A Density-Peak-Based Clustering Method for Multiple Densities Dataset
Abstract
:1. Introduction
2. Related Work of Density Peak Clustering Algorithm
3. Logical Flow of Proposed Method
3.1. Definition of the Relevant Concepts
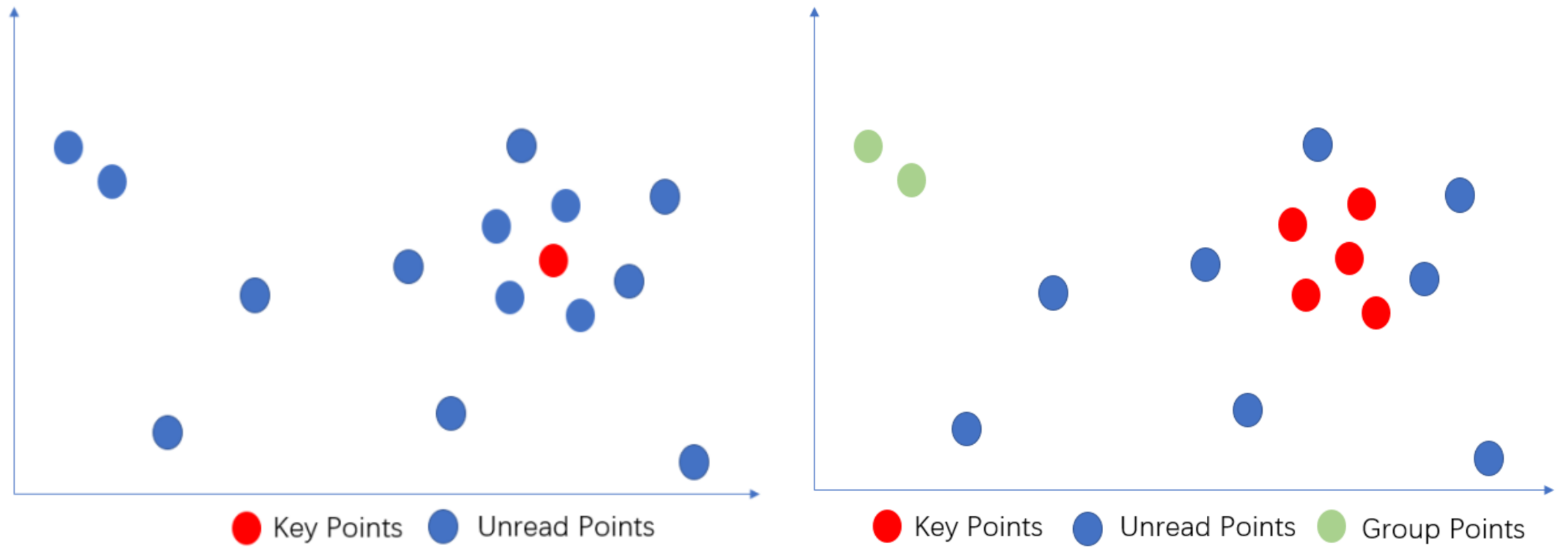
3.1.1. Point Definitions
- (1)
- Key points
- (2)
- Group points
- (3)
- Unread points
3.1.2. Silhouette Coefficient Calculation
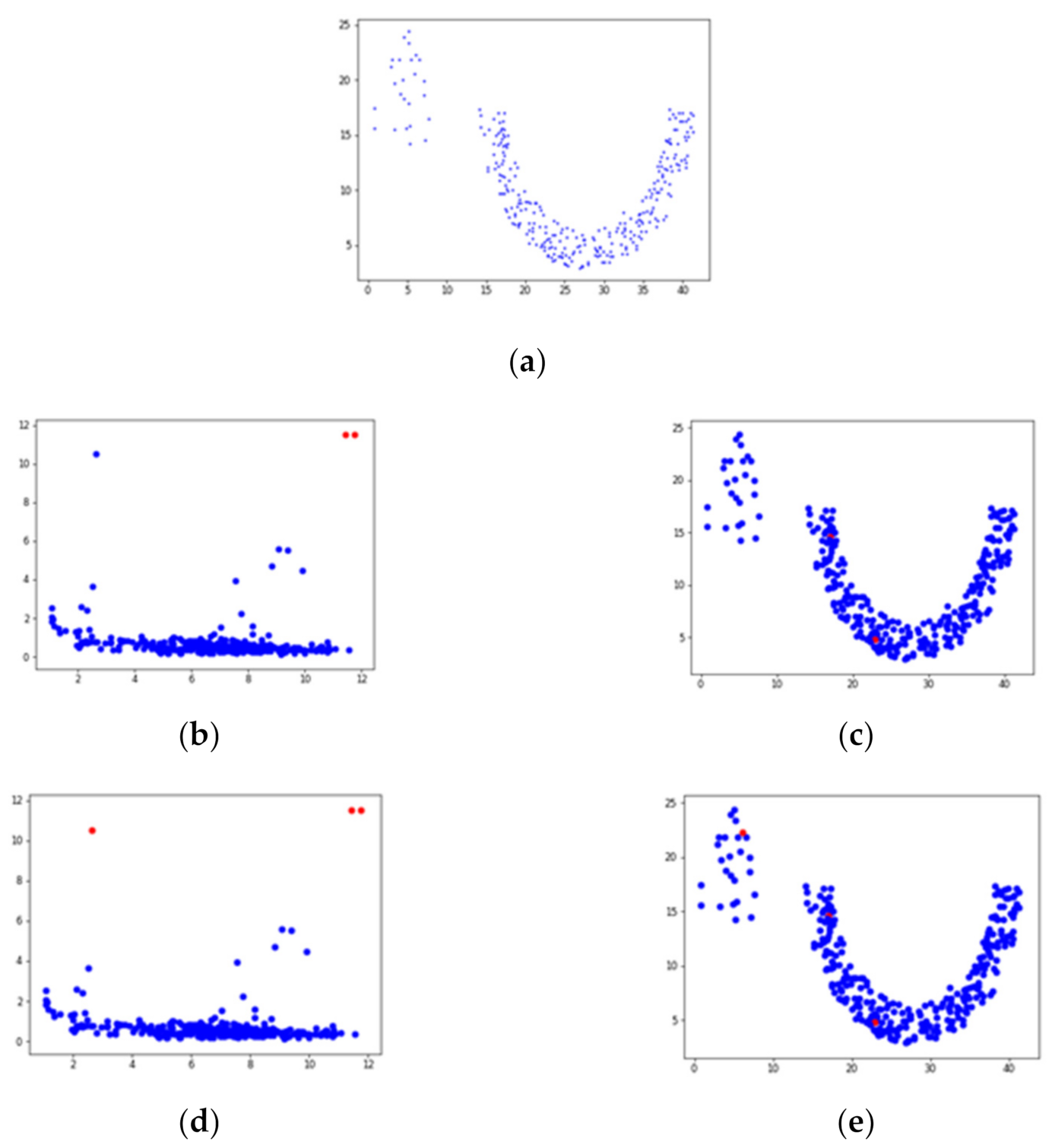
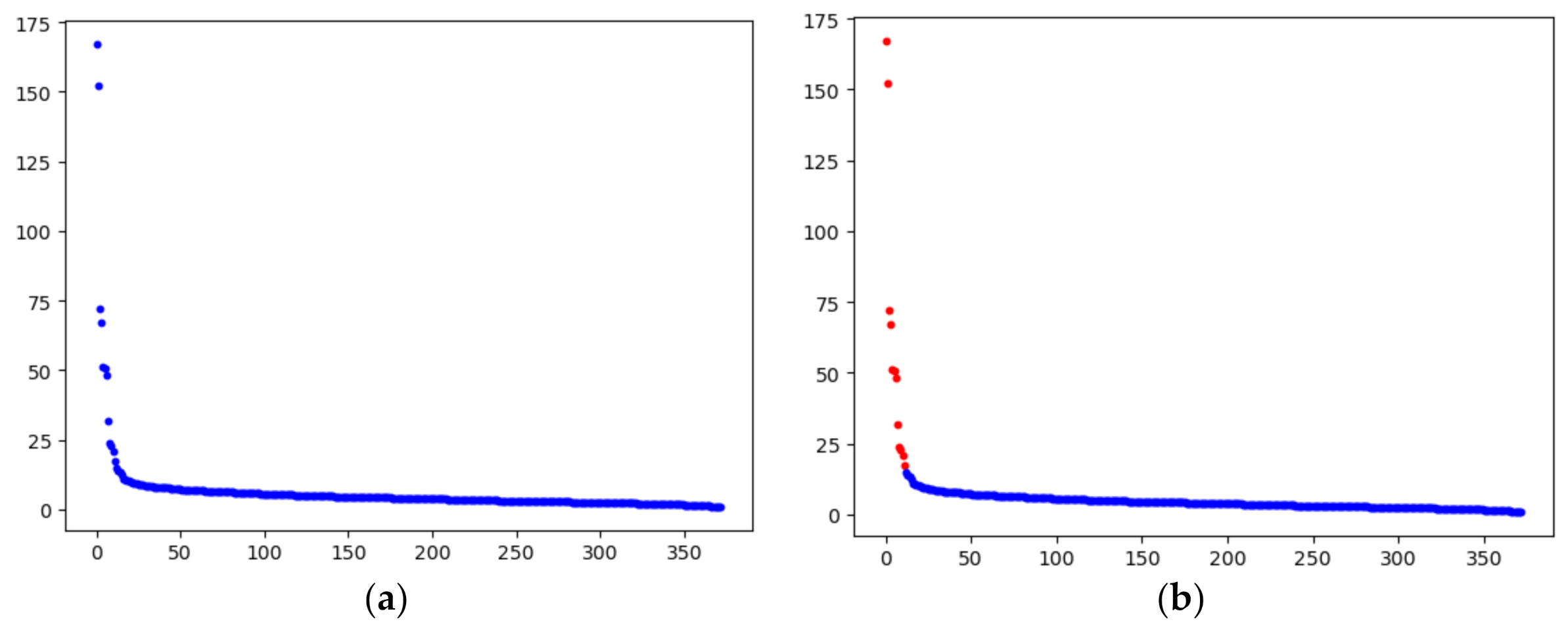
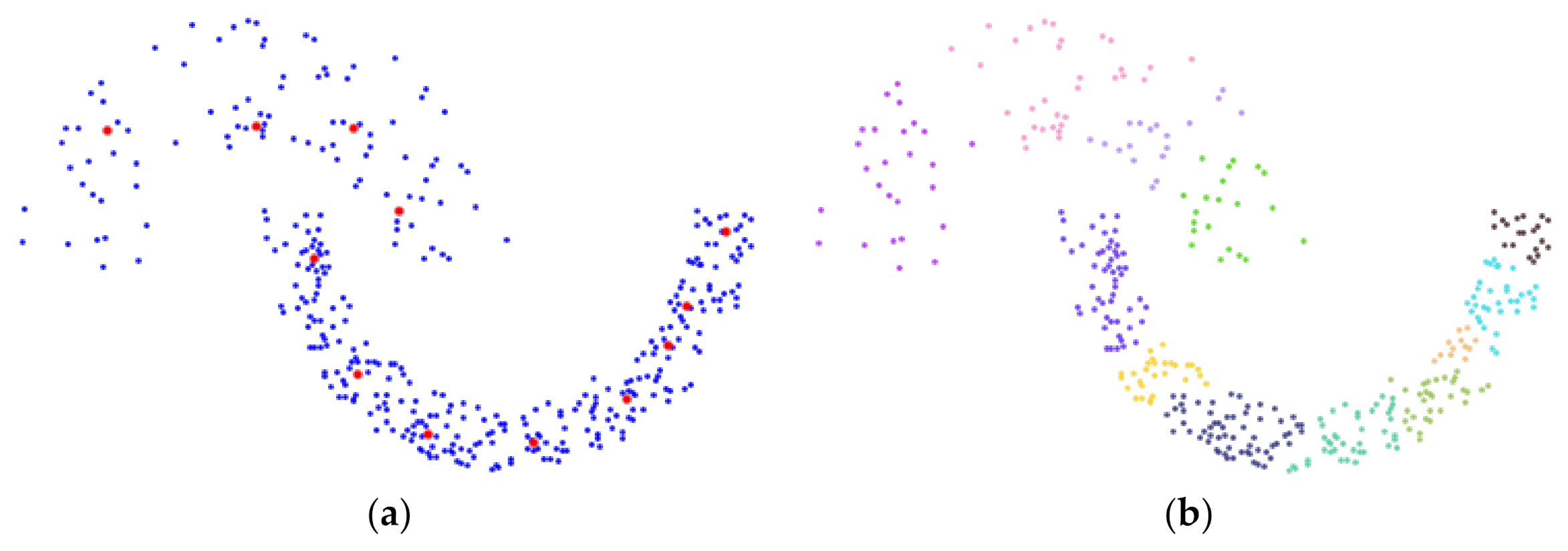
3.2. Detect and Select Key Points
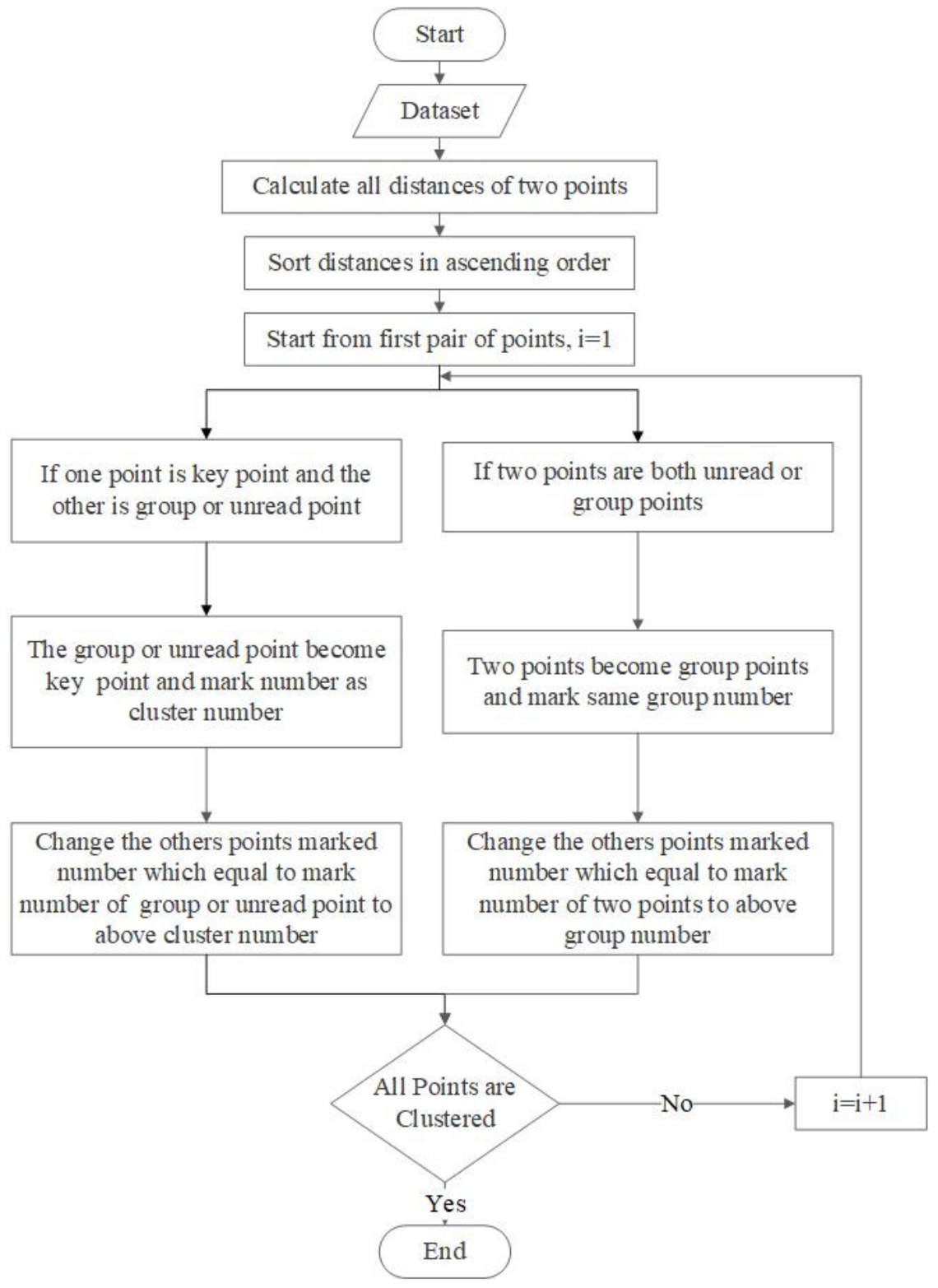
3.3. Detect Preliminary Clusters
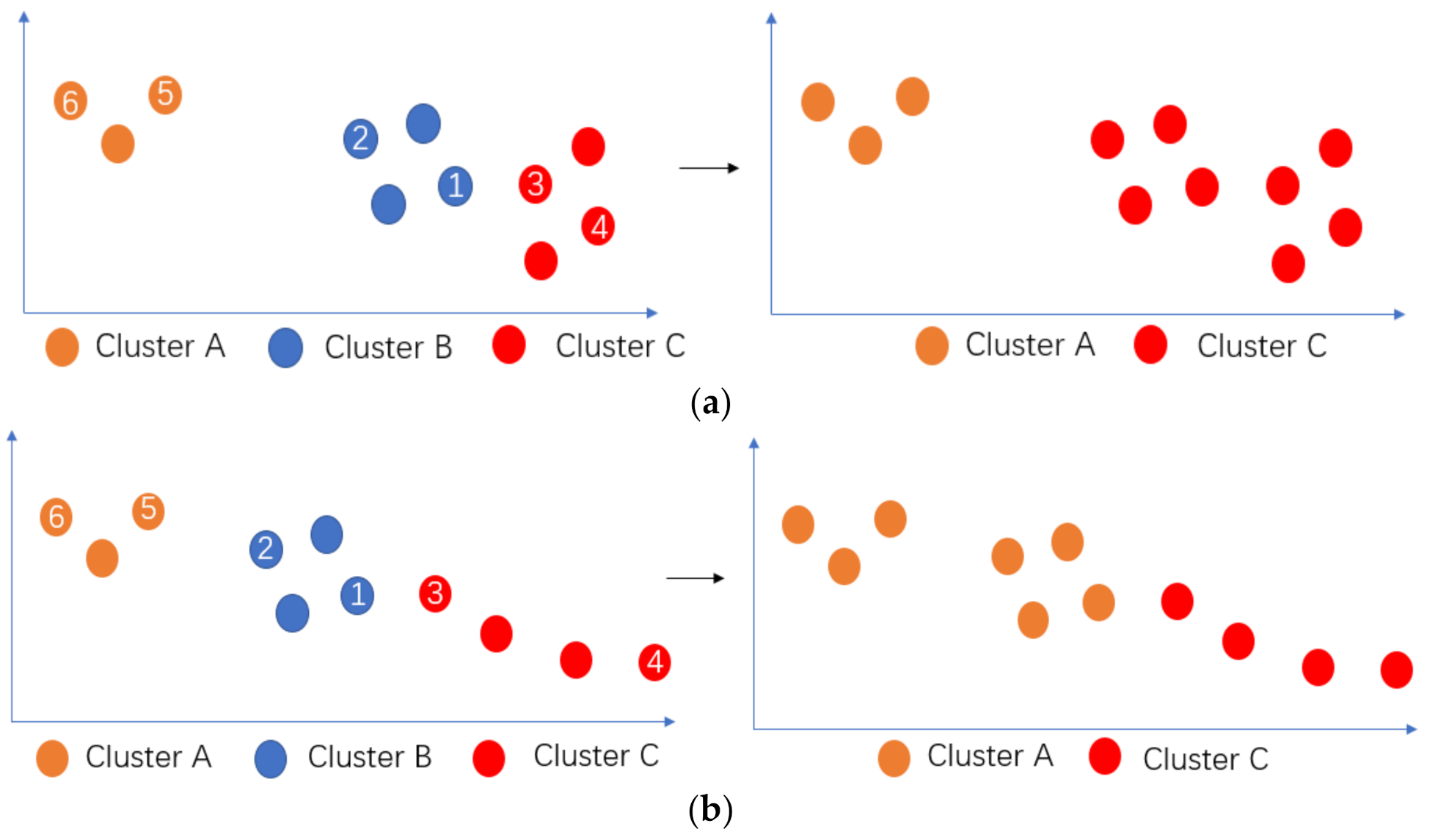
3.4. Merge Adjacent Clusters
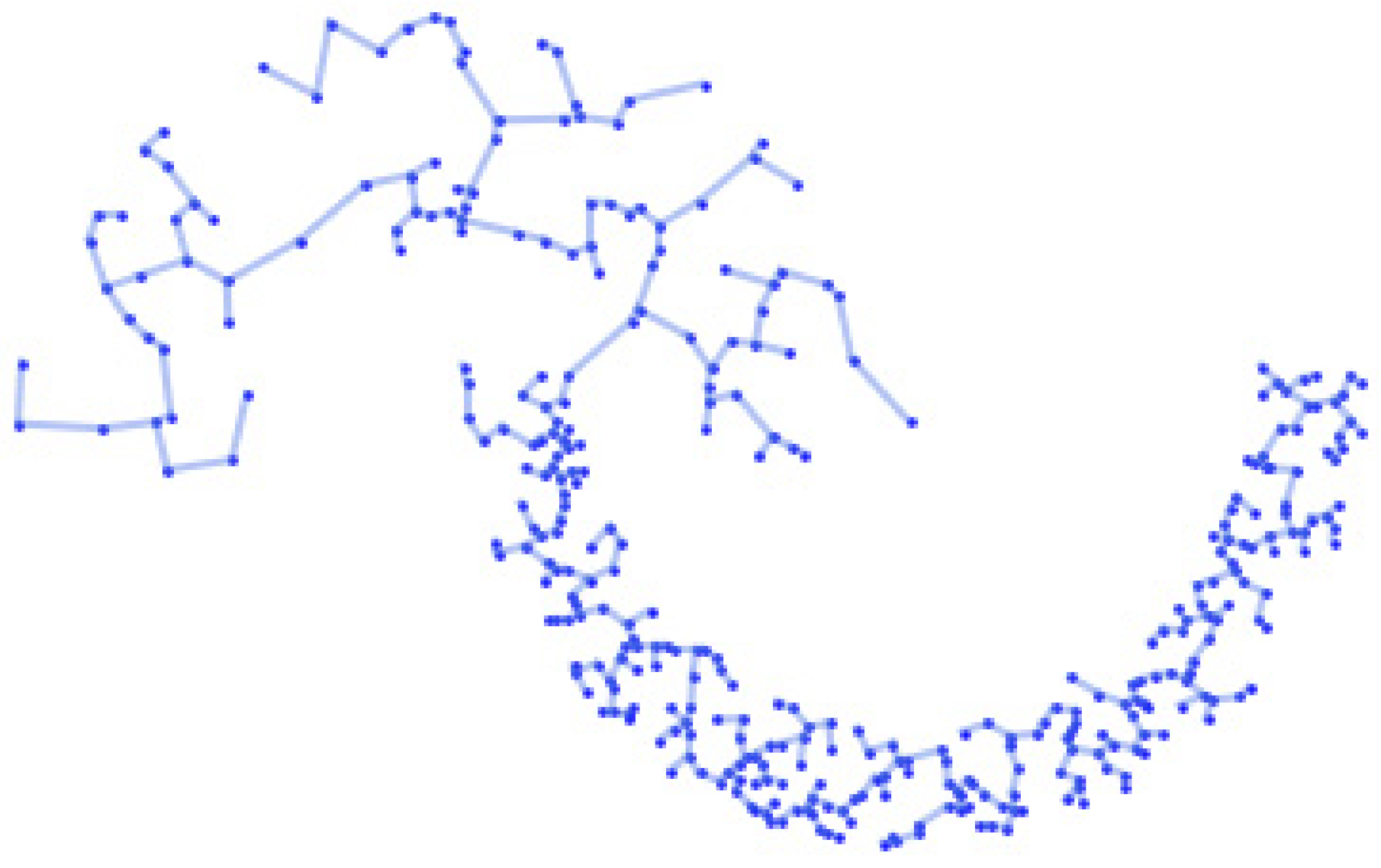
3.5. Construct Network
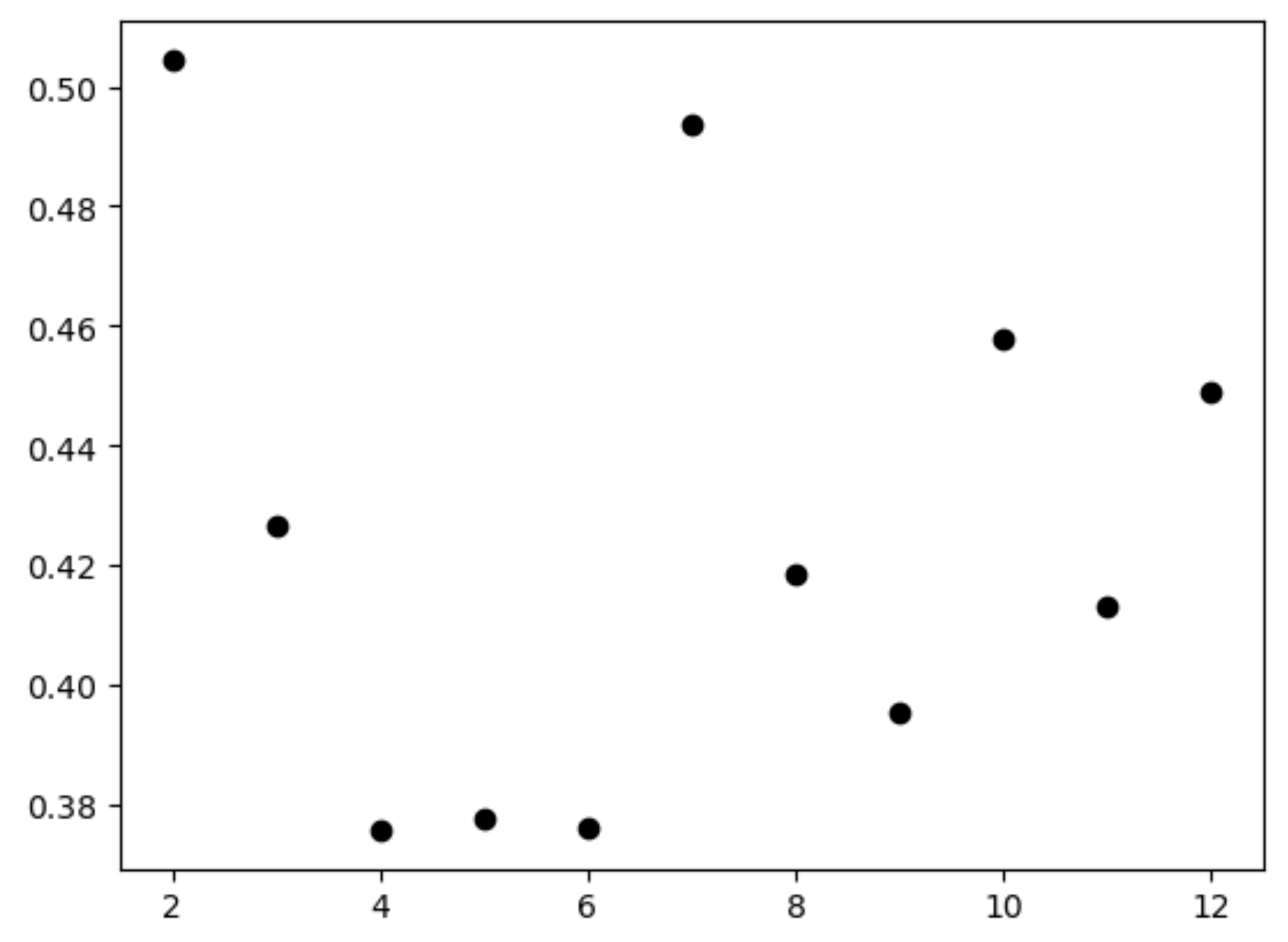
3.6. Identify the Optimal Cluster Result
4. Experimental Results
5. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bradlow, E.T.; Gangwar, M.; Kopalle, P.; Voleti, S. The role of big data and predictive analytics in retailing. J. Retail. 2017, 93, 79–95. [Google Scholar] [CrossRef] [Green Version]
- González, M.C.; Hidalgo, C.A.; Barabási, A.-L. Understanding individual human mobility patterns. Nature 2008, 453, 779–782. [Google Scholar] [CrossRef]
- Owda, A.; Balsa-Barreiro, J.; Fritsch, D. Methodology for Digital Preservation of the Cultural and Patrimonial Heritage: Generation of a 3D Model of the Church St. Peter and Paul (Calw, Germany) by Using Laser Scanning and Digital Photo-Grammetry; Emerald Publishing—Sensor Review: Bingley, UK, 2018. [Google Scholar]
- Balsa-Barreiro, J.; Lerma, L.J. A new methodology to estimate the discrete-return point density on airborne LiDAR sur-veys. Int. J. Remote Sens. 2014, 35, 1496–1510. [Google Scholar] [CrossRef]
- Balsa-Barreiro, J.; Lerma, J.L. Empirical study of variation in lidar point density over different land covers. Int. J. Remote Sens. 2014, 35, 3372–3383. [Google Scholar] [CrossRef]
- Balsa-Barreiro, J.; Avariento Vicent, J.P.; Lerma García, L.J. Airborne light detection and ranging (LiDAR) point density analysis. Sci. Res. Essays 2012, 7, 3010–3019. [Google Scholar] [CrossRef]
- Han, J.; Pei, J.; Kamber, M. Data Mining: Concepts and Techniques; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Raykov, Y.P.; Boukouvalas, A.; Baig, F.; Little, M.A. What to do when k-means clustering fails: A simple yet principled alternative Algorithm. PLoS ONE 2016, 11, e0162259. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-S.; Jun, C.-H. A simple and fast algorithm for K-medoids clustering. Expert Syst. Appl. 2009, 36, 3336–3341. [Google Scholar] [CrossRef]
- Zhang, T.; Ramakrishnan, R.; Livny, M. BIRCH: An efficient data clustering method for very large databases. ACM Sigmod Rec. 1996, 25, 103–114. [Google Scholar] [CrossRef]
- Guha, S.; Rastogi, R.; Shim, K. Cure: An efficient clustering algorithm for large databases. Inf. Syst. 2001, 26, 35–58. [Google Scholar] [CrossRef]
- Karypis, G.; Han, E.-H.; Kumar, V. Chameleon: Hierarchical clustering using dynamic modeling. Computer 1999, 32, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining KDD-96, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Ankerst, M.; Breunig, M.M.; Kriegel, H.-P.; Sander, J. OPTICS: Ordering points to identify the clustering structure. ACM Sigmod Rec. 1999, 28, 49–60. [Google Scholar] [CrossRef]
- Hinneburg, A.; Gabriel, H.-H. DENCLUE 2.0: Fast clustering based on kernel density estimation. In OpenMP in the Era of Low Power Devices and Accelerators; Springer Science and Business Media LLC.: Berlin/Heidelberg, Germany, 2007; pp. 70–80. [Google Scholar]
- Wang, W.; Yang, J.; Muntz, R. STING: A statistical information grid approach to spatial data mining. VLDB 1997, 97, 186–195. [Google Scholar]
- Uncu, Ö.; Gruver, W.A.; Kotak, D.B.; Sabaz, D.; Alibhai, Z.; Ng, C. GRIDBSCAN: GRId density-based spatial clustering of applications with noise. In Proceedings of the 2006 IEEE International Conference on Systems, Man and Cybernetics, Taipei, Taiwan, 8–11 October 2006; 2006; Volume 4, pp. 2976–2981. [Google Scholar]
- Rodriguez, A.; Laio, A. Clustering by fast search and find of density peaks. Science 2014, 344, 1492–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, K.-H.; Sester, M. Parameter-free cluster detection in spatial databases and its application to typification. Int. Arch. Photogramm. Remote Sens. 2000, 33, 75–83. [Google Scholar]
- Ding, J.; He, X.; Yuan, J.; Jiang, B. Automatic clustering based on density peak detection using generalized extreme value distribution. Soft Comput. 2018, 22, 2777–2796. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, Z.; Yu, F. Adaptive density peak clustering based on K-nearest neighbors with aggregating strategy. Knowl.-Based Syst. 2017, 133, 208–220. [Google Scholar]
- Xie, J.; Gao, H.; Xie, W.; Liu, X.; Grant, P.W. Robust clustering by detecting density peaks and assigning points based on fuzzy weighted K-nearest neighbors. Inf. Sci. 2016, 354, 19–40. [Google Scholar] [CrossRef]
- Du, M.; Ding, S.; Jia, H. Study on density peaks clustering based on k-nearest neighbors and principal component analysis. Knowl.-Based Syst. 2016, 99, 135–145. [Google Scholar] [CrossRef]
- Jinyin, C.; Xiang, L.; Haibing, Z.; Xintong, B. A novel cluster center fast determination clustering algorithm. Appl. Soft Comput. 2017, 57, 539–555. [Google Scholar] [CrossRef]
- Ruan, S.; Mehmood, R.; Daud, A.; Dawood, H.; Alowibdi, J.S. An adaptive method for clustering by fast search-and-find of density peaks: Adaptive-dp. In Proceedings of the 26th International Conference on World Wide Web Companion, Perth, Australia, 3–7 April 2017; pp. 119–127. [Google Scholar]
- Wang, G.; Song, Q. Automatic clustering via outward statistical testing on density metrics. IEEE Trans. Knowl. Data Eng. 2016, 28, 1971–1985. [Google Scholar] [CrossRef]
- Xu, J.; Wang, G.; Deng, W. DenPEHC: Density peak based efficient hierarchical clustering. Inf. Sci. 2016, 373, 200–218. [Google Scholar] [CrossRef]
- Wang, M.; Zuo, W.; Wang, Y. An improved density peaks-based clustering method for social circle discovery in social networks. Neurocomputing 2016, 179, 219–227. [Google Scholar] [CrossRef]
- Parmar, M.; Wang, D.; Zhang, X.; Tan, A.H.; Miao, C.; Jiang, J.; Zhou, Y. REDPC: A residual error-based density peak clustering algorithm. Neurocomputing 2019, 348, 82–96. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 21 June–18 July 1965; Volume 1, pp. 281–297. [Google Scholar]
- Lin, J.-L.; Kuo, J.-C.; Chuang, H.-W. Improving density peak clustering by automatic peak selection and single linkage clustering. Symmetry 2020, 12, 1168. [Google Scholar] [CrossRef]
- Ren, C.; Sun, L.; Yu, Y.; Wu, Q. Effective density peaks clustering algorithm based on the layered k-nearest neighbors and subcluster merging. IEEE Access 2020, 8, 123449–123468. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Lazarenko, D.; Bonald, T. Pairwise adjusted mutual information. arXiv 2021, arXiv:2103.12641. [Google Scholar]
- Campello, R. A fuzzy extension of the Rand index and other related indexes for clustering and classification assessment. Pattern Recognit. Lett. 2007, 28, 833–841. [Google Scholar] [CrossRef]




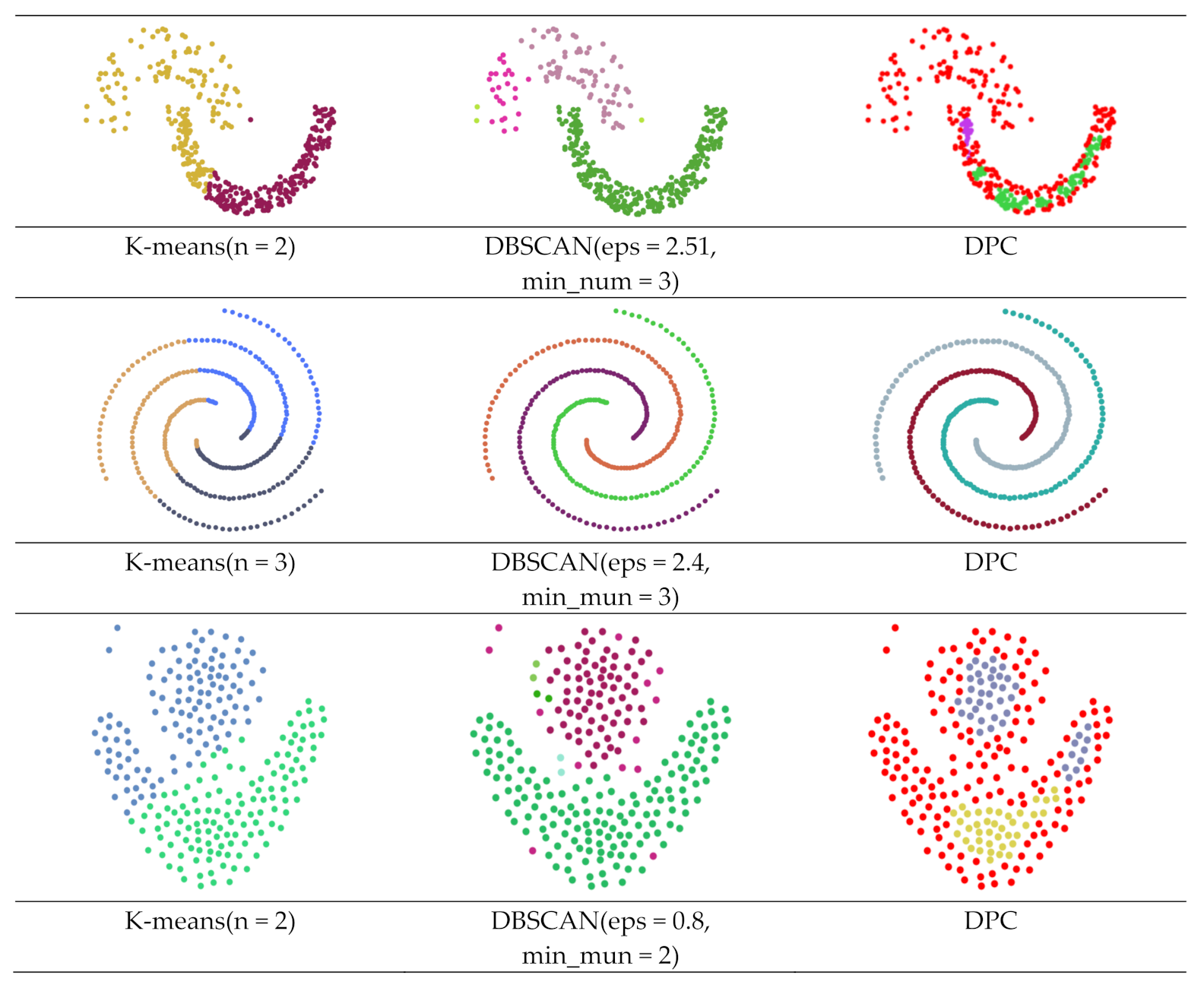
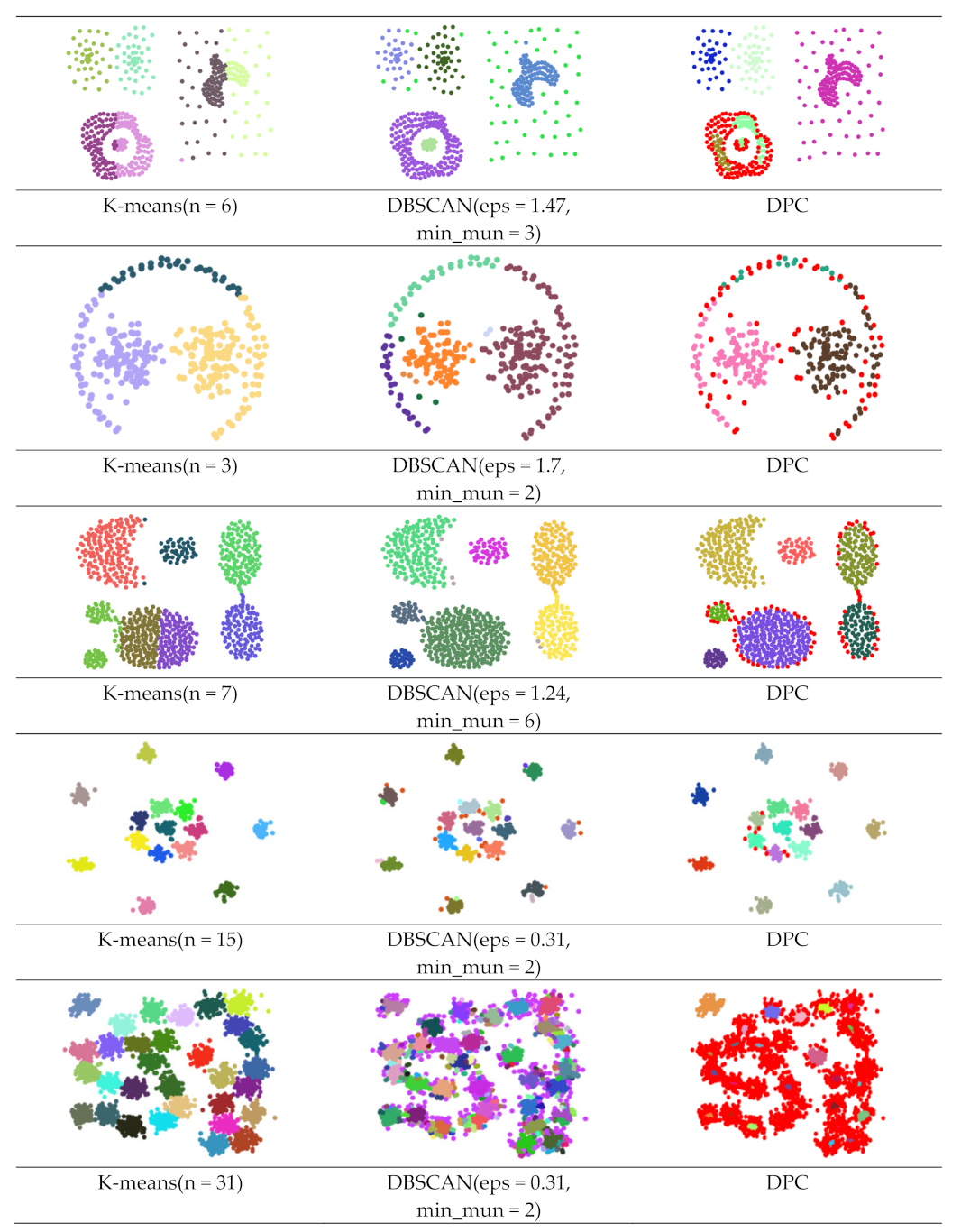
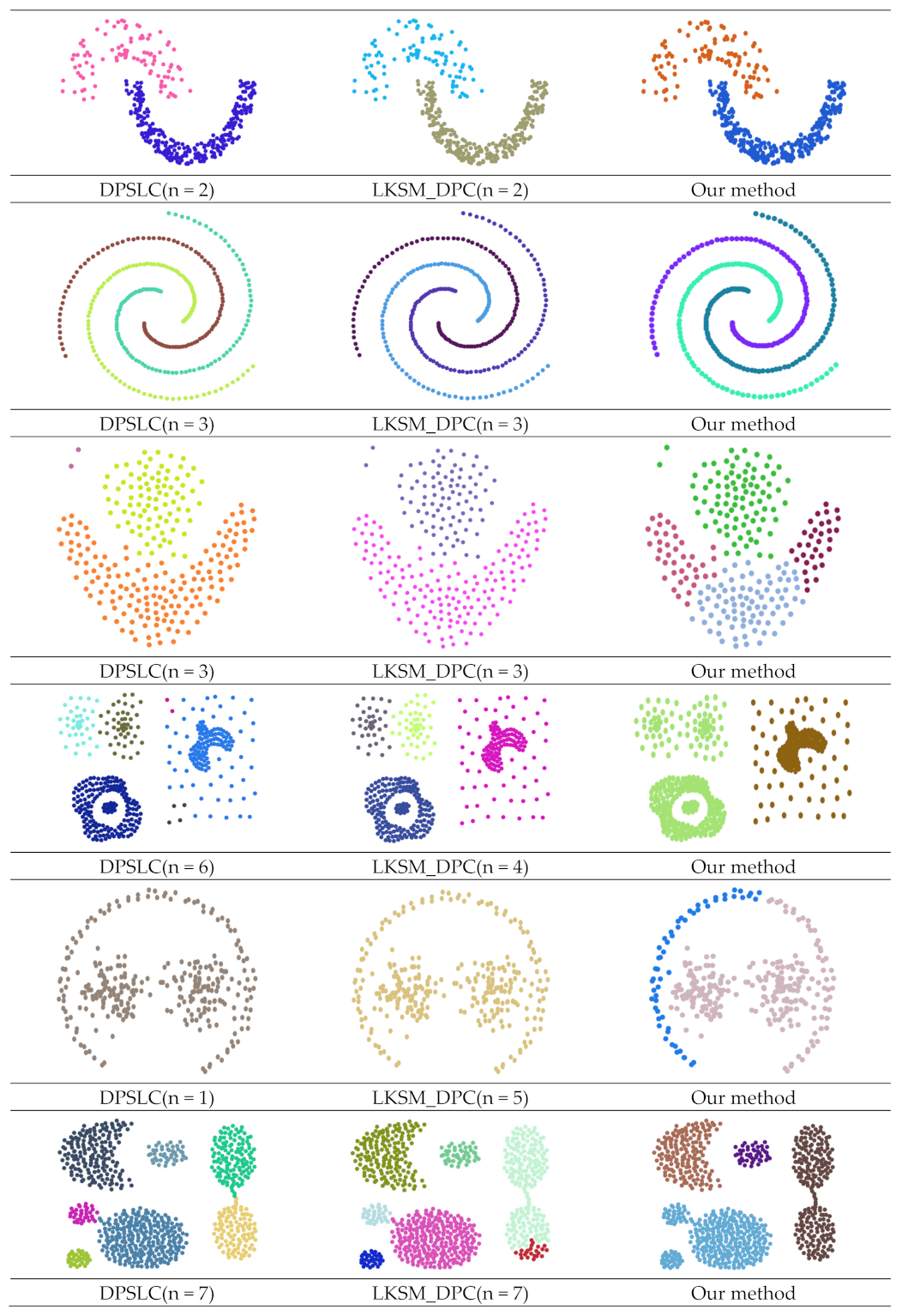
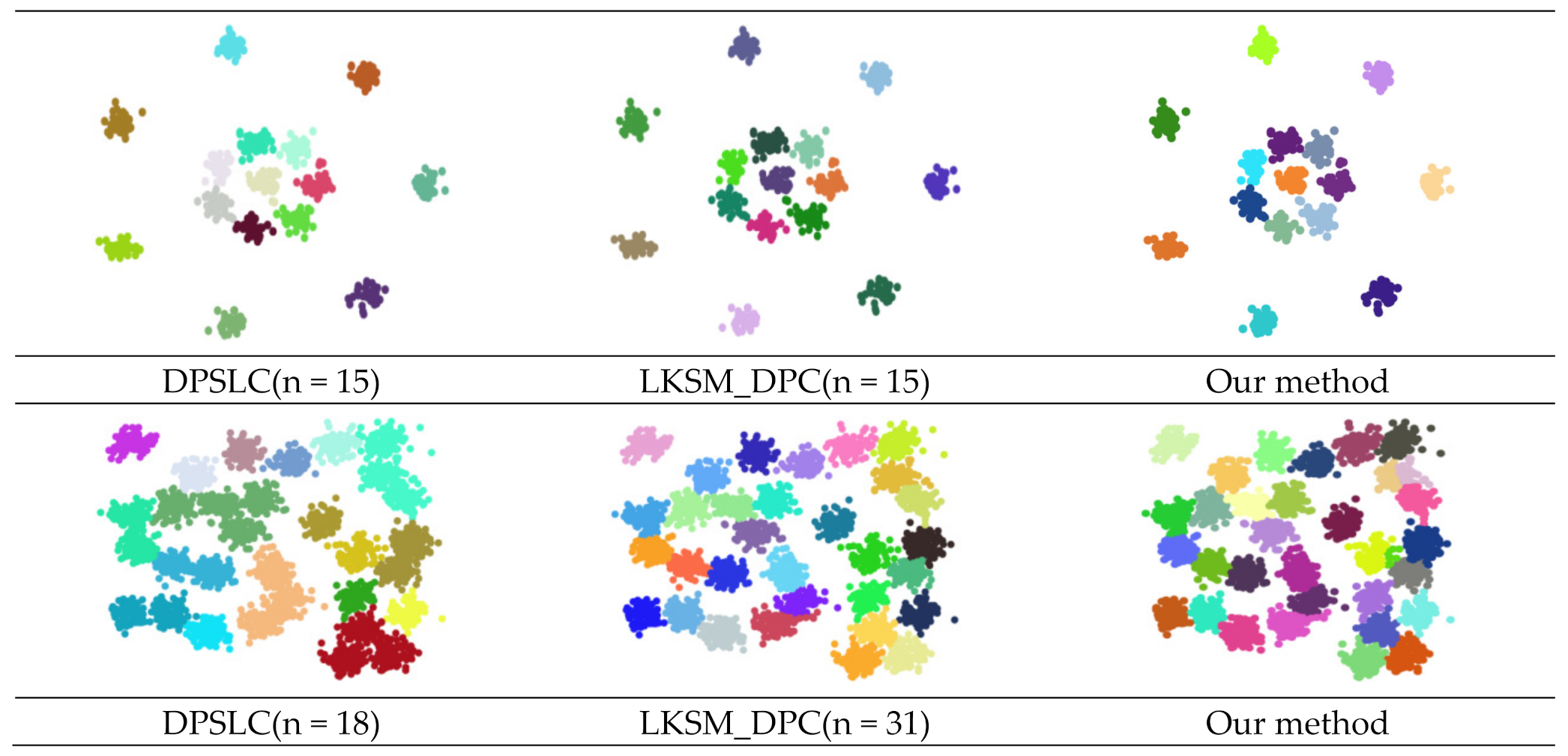
| Dataset | Method | ARI | AMI | FMI |
|---|---|---|---|---|
| Dataset (a) | K-means | 0.318 | 0.364 | 0.698 |
| DBSCAN | 0.941 | 0.864 | 0.977 | |
| DPC | −0.051 | 0.177 | 0.550 | |
| DPSLC | 1.000 | 1.000 | 1.000 | |
| LKSM_DPC | 1.000 | 1.000 | 1.000 | |
| Our method | 1.000 | 1.000 | 1.000 | |
| Dataset (b) | K-means | −0.006 | −0.005 | 0.328 |
| DBSCAN | 1.000 | 1.000 | 1.000 | |
| DPC | 1.000 | 1.000 | 1.000 | |
| DPSLC | 1.000 | 1.000 | 1.000 | |
| LKSM_DPC | 1.000 | 1.000 | 1.000 | |
| Our method | 1.000 | 1.000 | 1.000 | |
| Dataset (c) | K-means | 0.453 | 0.397 | 0.736 |
| DBSCAN | 0.878 | 0.791 | 0.941 | |
| DPC | 0.080 | 0.171 | 0.551 | |
| DPSLC | 0.988 | 0.970 | 0.994 | |
| LKSM_DPC | 1.000 | 1.000 | 1.000 | |
| Our method | 0.521 | 0.668 | 0.735 | |
| Dataset (d) | K-means | 0.538 | 0.713 | 0.642 |
| DBSCAN | 0.976 | 0.950 | 0.982 | |
| DPC | 0.578 | 0.754 | 0.680 | |
| DPSLC | 0.788 | 0.846 | 0.856 | |
| LKSM_DPC | 0.783 | 0.858 | 0.855 | |
| Our method | 0.437 | 0.585 | 0.676 | |
| Dataset (e) | K-means | 0.461 | 0.543 | 0.662 |
| DBSCAN | 0.529 | 0.640 | 0.687 | |
| DPC | 0.438 | 0.455 | 0.622 | |
| DPSLC | 0.000 | 0.000 | 0.577 | |
| LKSM_DPC | 0.000 | 0.000 | 0.000 | |
| Our method | 0.126 | 0.290 | 0.554 | |
| Dataset (f) | K-means | 0.762 | 0.878 | 0.816 |
| DBSCAN | 0.980 | 0.971 | 0.984 | |
| DPC | 0.851 | 0.876 | 0.884 | |
| DPSLC | 0.998 | 0.996 | 0.998 | |
| LKSM_DPC | 0.890 | 0.924 | 0.917 | |
| Our method | 0.734 | 0.835 | 0.819 | |
| Dataset (g) | K-means | 0.993 | 0.994 | 0.993 |
| DBSCAN | 0.921 | 0.936 | 0.927 | |
| DPC | 0.975 | 0.980 | 0.976 | |
| DPSLC | 0.993 | 0.994 | 0.993 | |
| LKSM_DPC | 0.986 | 0.989 | 0.987 | |
| Our method | 0.986 | 0.989 | 0.987 | |
| Dataset (h) | K-means | 0.953 | 0.966 | 0.955 |
| DBSCAN | 0.550 | 0.768 | 0.565 | |
| DPC | 0.031 | 0.445 | 0.176 | |
| DPSLC | 0.585 | 0.869 | 0.653 | |
| LKSM_DPC | 0.935 | 0.956 | 0.938 | |
| Our method | 0.928 | 0.951 | 0.930 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.; Ma, D.; Yan, X.; Zhu, W.; Zhao, Z. A Density-Peak-Based Clustering Method for Multiple Densities Dataset. ISPRS Int. J. Geo-Inf. 2021, 10, 589. https://doi.org/10.3390/ijgi10090589
Shi Z, Ma D, Yan X, Zhu W, Zhao Z. A Density-Peak-Based Clustering Method for Multiple Densities Dataset. ISPRS International Journal of Geo-Information. 2021; 10(9):589. https://doi.org/10.3390/ijgi10090589
Chicago/Turabian StyleShi, Zhicheng, Ding Ma, Xue Yan, Wei Zhu, and Zhigang Zhao. 2021. "A Density-Peak-Based Clustering Method for Multiple Densities Dataset" ISPRS International Journal of Geo-Information 10, no. 9: 589. https://doi.org/10.3390/ijgi10090589






