kumar, B.; Green, S.A.; Prummel, K.D.; Hess, C.; Kaufman, C.K.; Mosimann, C.; Wiedemann, L.M.; Bronner, M.E.; Krumlauf, R. A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat. Commun. 2019, 10, 1189. [Google Scholar] [CrossRef] [PubMed]
- Tümpel, S.; Maconochie, M.; Wiedemann, L.M.; Krumlauf, R. Conservation and diversity in the cis-regulatory networks that integrate information controlling expression of Hoxa2 in hindbrain and cranial neural crest cells in vertebrates. Dev. Biol. 2002, 246, 45–56. [Google Scholar] [CrossRef]
- Schmidt, D.; Wilson, M.D.; Ballester, B.; Schwalie, P.C.; Brown, G.D.; Marshall, A.; Kutter, C.; Watt, S.; Martinez-Jimenez, C.P.; Mackay, S.; et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 2010, 328, 1036–1040. [Google Scholar] [CrossRef]
- Paris, M.; Kaplan, T.; Li, X.Y.; Villalta, J.E.; Lott, S.E.; Eisen, M.B. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet. 2013, 9, e1003748. [Google Scholar] [CrossRef]
- Farley, E.K.; Olson, K.M.; Zhang, W.; Brandt, A.J.; Rokhsar, D.S.; Levine, M.S. Suboptimization of developmental enhancers. Science 2015, 350, 325–328. [Google Scholar] [CrossRef]
- Crocker, J.; Abe, N.; Rinaldi, L.; McGregor, A.P.; Frankel, N.; Wang, S.; Alsawadi, A.; Valenti, P.; Plaza, S.; Payre, F.; et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 2015, 160, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lysakovskaia, K.; Stik, G.; Demel, C.; Söding, J.; Tian, T.V.; Graf, T.; Cramer, P. Evidence for additive and synergistic action of mammalian enhancers during cell fate determination. eLife 2021, 10, e65381. [Google Scholar] [CrossRef]
- Marsman, J.; Horsfield, J.A. Long distance relationships: Enhancer-promoter communication and dynamic gene transcription. Biochim. Biophys. Acta 2012, 1819, 1217–1227. [Google Scholar] [CrossRef]
- Lettice, L.A.; Heaney, S.J.; Purdie, L.A.; Li, L.; de Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; de Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735. [Google Scholar] [CrossRef]
- Tümpel, S.; Cambronero, F.; Sims, C.; Krumlauf, R.; Wiedemann, L.M. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc. Natl. Acad. Sci. USA 2008, 105, 20077–20082. [Google Scholar] [CrossRef]
- Lampe, X.; Samad, O.A.; Guiguen, A.; Matis, C.; Remacle, S.; Picard, J.J.; Rijli, F.M.; Rezsohazy, R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008, 36, 3214–3225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, M.F.; Kheradpour, P.; Washietl, S.; Parker, B.J.; Pedersen, J.S.; Kellis, M. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res. 2011, 21, 1916–1928. [Google Scholar] [CrossRef]
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2020, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.; Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455. [Google Scholar] [CrossRef]
- Lai, F.; Gardini, A.; Zhang, A.; Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 2015, 525, 399–403. [Google Scholar] [CrossRef]
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 2019, 76, 473–484.e7. [Google Scholar] [CrossRef] [PubMed]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef]
- Long, H.K.; Prescott, S.L.; Wysocka, J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 2016, 167, 1170–1187. [Google Scholar] [CrossRef] [PubMed]
- De Kumar, B.; Parrish, M.E.; Slaughter, B.D.; Unruh, J.R.; Gogol, M.; Seidel, C.; Paulson, A.; Li, H.; Gaudenz, K.; Peak, A.; et al. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015, 25, 1229–1243. [Google Scholar] [CrossRef] [PubMed]
- Maconochie, M.K.; Nonchev, S.; Manzanares, M.; Marshall, H.; Krumlauf, R. Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev. Biol. 2001, 233, 468–481. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Popperl, H.; Bienz, M.; Studer, M.; Chan, S.K.; Aparicio, S.; Brenner, S.; Mann, R.S.; Krumlauf, R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 1995, 81, 1031–1042. [Google Scholar] [CrossRef]
- Manzanares, M.; Cordes, S.; Kwan, C.-T.; Sham, M.-H.; Barsh, G.; Krumlauf, R. Segmental regulation of Hoxb3 by kreisler. Nature 1997, 387, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Mann, R.S. To Be Specific or Not: The Critical Relationship Between Hox And TALE Proteins. Trends Genet. 2016, 32, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.S.; Lelli, K.M.; Joshi, R. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009, 88, 63–101. [Google Scholar] [CrossRef]
- Berger, M.F.; Badis, G.; Gehrke, A.R.; Talukder, S.; Philippakis, A.A.; Pena-Castillo, L.; Alleyne, T.M.; Mnaimneh, S.; Botvinnik, O.B.; Chan, E.T.; et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 2008, 133, 1266–1276. [Google Scholar] [CrossRef]
- Noyes, M.B.; Christensen, R.G.; Wakabayashi, A.; Stormo, G.D.; Brodsky, M.H.; Wolfe, S.A. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 2008, 133, 1277–1289. [Google Scholar] [CrossRef]
- Nitta, K.R.; Jolma, A.; Yin, Y.; Morgunova, E.; Kivioja, T.; Akhtar, J.; Hens, K.; Toivonen, J.; Deplancke, B.; Furlong, E.E.; et al. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 2015, 4, e04837. [Google Scholar] [CrossRef]
- Loker, R.; Sanner, J.E.; Mann, R.S. Cell-type-specific Hox regulatory strategies orchestrate tissue identity. Curr. Biol. 2021, 31, 4246–4255.e4. [Google Scholar] [CrossRef]
- Carroll, S.B. Evolution at two levels: On genes and form. PLoS Biol. 2005, 3, e245. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.; Muller, M.; Affolter, M.; Percival-Smith, A.; Billeter, M.; Qian, Y.; Otting, G.; Wuthrich, K. The structure of the homeodomain and its functional implications. Trends Genet. 1990, 6, 323–329. [Google Scholar] [CrossRef]
- Jabet, C.; Gitti, R.; Summers, M.F.; Wolberger, C. NMR studies of the pbx1 TALE homeodomain protein free in solution and bound to DNA: Proposal for a mechanism of HoxB1-Pbx1-DNA complex assembly. J. Mol. Biol. 1999, 291, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Kissinger, C.; Liu, B.; Martin-Blanco, E.; Kronberg, T.; Pabo, C. Crystal structure of an engrailed homeodomain-DNA complex at 2.8A resolution: A framework for understanding homeodomain-DNA interactions. Cell 1990, 63, 579–590. [Google Scholar] [CrossRef]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.P.; De Kumar, B.; Paulson, A.; Parrish, M.E.; Scott, C.; Zhang, Y.; Florens, L.; Krumlauf, R. Genome-Wide Binding Analyses of HOXB1 Revealed a Novel DNA Binding Motif Associated with Gene Repression. J. Dev. Biol. 2021, 9, 6. [Google Scholar] [CrossRef]
- Singh, N.P.; De Kumar, B.; Paulson, A.; Parrish, M.E.; Zhang, Y.; Florens, L.; Conaway, J.W.; Si, K.; Krumlauf, R. A six-amino-acid motif is a major determinant in functional evolution of HOX1 proteins. Genes Dev. 2020, 34, 1680–1696. [Google Scholar] [CrossRef]
- Studer, M.; Gavalas, A.; Marshall, H.; Ariza-McNaughton, L.; Rijli, F.M.; Chambon, P.; Krumlauf, R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 1998, 125, 1025–1036. [Google Scholar] [CrossRef]
- De Kumar, B.; Parker, H.J.; Paulson, A.; Parrish, M.E.; Pushel, I.; Singh, N.P.; Zhang, Y.; Slaughter, B.D.; Unruh, J.R.; Florens, L.; et al. HOXA1 and TALE proteins display cross-regulatory interactions and form a combinatorial binding code on HOXA1 targets. Genome Res. 2017, 27, 1501–1512. [Google Scholar] [CrossRef]
- Manzanares, M.; Bel-Vialer, S.; Ariza-McNaughton, L.; Ferretti, E.; Marshall, H.; Maconochie, M.K.; Blasi, F.; Krumlauf, R. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involves auto and cross-regulatory mechanisms. Development 2001, 128, 3595–3607. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Kambris, Z.; Capovilla, M.; Berenger, H.; Pradel, J.; Graba, Y. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev. Cell 2003, 4, 761–768. [Google Scholar] [CrossRef]
- Dard, A.; Reboulet, J.; Jia, Y.; Bleicher, F.; Duffraisse, M.; Vanaker, J.M.; Forcet, C.; Merabet, S. Human HOX Proteins Use Diverse and Context-Dependent Motifs to Interact with TALE Class Cofactors. Cell Rep. 2018, 22, 3058–3071. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Litim-Mecheri, I.; Karlsson, D.; Dixit, R.; Saadaoui, M.; Monier, B.; Brun, C.; Thor, S.; Vijayraghavan, K.; Perrin, L.; et al. Insights into Hox protein function from a large scale combinatorial analysis of protein domains. PLoS Genet. 2011, 7, e1002302. [Google Scholar] [CrossRef] [PubMed]
- Zandvakili, A.; Gebelein, B. Mechanisms of Specificity for Hox Factor Activity. J. Dev. Biol. 2016, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Zeiske, T.; Baburajendran, N.; Kaczynska, A.; Brasch, J.; Palmer, A.G., 3rd; Shapiro, L.; Honig, B.; Mann, R.S. Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites. Cell Rep. 2018, 24, 2221–2230. [Google Scholar] [CrossRef]
- Merabet, S.; Hudry, B.; Saadaoui, M.; Graba, Y. Classification of sequence signatures: A guide to Hox protein function. Bioessays 2009, 31, 500–511. [Google Scholar] [CrossRef]
- Rivas, M.L.; Espinosa-Vazquez, J.M.; Sambrani, N.; Greig, S.; Merabet, S.; Graba, Y.; Hombria, J.C. Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function. PLoS Genet. 2013, 9, e1003252. [Google Scholar] [CrossRef]
- Berthelsen, J.; Zappavigna, V.; Ferretti, E.; Mavilio, F.; Blasi, F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998, 17, 1434–1445. [Google Scholar] [CrossRef]
- Ferretti, E.; Cambronero, F.; Tümpel, S.; Longobardi, E.; Wiedemann, L.M.; Blasi, F.; Krumlauf, R. Hoxb1 enhancer and control of rhombomere 4 expression: Complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell. Biol. 2005, 25, 8541–8552. [Google Scholar] [CrossRef]
- Longobardi, E.; Penkov, D.; Mateos, D.; De Florian, G.; Torres, M.; Blasi, F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev. Dyn. 2014, 243, 59–75. [Google Scholar] [CrossRef]
- Penkov, D.; Tanaka, S.; Di Rocco, G.; Berthelsen, J.; Blasi, F.; Ramirez, F. Cooperative interactions between PBX, PREP, and HOX proteins modulate the activity of the alpha 2(V) collagen (COL5A2) promoter. J. Biol. Chem. 2000, 275, 16681–16689. [Google Scholar] [CrossRef]
- De Kumar, B.; Parker, H.J.; Parrish, M.E.; Lange, J.J.; Slaughter, B.D.; Unruh, J.R.; Paulson, A.; Krumlauf, R. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc. Natl. Acad. Sci. USA 2017, 114, 5838–5845. [Google Scholar] [CrossRef]
- De Kumar, B.; Parker, H.J.; Paulson, A.; Parrish, M.E.; Zeitlinger, J.; Krumlauf, R. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev. Biol. 2017, 432, 151–164. [Google Scholar] [CrossRef]
- Gavalas, A.; Trainor, P.; Ariza-McNaughton, L.; Krumlauf, R. Synergy between Hoxa1 and Hoxb1: The relationship between arch patterning and the generation of cranial neural crest. Development 2001, 128, 3017–3027. [Google Scholar] [CrossRef]
- Manley, N.R.; Capecchi, M.R. Hox group 3 paralogous genes act synergistically in the formation of somitic and neural crest-derived structures. Dev. Biol. 1997, 192, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Rossel, M.; Capecchi, M.R. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 1999, 126, 5027–5040. [Google Scholar] [CrossRef]
- Wellik, D.M.; Capecchi, M.R. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 2003, 301, 363–367. [Google Scholar] [CrossRef]
- Horan, G.S.; Ramirez-Solis, R.; Featherstone, M.S.; Wolgemuth, D.J.; Bradley, A.; Behringer, R.R. Compound mutants for the paralogous Hoxa-4, Hoxb-4, and Hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995, 9, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Svingen, T.; Tonissen, K.F. Hox transcription factors and their elusive mammalian gene targets. Heredity 2006, 97, 88–96. [Google Scholar] [CrossRef]
- Bridoux, L.; Zarrineh, P.; Mallen, J.; Phuycharoen, M.; Latorre, V.; Ladam, F.; Losa, M.; Baker, S.M.; Sagerstrom, C.; Mace, K.A.; et al. HOX paralogs selectively convert binding of ubiquitous transcription factors into tissue-specific patterns of enhancer activation. PLoS Genet. 2020, 16, e1009162. [Google Scholar] [CrossRef] [PubMed]
- Bulajić, M.; Srivastava, D.; Dasen, J.S.; Wichterle, H.; Mahony, S.; Mazzoni, E.O. Differential abilities to engage inaccessible chromatin diversify vertebrate Hox binding patterns. Development 2020, 147, dev194761. [Google Scholar] [CrossRef]
- Banigan, E.J.; Mirny, L.A. Loop extrusion: Theory meets single-molecule experiments. Curr. Opin. Cell Biol. 2020, 64, 124–138. [Google Scholar] [CrossRef]
- Mirny, L.; Dekker, J. Mechanisms of Chromosome Folding and Nuclear Organization: Their Interplay and Open Questions. Cold Spring Harb. Perspect. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Narendra, V.; Bulajić, M.; Dekker, J.; Mazzoni, E.O.; Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016, 30, 2657–2662. [Google Scholar] [CrossRef]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007, 128, 1231–1245. [Google Scholar] [CrossRef]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Narendra, V.; Rocha, P.P.; An, D.; Raviram, R.; Skok, J.A.; Mazzoni, E.O.; Reinberg, D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 2015, 347, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Bolt, C.C.; Duboule, D. The regulatory landscapes of developmental genes. Development 2020, 147, dev171736. [Google Scholar] [CrossRef] [PubMed]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, Q.; Canzio, D.; Shou, J.; Li, J.; Gorkin, D.U.; Jung, I.; Wu, H.; Zhai, Y.; Tang, Y.; et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 2015, 162, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Kim, C.Y.; Lee, J.Y.; Kim, M.H. Retinoic acid and CTCF play key roles in inducing the collinear expression of the Hoxa cluster. Acta Biochim. Biophys. Sin. 2018, 50, 555–559. [Google Scholar] [CrossRef]
- Luo, H.; Wang, F.; Zha, J.; Li, H.; Yan, B.; Du, Q.; Yang, F.; Sobh, A.; Vulpe, C.; Drusbosky, L.; et al. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood 2018, 132, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liao, Z.; Luo, H.; Benyoucef, A.; Kang, Y.; Lai, Q.; Dovat, S.; Miller, B.; Chepelev, I.; Li, Y.; et al. Alteration of CTCF-associated chromatin neighborhood inhibits TAL1-driven oncogenic transcription program and leukemogenesis. Nucleic Acids Res. 2020, 48, 3119–3133. [Google Scholar] [CrossRef]
- Qiu, Y.; Huang, S. CTCF-mediated genome organization and leukemogenesis. Leukemia 2020, 34, 2295–2304. [Google Scholar] [CrossRef]
- Morcillo, P.; Rosen, C.; Baylies, M.K.; Dorsett, D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997, 11, 2729–2740. [Google Scholar] [CrossRef]
- Kong, S.; Bohl, D.; Li, C.; Tuan, D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol. Cell. Biol. 1997, 17, 3955–3965. [Google Scholar] [CrossRef]
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012, 149, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Dunn, T.M.; Hahn, S.; Ogden, S.; Schleif, R.F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: Addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. USA 1984, 81, 5017–5020. [Google Scholar] [CrossRef] [PubMed]
- Takaki, R.; Dey, A.; Shi, G.; Thirumalai, D. Theory and simulations of condensin mediated loop extrusion in DNA. Nat. Commun. 2021, 12, 5865. [Google Scholar] [CrossRef] [PubMed]
- Chambeyron, S.; Bickmore, W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004, 18, 1119–1130. [Google Scholar] [CrossRef]
- Symmons, O.; Uslu, V.V.; Tsujimura, T.; Ruf, S.; Nassari, S.; Schwarzer, W.; Ettwiller, L.; Spitz, F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014, 24, 390–400. [Google Scholar] [CrossRef]
- Gambetta, M.C.; Furlong, E.E.M. The Insulator Protein CTCF Is Required for Correct. Genetics 2018, 210, 129–136. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef]
- de Wit, E.; Vos, E.S.; Holwerda, S.J.; Valdes-Quezada, C.; Verstegen, M.J.; Teunissen, H.; Splinter, E.; Wijchers, P.J.; Krijger, P.H.; de Laat, W. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 2015, 60, 676–684. [Google Scholar] [CrossRef]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef]
- Ghavi-Helm, Y.; Klein, F.A.; Pakozdi, T.; Ciglar, L.; Noordermeer, D.; Huber, W.; Furlong, E.E. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014, 512, 96–100. [Google Scholar] [CrossRef]
- Espinola, S.M.; Götz, M.; Bellec, M.; Messina, O.; Fiche, J.-B.; Houbron, C.; Dejean, M.; Reim, I.; Cardozo Gizzi, A.M.; Lagha, M.; et al. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 2021, 53, 477–486. [Google Scholar] [CrossRef]
- Chen, H.; Levo, M.; Barinov, L.; Fujioka, M.; Jaynes, J.B.; Gregor, T. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 2018, 50, 1296–1303. [Google Scholar] [CrossRef]
- Bartman, C.R.; Hsu, S.C.; Hsiung, C.C.; Raj, A.; Blobel, G.A. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol. Cell 2016, 62, 237–247. [Google Scholar] [CrossRef]
- Larsson, A.J.M.; Johnsson, P.; Hagemann-Jensen, M.; Hartmanis, L.; Faridani, O.R.; Reinius, B.; Segerstolpe, Å.; Rivera, C.M.; Ren, B.; Sandberg, R. Genomic encoding of transcriptional burst kinetics. Nature 2019, 565, 251–254. [Google Scholar] [CrossRef]
- Alexander, J.M.; Guan, J.; Li, B.; Maliskova, L.; Song, M.; Shen, Y.; Huang, B.; Lomvardas, S.; Weiner, O.D. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 2019, 8, e41769. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.W.; Hendrix, D.A.; Levine, M.S. Shadow enhancers as a source of evolutionary novelty. Science 2008, 321, 1314. [Google Scholar] [CrossRef]
- Perry, M.W.; Boettiger, A.N.; Bothma, J.P.; Levine, M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010, 20, 1562–1567. [Google Scholar] [CrossRef]
- Waymack, R.; Fletcher, A.; Enciso, G.; Wunderlich, Z. Shadow enhancers can suppress input transcription factor noise through distinct regulatory logic. eLife 2020, 9, e59351. [Google Scholar] [CrossRef]
- Fukaya, T.; Lim, B.; Levine, M. Enhancer Control of Transcriptional Bursting. Cell 2016, 166, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Anastasakis, D.G.; Rodriguez, J.; Palangat, M.; Gudla, P.; Zaki, G.; Tandon, M.; Pegoraro, G.; Chow, C.C.; Hafner, M.; et al. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. Cell 2021, 184, 2878–2895.e20. [Google Scholar] [CrossRef] [PubMed]
- Bothma, J.P.; Garcia, H.G.; Esposito, E.; Schlissel, G.; Gregor, T.; Levine, M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. USA 2014, 111, 10598–10603. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Zeitlinger, J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017, 49, 1045–1051. [Google Scholar] [CrossRef]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef]
- Lim, B.; Heist, T.; Levine, M.; Fukaya, T. Visualization of Transvection in Living Drosophila Embryos. Mol. Cell 2018, 70, 287–296.e6. [Google Scholar] [CrossRef]
- Delpretti, S.; Montavon, T.; Leleu, M.; Joye, E.; Tzika, A.; Milinkovitch, M.; Duboule, D. Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. 2013, 5, 137–150. [Google Scholar] [CrossRef]
- Andrey, G.; Montavon, T.; Mascrez, B.; Gonzalez, F.; Noordermeer, D.; Leleu, M.; Trono, D.; Spitz, F.; Duboule, D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 2013, 340, 1234167. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Mendelson Cohen, N.; Szabo, Q.; Fritsch, L.; Papadopoulos, G.L.; Lubling, Y.; Xu, X.; Lv, X.; Hugnot, J.P.; Tanay, A.; et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 2017, 171, 557–572.e24. [Google Scholar] [CrossRef] [PubMed]
- Cardozo Gizzi, A.M.; Cattoni, D.I.; Fiche, J.B.; Espinola, S.M.; Gurgo, J.; Messina, O.; Houbron, C.; Ogiyama, Y.; Papadopoulos, G.L.; Cavalli, G.; et al. Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol. Cell 2019, 74, 212–222.e5. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Ando, T.; Priest, D.G.; Taniguchi, Y. Hi-CO: 3D genome structure analysis with nucleosome resolution. Nat. Protoc. 2021, 16, 3439–3469. [Google Scholar] [CrossRef]
- Akgol Oksuz, B.; Yang, L.; Abraham, S.; Venev, S.V.; Krietenstein, N.; Parsi, K.M.; Ozadam, H.; Oomen, M.E.; Nand, A.; Mao, H.; et al. Systematic evaluation of chromosome conformation capture assays. Nat. Methods 2021, 18, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Mateo, L.J.; Murphy, S.E.; Hafner, A.; Cinquini, I.S.; Walker, C.A.; Boettiger, A.N. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 2019, 568, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Tyagi, S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 2010, 472, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, S.M.; Wu, M.T.; Levesque, M.J.; Raj, A. Turbo FISH: A method for rapid single molecule RNA FISH. PLoS ONE 2013, 8, e75120. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.M.T.; Schwarzkopf, M.; Fornace, M.E.; Acharya, A.; Artavanis, G.; Stegmaier, J.; Cunha, A.; Pierce, N.A. Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 2018, 145, dev165753. [Google Scholar] [CrossRef] [PubMed]
- Garcia, H.G.; Gregor, T. Live Imaging of mRNA Synthesis in Drosophila. Methods Mol. Biol. 2018, 1649, 349–357. [Google Scholar] [CrossRef]
- Lim, B.; Fukaya, T.; Heist, T.; Levine, M. Temporal dynamics of pair-rule stripes in living Drosophila embryos. Proc. Natl. Acad. Sci. USA 2018, 115, 8376–8381. [Google Scholar] [CrossRef] [PubMed]
- Bothma, J.P.; Garcia, H.G.; Ng, S.; Perry, M.W.; Gregor, T.; Levine, M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife 2015, 4, e07956. [Google Scholar] [CrossRef]
- Bothma, J.P.; Norstad, M.R.; Alamos, S.; Garcia, H.G. LlamaTags: A Versatile Tool to Image Transcription Factor Dynamics in Live Embryos. Cell 2018, 173, 1810–1822.e16. [Google Scholar] [CrossRef]
- Wissink, E.M.; Vihervaara, A.; Tippens, N.D.; Lis, J.T. Nascent RNA analyses: Tracking transcription and its regulation. Nat. Rev. Genet. 2019, 20, 705–723. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e16. [Google Scholar] [CrossRef]
- Boube, M.; Hudry, B.; Immarigeon, C.; Carrier, Y.; Bernat-Fabre, S.; Merabet, S.; Graba, Y.; Bourbon, H.M.; Cribbs, D.L. Drosophila melanogaster Hox transcription factors access the RNA polymerase II machinery through direct homeodomain binding to a conserved motif of mediator subunit Med19. PLoS Genet. 2014, 10, e1004303. [Google Scholar] [CrossRef]
- Soshnikova, N.; Duboule, D. Epigenetic temporal control of mouse Hox genes in vivo. Science 2009, 324, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, D.; Leleu, M.; Schorderet, P.; Joye, E.; Chabaud, F.; Duboule, D. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife 2014, 3, e02557. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carballo, E.; Lopez-Delisle, L.; Yakushiji-Kaminatsui, N.; Ullate-Agote, A.; Duboule, D. Impact of genome architecture on the functional activation and repression of Hox regulatory landscapes. BMC Biol. 2019, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Gonzalez, F.; Duboule, D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 2003, 113, 405–417. [Google Scholar] [CrossRef]
- Gonzalez, F.; Duboule, D.; Spitz, F. Transgenic analysis of Hoxd gene regulation during digit development. Dev. Biol. 2007, 306, 847–859. [Google Scholar] [CrossRef]
- Neijts, R.; Amin, S.; van Rooijen, C.; Tan, S.; Creyghton, M.P.; de Laat, W.; Deschamps, J. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 2016, 30, 1937–1942. [Google Scholar] [CrossRef]
- Berlivet, S.; Paquette, D.; Dumouchel, A.; Langlais, D.; Dostie, J.; Kmita, M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013, 9, e1004018. [Google Scholar] [CrossRef]
- Eskeland, R.; Freyer, E.; Leeb, M.; Wutz, A.; Bickmore, W.A. Histone acetylation and the maintenance of chromatin compaction by Polycomb repressive complexes. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 71–78. [Google Scholar] [CrossRef]
- De Kumar, B.; Krumlauf, R. HOXs and lincRNAs: Two sides of the same coin. Sci. Adv. 2016, 2, e1501402. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Deng, C.; Li, Y.; Zhou, L.; Cho, J.; Patel, B.; Terada, N.; Li, Y.; Bungert, J.; Qiu, Y.; Huang, S. HoxBlinc RNA Recruits Set1/MLL Complexes to Activate Hox Gene Expression Patterns and Mesoderm Lineage Development. Cell Rep. 2016, 14, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Gentile, C.; Kmita, M. The remote transcriptional control of Hox genes. Int. J. Dev. Biol. 2018, 62, 685–692. [Google Scholar] [CrossRef]
- Parker, H.J.; Bronner, M.E.; Krumlauf, R. The vertebrate Hox gene regulatory network for hindbrain segmentation: Evolution and diversification: Coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. Bioessays 2016, 38, 526–538. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.J.; Krumlauf, R. A Hox gene regulatory network for hindbrain segmentation. Curr. Top. Dev. Biol. 2020, 139, 169–203. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afzal, Z.; Krumlauf, R. Transcriptional Regulation and Implications for Controlling Hox Gene Expression. J. Dev. Biol. 2022, 10, 4. https://doi.org/10.3390/jdb10010004
Afzal Z, Krumlauf R. Transcriptional Regulation and Implications for Controlling Hox Gene Expression. Journal of Developmental Biology. 2022; 10(1):4. https://doi.org/10.3390/jdb10010004
Chicago/Turabian StyleAfzal, Zainab, and Robb Krumlauf. 2022. "Transcriptional Regulation and Implications for Controlling Hox Gene Expression" Journal of Developmental Biology 10, no. 1: 4. https://doi.org/10.3390/jdb10010004
APA StyleAfzal, Z., & Krumlauf, R. (2022). Transcriptional Regulation and Implications for Controlling Hox Gene Expression. Journal of Developmental Biology, 10(1), 4. https://doi.org/10.3390/jdb10010004






