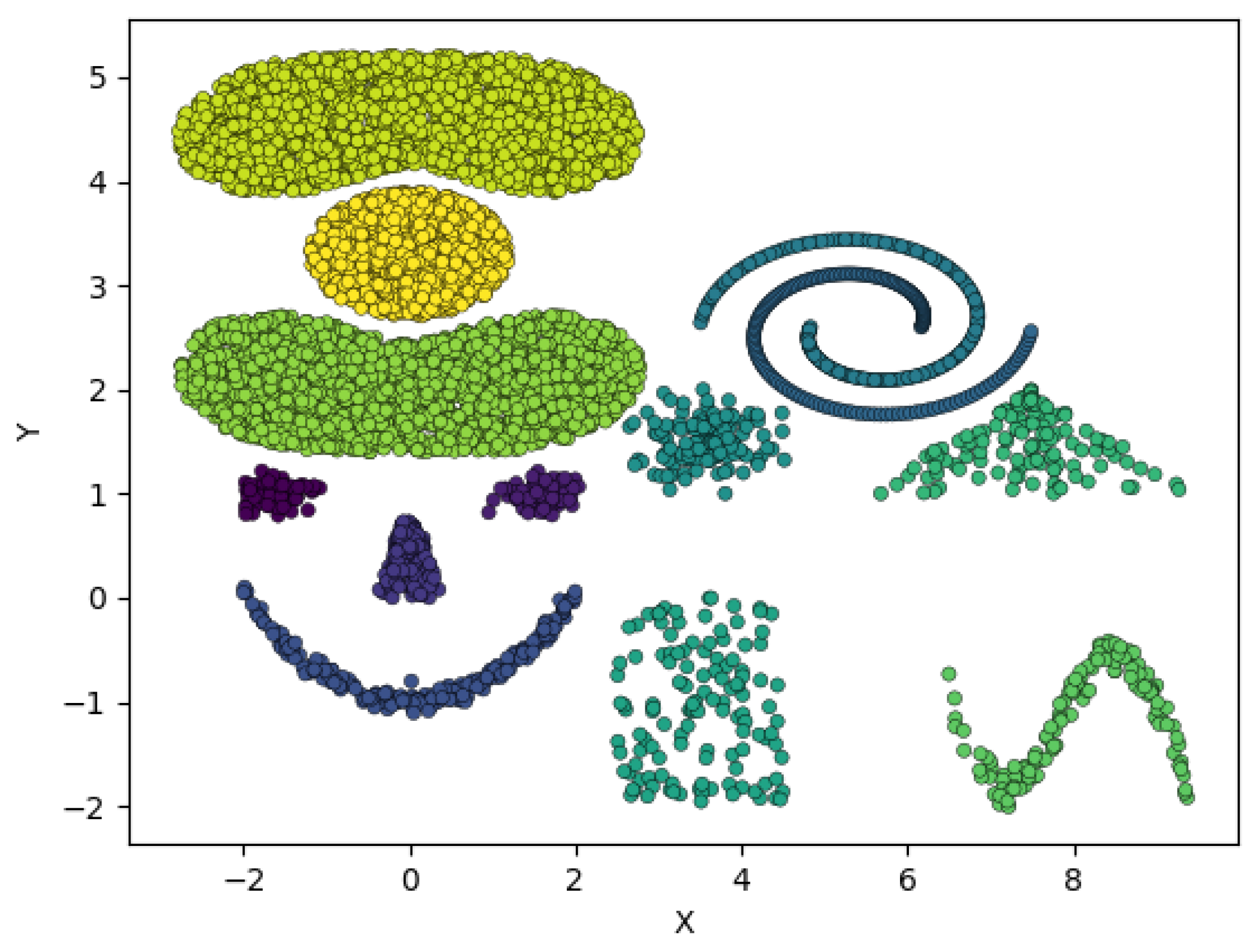
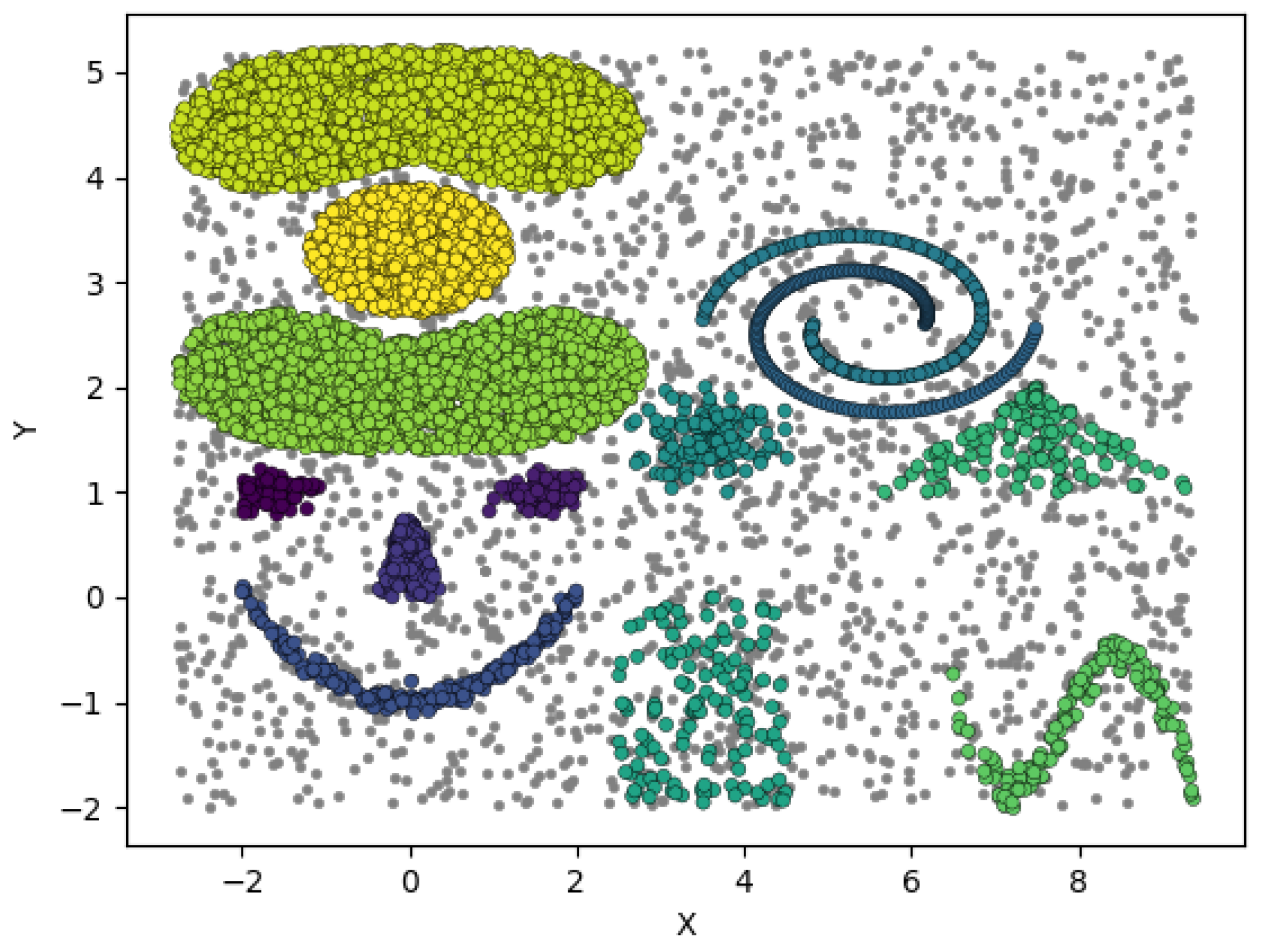
4.2. MD Re-Clustering
Observation 2 leads to the recognition of cases where data with high-dimensional skew cannot be clustered effectively with MD. However, some cases can still be clustered accurately if identified clusters are iteratively re-clustered in a way in which the dimensional skew is lessened. For example,
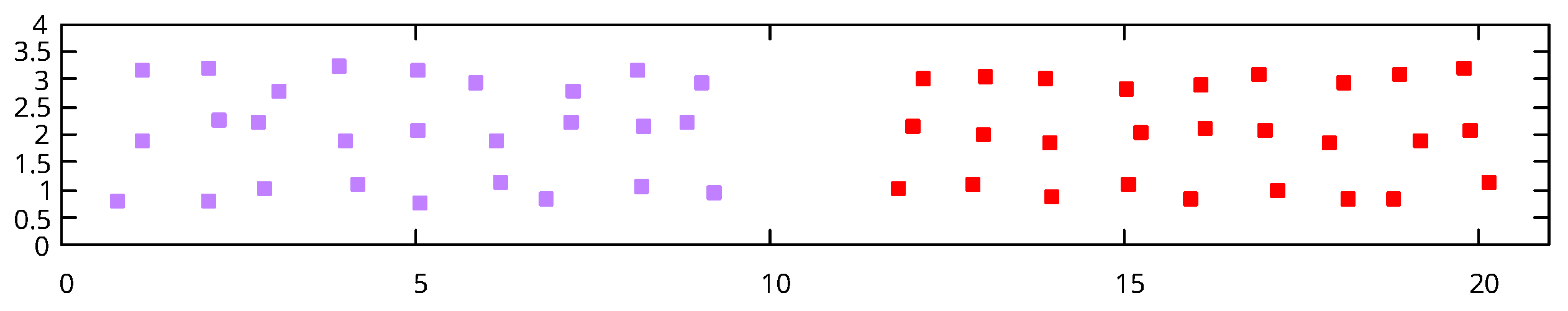
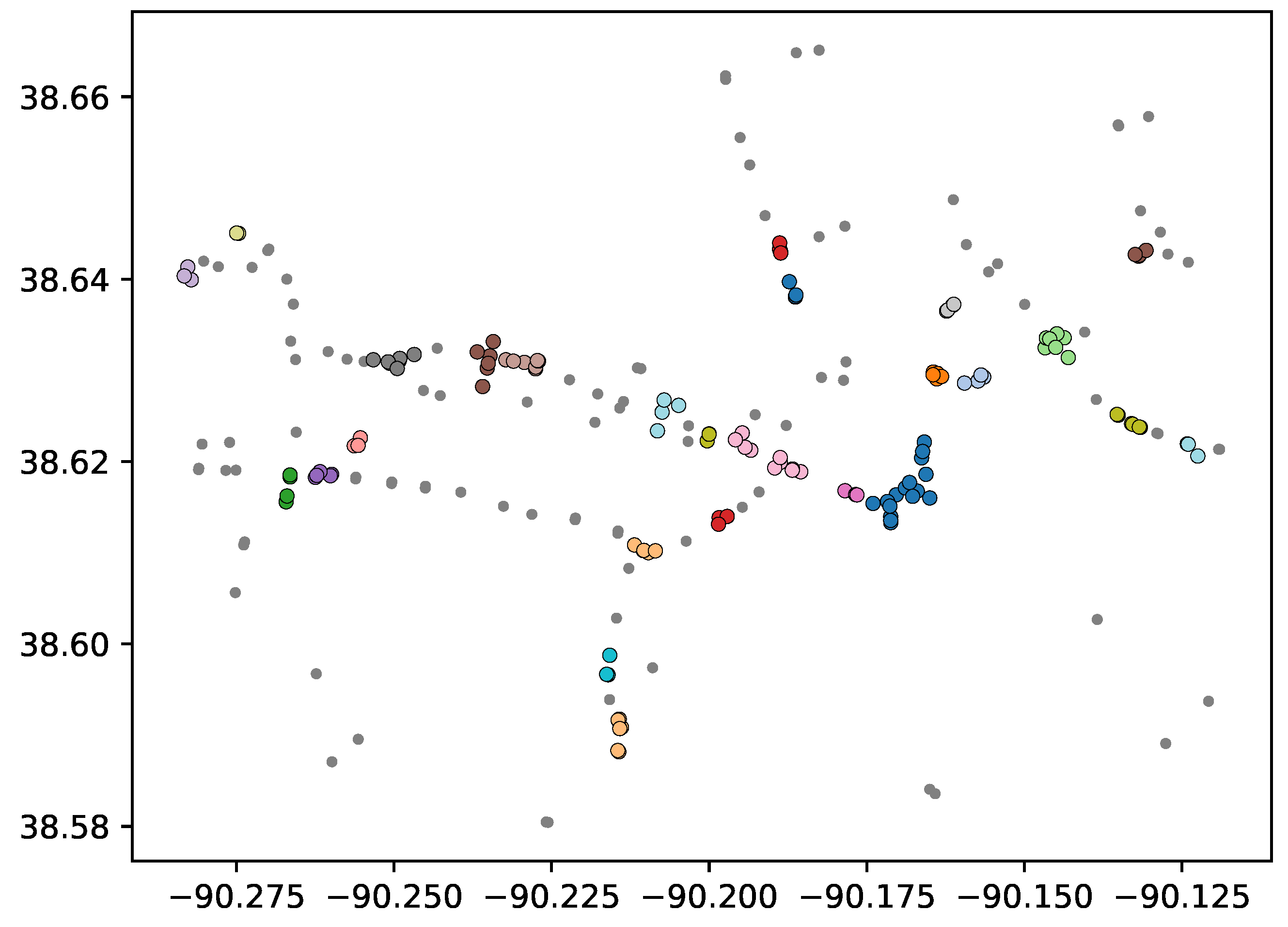
Figure 2 depicts a situation where the variance of the overall data is much greater in the
y direction than the
x direction. Again, because depth values are scaled based on the variance of the data, a unit of Euclidean distance in the
x direction yields a depth value of
and unit of Euclidean distance in the
y direction yields a depth value of
, thus, using a DBSCAN algorithm with MD cannot distinguish the two horizontal clusters without breaking apart the large vertically oriented cluster. In this case, such an algorithm can identify two clusters, the larger, vertically oriented cluster, and the other containing both horizontal point configurations on the left. Because the two horizontally oriented clusters have drastically different shape that the overall data set (their variance is much greater in the
x direction as opposed to the overall data set, which has high dimensional skew in the
y direction), it is possible to re-cluster those points taking into account the
local shape of the cluster, a process we refer to as the
Re-Cluster MD Algorithm.
The Re-Cluster MD Algorithm will first perform clustering on the overall data set, then examine each cluster and decide if it should be re-clustered based on local information. To re-cluster a cluster, we recompute the covariance matrix using only the points in the cluster, and then run the original clustering algorithm on those points using their covariance matrix for depth computations. This leads to two problems:
Problem 1: The value used for the original clustering will not be relevant on the new cluster since the points will typically cover a smaller area than the original point set, and
Problem 2: We must know when to re-cluster, and when not to.
Problem 1: Because depth values under MD are scaled by the data, if we compute depth values based on a subset of the original data, the depth values will be scaled to that subset. For example, in
Figure 2, an original clustering can identify 2 clusters, the larger vertical cluster, whose points we denote
, and the two horizontal lines in a second cluster, whose points we denote
. Re-clustering
involves computing the covariance matrix over
, and then computing depth values on those points. Depth values will then be computed by scaling Euclidean distance by this new covariance matrix; the range and density of points in
with respect to their covariance matrix is different than the range and density of the original point set with respect to the original covariance matrix, thus a unit of depth in
may cover a much different Euclidean distance than a unit of depth in the original data set, even if the variances of the subcluster are proportional to the variances of the original data set. Therefore, the
value from the original clustering will not necessarily provide a good clustering when re-clustering
.
To address the issue of local depth values differing from global depth values when re-clustering, we suggest two remedies. The first is to simply choose a new value of for each re-clustering instance. This may prove useful in exploratory analysis, or if the densities of clusters vary greatly, but requires more fine tuning by the user. A second approach is to alter the depth computation such that the global value for maintains relevance when re-clustering. We use the second approach to provide a general algorithm with fewer parameters to tune. Our goal is to scale the covariance matrix of the smaller cluster based on the full point set to achieve meaningful clustering with the same depth value.
A covariance matrix for a data set can be thought of as a linear transformation from non-correlated data to the data set (or vice versa). Furthermore, a covariance matrix
K for a data set is equivalent to a combination of its eigenvalues and eigenvectors; let
L be a matrix with the eigenvalues of
K in the diagonal, and
V be the matrix with the corresponding eigenvalues in its columns:
Intuitively, in 2-dimensional data, the eigenvectors scaled by the eignenvalues of the covariance matrix define a representative ellipse bounding (almost) the entire data set. The same concept extends to higher dimensions. The value used to cluster the data set is relative to depth values computed by scaling Euclidean distance along those eigenvectors with respect to those eigenvalues. When re-clustering a subset of the original data, the eigenvectors of the covariance matrix of the subset may, naturally, be different than those of the entire data set, and the eigenvalues of those eigenvectors will typically represent a smaller ellipse (in terms of area/volume), which means the depth values of two points may change drastically based on the new covariance matrix.
To remedy the change in relative depth values between points in a data set vs. points in a subset of that data when re-clustering, we take advantage of Equation (
9) to scale the eigenvalues of the covariance matrix of the subset of data while maintaining the proportion of variance explained by each eigenvector. In 2-dimensional data, this (intuitively) scales the representative ellipse defined by the eigenvectors and eigenvalues of the subset of data to an ellipse with the same area of the representative ellipse defined by the original data set (or volume in higher dimensions). More generally, we propose to scale each of the eigenvalues of the covariance matrix of the subset of data by a value
s such that the product of such eigenvalues is equivalent to the product of the eigenvalues of the covariance matrix of the original data set. Let
D be a data set of dimension
d and
,
K and
be the respective covariance matrices, and
and
be the eigenvalues of the their respective covariance matrices. We use the following formula to compute
s with the goal of using the same
on the subset of data:
To make the
value used for the original clustering relevant on the smaller subset of data, we scale the eigenvalues in Equation (
9) by
s in Equation (
10). Using this technique allows us to use the same
value when re-examining clusters after an initial clustering.
Problem 2: In the case of MD-based clustering techniques, one does not need to re-cluster a cluster that has similar shape to the overall data set, since the Euclidean distance covered by depth values in all directions will be similar to that in the overall data set (especially once the covariance matrix for the local cluster is scaled such that the same value may be used). We use the term shape of the data to mean the representative ellipsoid of the data defined by the eigenvectors of its covariance matrix; for the purposes of this section, we are not concerned with scale, so the shape of two data sets may be considered identical even if their representative ellipsoids have different area/volume.
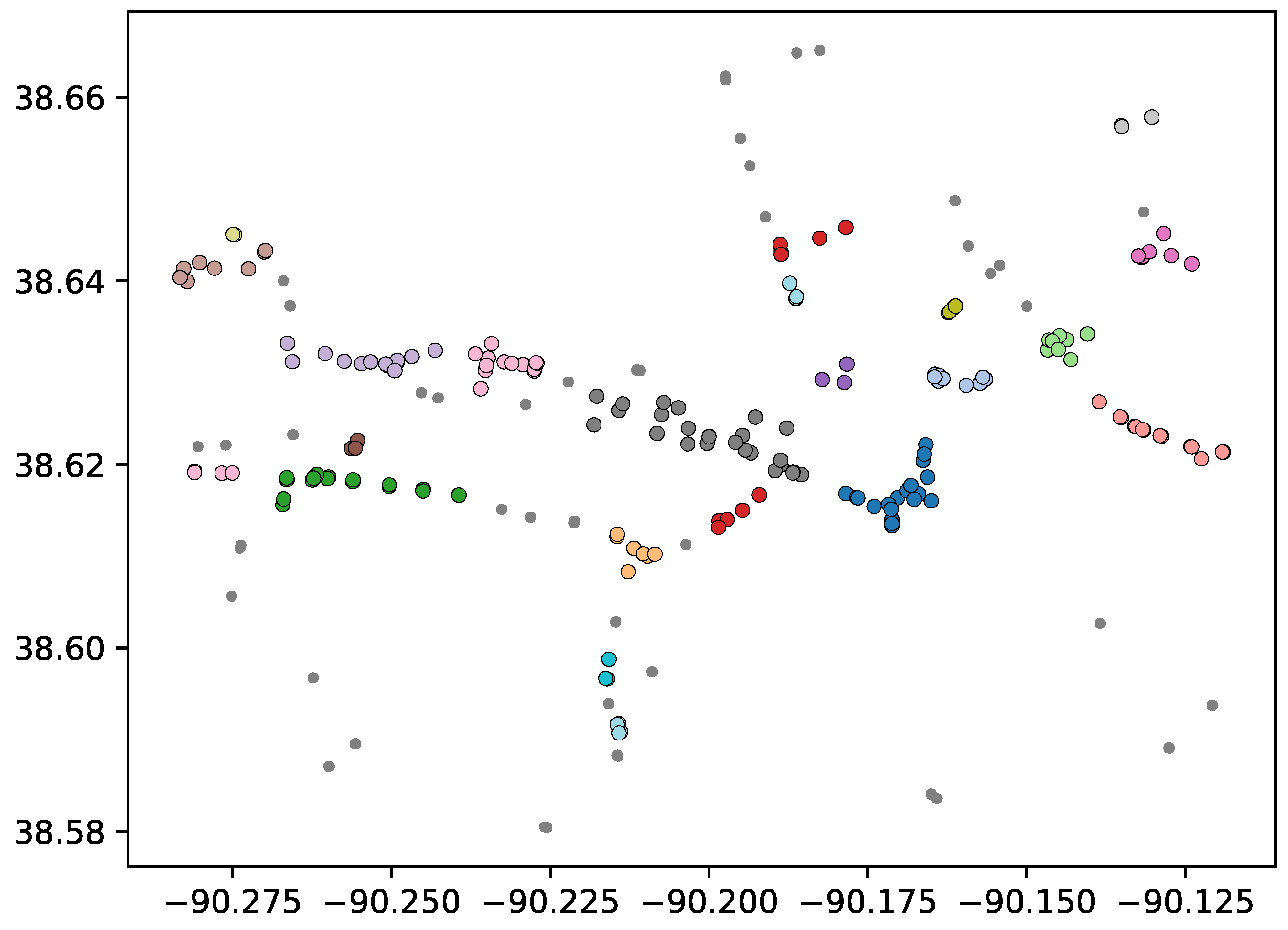
Re-clustering a subset of points can lead to poor results if a cluster has similar shape to the original data set. For example, using the data set shown in
Figure 2, an initial clustering step using MD will discover 2 clusters, the vertically oriented points to the right form one cluster, and the points forming the two horizontal structures on the left of the figure. The two horizontal structures should each be identified as their own cluster (forming 3 clusters total), but because the point set is distributed in a fashion in which there is significantly higher amount of variance along the first principle component (vertical) than the second (horizontal), the two horizontal structures cannot be differentiated using MD on the entire data set. If we re-cluster the points forming the horizontal structures using MD, with the covariance matrix scaled as discussed above, we do identify both clusters using the same
value as the original clustering. However, a problem arises in that the variances of the principle components of the vertical cluster are now even more extreme, relative to each other; the result is that a unit of depth in the horizontal direction covers so little Euclidean distance that the vertical cluster re-clusters to three clusters (respectively, the points with
x an coordinate of 7, the points with an
x coordinate of 8, and the points with an
x coordinate of 9). Therefore, we require a mechanism to determine when a cluster should be re-clustered, and when it should not be.
We propose a mechanism to determine when a cluster should be re-clustered based on the shape of the data in a cluster as defined by its principle components relative to the shape of the overall data as defined by its principle components. Intuitively, the vertical cluster in
Figure 2 should not go through a re-clustering step because it’s shape is similar to the overall shape of the data (as defined by the covariance matrices); thus, it is unlikely that the overall shape of the data is obscuring substructures in that cluster. The cluster containing the horizontal structures in
Figure 2, on the other hand, has a much different shape (specifically the orientation of the principal components) than the overall data. There are various methods to compare shape of data sets in the literature (see [
17,
18] for a review). In order to compare the shape of data sets, we use the statistics defined in [
18] that are meant to compare the shape of multivariate data sets via their covariance matrices. We choose this method because it is computationally simple, easy to interpret, produces a value as opposed to a strict classification, and does not depend on distributional assumptions. We summarize the statistics in the remainder of this section.
Let
and
be two multivariate point sets and
and
be matrices, respectively, containing the eigenvectors of the covariance matrices of
and
.
and
are organized such that the eigenvectors are in their columns. Let
be a function that computes the covariance matrix of a data set.
The variances in
are the eigenvalues of the of the principle components of the original data (likewise for
). In other words, those values indicate the amount of variance explained by the eigenvectors indicating the principle components of the original data. Similarly:
The variances in
indicate the amount of variance from data set
explained by the eigenvectors representing the principle components of
(likewise for
). For each dimension of the data, let:
where
is the amount of total variance in
explained by the
eigenvector in
,
is the amount of total variance in
explained by the
eigenvector in
, etc. Using these values, one may compute the following statistic for
n-dimensional data:
For our purposes, we do not require the sum be multiplied by 2, but we keep it such that the statistic fits within the framework defined in [
18]. Intuitively
S in Equation (
14) is a measure of the difference of the ability of the eigenvectors of the covariance matrix from one data set to explain the variation in the other data set, and vice versa. As constructed, Equation (
14) has two properties that are not desirable for our purposes. First, the statistic is sensitive to the magnitude of the eigenvectors used. In other words, a data set with small range and a data set with large range will compute as being different. Second, the value produced is not bounded in a way to make comparisons easy. Both of these properties are addressed if instead of using the raw variances for the various
v values in Equation (
13), we express each value as a proportion of total variance explained; thus, covariance matrix differences are computed on orientation of the eigenvectors, and proportion of the eigenvalues (meaning two covariance matrices covering the same data, but at difference scales, are considered equivalent).
Finally, the maximum value of
S in Equation (
14) when expressing variance explained as proportion of total variance is 8, and occurs in the case when single eigenvectors explain all the variance in each sample, and those eigenvectors are orthogonal. Likewise, a value of 0 occurs when
. It is useful to have similarity values range from
, and since we are in the context of depth calculations where a depth of 1 indicates identity, we will express the statistic from Equation (
14) as the following:
We incorporate covariance matrix similarity comparisons with DBSCAN using MD in order to achieve an algorithm able to re-cluster point clusters with covariance matrices that are different in shape than the matrix of the overall data and that is robust in its parameterization. We introduce a new user-defined variable, that represents a covariance matrix similarity threshold. A cluster will be re-clustered based on its scaled MD value if its covariance matrix similarity value as compared to the data set from which the cluster was identified is below . The algorithm is presented in Algorithm 2.
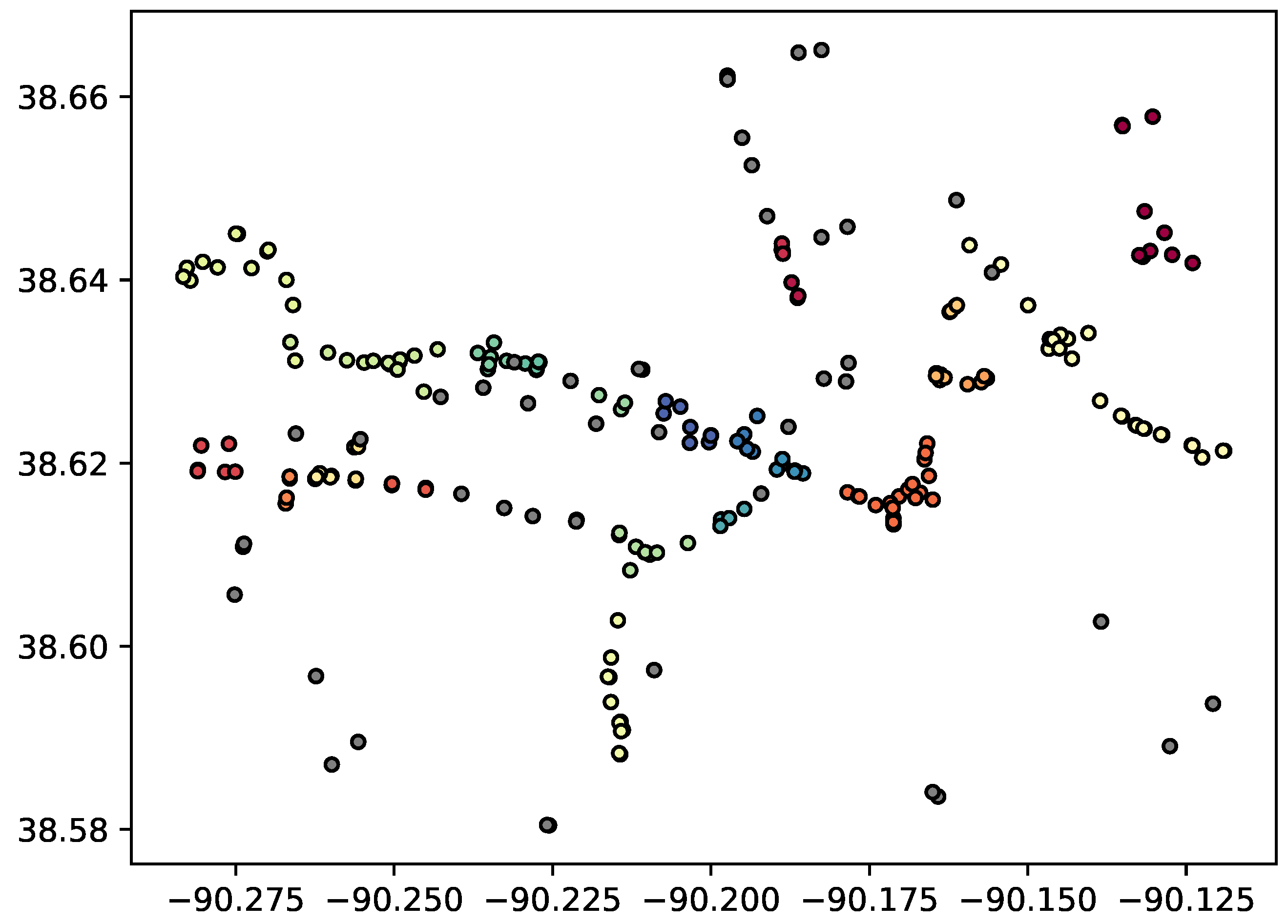
Algorithm 2 essentially performs repeated DBSCAN algorithms on increasingly smaller subsets of the data until no new clusters are found. The time complexity of optimal DBSCAN is
. In the worst possible case, the DBSCAN algorithm would result in 2 clusters, one consisting of the minimal number of points in a cluster, then the other containing the remaining points that must be re-clustered. This pattern could repeat, essentially leading to
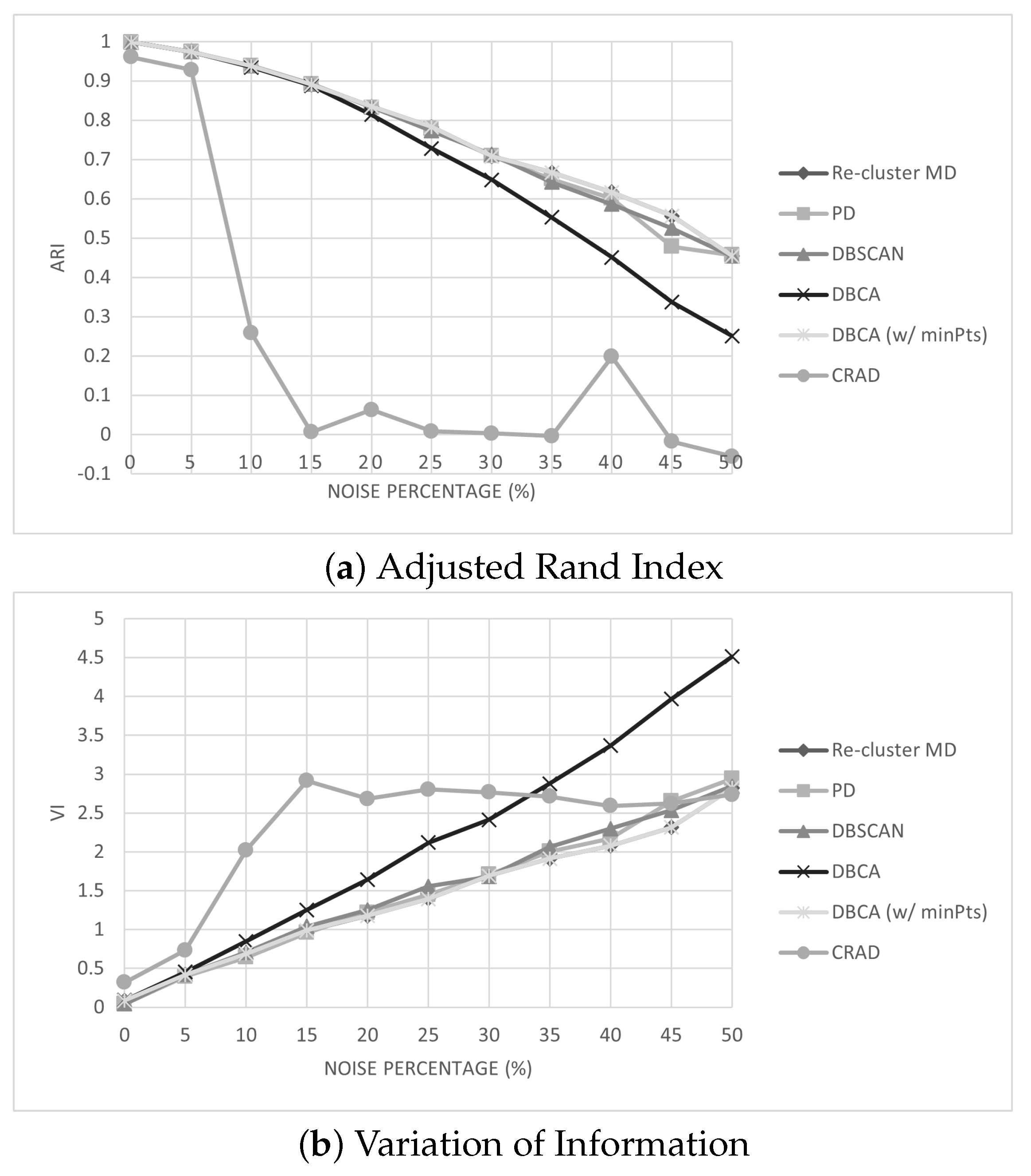
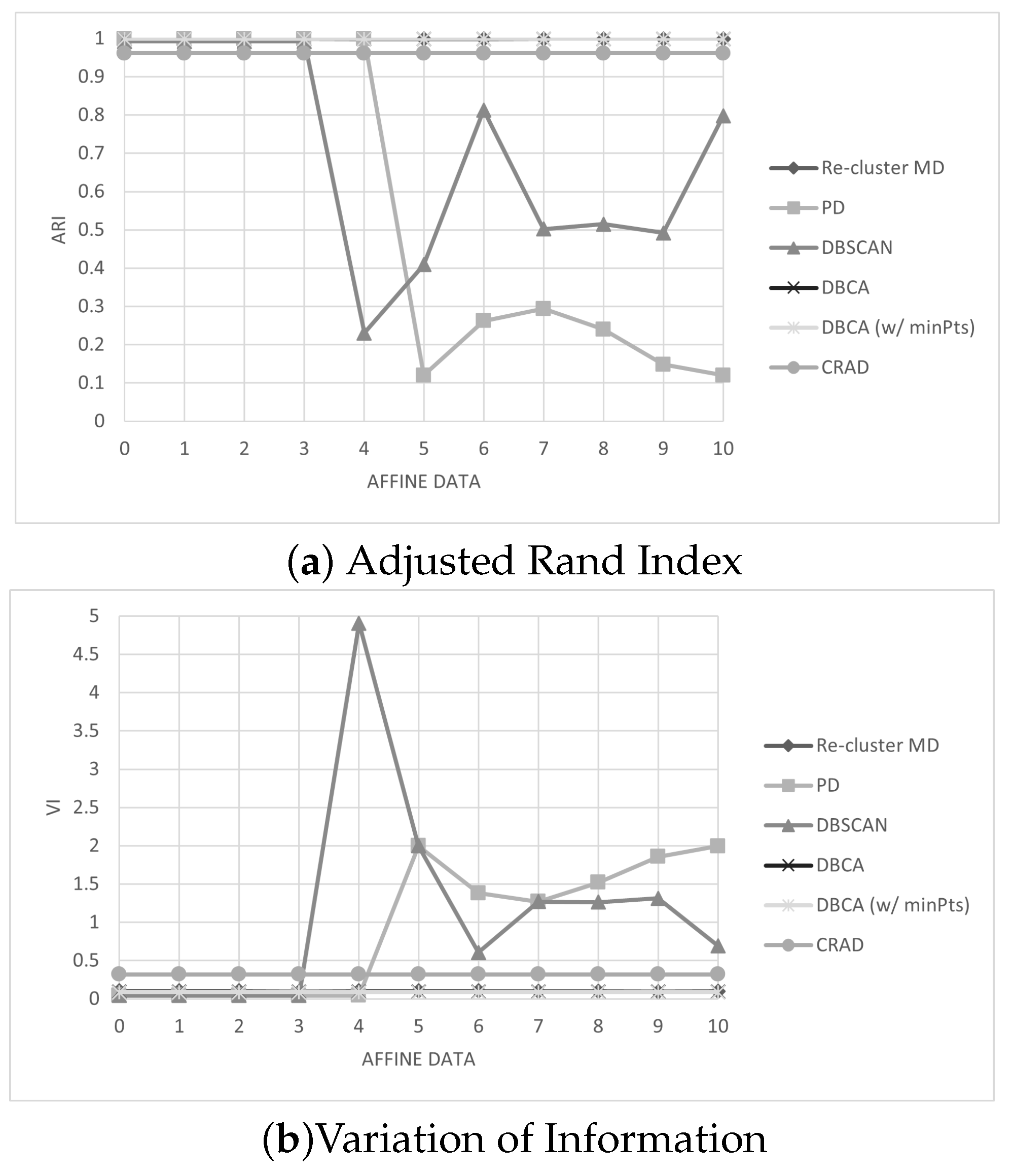
. In practice, such a degenerate case would require high dimensionality to be possible, since the shape of each successively smaller cluster would need to change enough to reveal clusters with respect to different dimensions, and is unlikely. In the experiments in this paper, only a single re-clustering step is necessary.
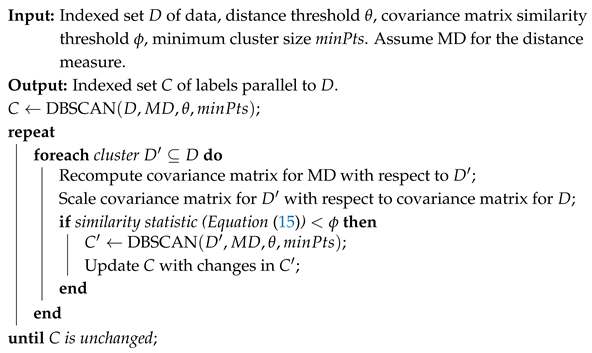
| Algorithm 2: Algorithm to re-cluster initial clusters from an MD-based DBSCAN |
![Ijgi 14 00298 i002]() |