Use of the Split Luciferase Complementation Assay to Identify Novel Small Molecules That Disrupt Essential Protein–Protein Interactions of Viruses
Abstract
1. Introduction
2. SLCA Applications in Viruses
2.1. Human Immunodeficiency Virus Type-1 (HIV-1)
2.2. Dengue
2.3. Eastern Equine Encephalitis Virus (EEEV)
2.4. Western Equine Encephalitis Virus (WEEV)
2.5. Epstein–Barr Virus (EBV)
2.6. Rabies Virus
3. SLCA
4. FDA-Approved Inhibitors
5. Conclusions
6. Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ulgheri, F.M.; Bernardes, B.G.; Lancellotti, M. Decoding Dengue: A Global Perspective, History, Role, and Challenges. Pathogens 2025, 14, 954. [Google Scholar] [CrossRef]
- World Health Organization. Dengue. 2025. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 25 September 2025).
- Byk, L.A.; Gamarnik, A.V. Properties and Functions of the Dengue Virus Capsid Protein. Annu. Rev. Virol. 2016, 3, 263–281. [Google Scholar] [CrossRef]
- Zhang, J.; Tamilarasu, N.; Hwang, S.; Garber, M.E.; Huq, I.; Jones, K.A.; Rana, T.M. HIV-1 TAR RNA Enhances the Interaction between Tat and Cyclin T1. J. Biol. Chem. 2000, 275, 34314–34319. [Google Scholar] [CrossRef]
- Zhang, H. Id protein-firefly luciferase N-fragment & firefly luciferase C-fragment-myod protein. In Molecular Imaging and Contrast Agent Database (MICAD); U.S. National Library of Medicine: Bethesda, MD, USA, 2008. Available online: https://www.ncbi.nlm.nih.gov/books/NBK23175/ (accessed on 25 September 2025).
- Misawa, N.; Kafi, A.K.; Hattori, M.; Miura, K.; Masuda, K.; Ozawa, T. Rapid and high-sensitivity cell-based assays of protein-protein interactions using split click beetle luciferase complementation: An approach to the study of G-protein-coupled receptors. Anal. Chem. 2010, 82, 2552–2560. [Google Scholar] [CrossRef]
- Lang, Y.; Li, Z.; Li, H. Analysis of Protein-Protein Interactions by Split Luciferase Complementation Assay. Curr. Protoc. Toxicol. 2019, 82, e90. [Google Scholar] [CrossRef]
- Kawamura, G.; Ozawa, T. Luciferase complementation for cellular assays beyond protein-protein interactions. Anal. Sci. 2025, 41, 571–583. [Google Scholar] [CrossRef]
- Liang, Y.; Li, Z. Split-Luciferase Complementation for Analysis of Virus-Host Protein Interactions. Methods Mol. Biol. 2022, 2400, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Villalobos, V.; Naik, S.; Piwnica-Worms, D. Detection of protein-protein interactions in live cells and animals with split firefly luciferase protein fragment complementation. Methods Mol. Biol. 2008, 439, 339–352. [Google Scholar] [CrossRef]
- Paulmurugan, R.; Gambhir, S.S. Firefly Luciferase Enzyme Fragment Complementation for Imaging in Cells and Living Animals. Anal. Chem. 2005, 77, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Jones, J. The split luciferase complementation assay. Methods Mol. Biol. 2010, 655, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.; Howell, M.H.; von Arnim, A.G. Structure-function studies on the active site of the coelenterazine-dependent luciferase from Renilla. Protein Sci. 2008, 17, 725–735. [Google Scholar] [CrossRef]
- Bignon, C.; Gruet, A.; Longhi, S. Split-GFP Reassembly Assay: Strengths and Caveats from a Multiparametric Analysis. Int. J. Mol. Sci. 2022, 23, 13167. [Google Scholar] [CrossRef]
- Magliery, T.J.; Wilson, C.G.M.; Pan, W.; Mishler, D.; Ghosh, I.; Hamilton, A.D.; Regan, L. Detecting Protein–Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism. J. Am. Chem. Soc. 2005, 127, 146–157. [Google Scholar] [CrossRef]
- Niu, X.; Ye, K.; Wang, L.; Lin, Y.; Du, D. A review on emerging principles and strategies for colorimetric and fluorescent detection of alkaline phosphatase activity. Anal. Chim. Acta 2019, 1086, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.Z.; Wang, Z.; Ning, K.; Ren, C.; Yang, D.; Hu, X.Y.; Xu, Q. Ultrasensitive alkaline phosphatase activity assay based on controllable signal probe production coupled with the cathodic photoelectrochemical analysis. Food Chem. 2023, 421, 136177. [Google Scholar] [CrossRef]
- Azad, T.; Tashakor, A.; Hosseinkhani, S. Split-luciferase complementary assay: Applications, recent developments, and future perspectives. Anal. Bioanal. Chem. 2014, 406, 5541–5560. [Google Scholar] [CrossRef]
- Chen, M.; Yan, C.; Qin, F.; Zhang, X.E. Near-Infrared Luciferase Complementation Assay with Enhanced Bioluminescence for Studying Protein-Protein Interactions and Drug Evaluation Under Physiological Conditions. Anal. Chem. 2022, 94, 13700–13709. [Google Scholar] [CrossRef]
- Hansen, T.; Baris, J.; Zhao, M.; Sutton, R.E. Cell-based and cell-free firefly luciferase complementation assay to quantify Human Immunodeficiency Virus type 1 Rev-Rev interaction. Virology 2022, 576, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.F.; Gan, C.Y.; Cui, J.; Luo, Y.Y.; Cai, X.F.; Yuan, Y.; Shen, J.; Li, Z.Y.; Zhang, W.L.; Long, Q.X.; et al. Identification of Compounds Targeting Hepatitis B Virus Core Protein Dimerization through a Split Luciferase Complementation Assay. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. HIV. 2025. Available online: https://cdn.who.int/media/docs/default-source/hq-hiv-hepatitis-and-stis-library/who-ias-hiv-statistics_2025-new.pdf (accessed on 25 September 2025).
- Moyo, E.; Moyo, P.; Murewanhema, G.; Mhango, M.; Chitungo, I.; Dzinamarira, T. Key populations and Sub-Saharan Africa’s HIV response. Front. Public Health 2023, 11, 1079990. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. The cell biology of HIV-1 latency and rebound. Retrovirology 2024, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Booth, D.S.; Jayaraman, B.; Cheng, Y.; Frankel, A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. USA 2010, 107, 12481–12486. [Google Scholar] [CrossRef]
- Campbell, E.M.; Hope, T.J. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015, 13, 471–483. [Google Scholar] [CrossRef]
- Canales, E.; Tse, W.; Schroeder, S.D.; Chou, C.H.; Liu, Q.; Zhang, J.; Lazerwith, S.E.; Morganelli, P.; Saito, R.D.; Brizgys, G.; et al. Discovery of Lenacapavir: First-in-Class Twice-Yearly Capsid Inhibitor for HIV-1 Treatment and Pre-exposure Prophylaxis. J. Med. Chem. 2025, 68, 21072–21094. [Google Scholar] [CrossRef]
- Neverette, N.C.; Dumond, J.B.; McMahon, D.K.; Devanathan, A.S. Lenacapavir: Playing the Long Game in the New Era of Antiretrovirals. Clin. Pharmacol. Ther. 2024, 117, 353–367. [Google Scholar] [CrossRef]
- Li, C.; Burdick, R.C.; Siddiqui, R.; Janaka, S.K.; Hsia, R.C.; Hu, W.S.; Pathak, V.K. Lenacapavir disrupts HIV-1 core integrity while stabilizing the capsid lattice. Proc. Natl. Acad. Sci. USA 2025, 122, e2420497122. [Google Scholar] [CrossRef]
- Eschbach, J.E.; Puray-Chavez, M.; Mohammed, S.; Wang, Q.; Xia, M.; Huang, L.C.; Shan, L.; Kutluay, S.B. HIV-1 capsid stability and reverse transcription are finely balanced to minimize sensing of reverse transcription products via the cGAS-STING pathway. mBio 2024, 15, e0034824. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.W.; Briganti, L.; Annamalai, A.S.; Greenwood, J.; Shkriabai, N.; Haney, R.; Armstrong, M.L.; Wempe, M.F.; Singh, S.P.; Francis, A.C.; et al. The primary mechanism for highly potent inhibition of HIV-1 maturation by lenacapavir. PLoS Pathog. 2025, 21, e1012862. [Google Scholar] [CrossRef]
- National Institutes of Health. DailyMed—SUNLENCA-Lenacapavir Sodium Tablet, Film Coated SUNLENCA-Lenacapavir Sodium Kit. 2025. Available online: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=e5652804-29c4-40d7-aeb2-0142ed2a7b5b (accessed on 25 September 2025).
- Briganti, L.; Annamalai, A.S.; Bester, S.M.; Wei, G.; Andino-Moncada, J.R.; Singh, S.P.; Kleinpeter, A.B.; Tripathi, M.; Nguyen, B.; Radhakrishnan, R.; et al. Structural and mechanistic bases for resistance of the M66I capsid variant to lenacapavir. mBio 2025, 16, e0361324. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Garber, M.E.; Fang, S.-M.; Fischer, W.H.; Jones, K.A. A Novel CDK9-Associated C-Type Cyclin Interacts Directly with HIV-1 Tat and Mediates Its High-Affinity, Loop-Specific Binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Shin, Y.; Kim, H.G.; Park, C.M.; Choi, M.S.; Kim, D.-E.; Choi, B.-S.; Kim, K.; Yoon, C.-H. Identification of novel compounds against Tat-mediated human immunodeficiency virus-1 transcription by high-throughput functional screening assay. Biochem. Biophys. Res. Commun. 2020, 523, 368–374. [Google Scholar] [CrossRef]
- Diamond, M.S.; Pierson, T.C. Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell 2015, 162, 488–492. [Google Scholar] [CrossRef]
- Tay, M.Y.; Saw, W.G.; Zhao, Y.; Chan, K.W.; Singh, D.; Chong, Y.; Forwood, J.K.; Ooi, E.E.; Grüber, G.; Lescar, J.; et al. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J. Biol. Chem. 2015, 290, 2379–2394. [Google Scholar] [CrossRef] [PubMed]
- Jablunovsky, A.; Jose, J. The Dynamic Landscape of Capsid Proteins and Viral RNA Interactions in Flavivirus Genome Packaging and Virus Assembly. Pathogens 2024, 13, 120. [Google Scholar] [CrossRef]
- Figueira-Mansur, J.; Aguilera, E.A.; Stoque, R.M.; Ventura, G.T.; Mohana-Borges, R. Mutations in the dimer interfaces of the dengue virus capsid protein affect structural stability and impair RNA-capsid interaction. Sci. Rep. 2019, 9, 2829. [Google Scholar] [CrossRef] [PubMed]
- Sangiambut, S.; Promphet, N.; Chaiyaloom, S.; Puttikhunt, C.; Avirutnan, P.; Kasinrerk, W.; Sittisombut, N.; Malasit, P. Increased capsid oligomerization is deleterious to dengue virus particle production. J. Gen. Virol. 2021, 102, 001635. [Google Scholar] [CrossRef]
- Hasan, S.S.; Dey, D.; Singh, S.; Martin, M. The Structural Biology of Eastern Equine Encephalitis Virus, an Emerging Viral Threat. Pathogens 2021, 10, 973. [Google Scholar] [CrossRef]
- Carey, B.D.; Bakovic, A.; Callahan, V.; Narayanan, A.; Kehn-Hall, K. New World alphavirus protein interactomes from a therapeutic perspective. Antivir. Res. 2019, 163, 125–139. [Google Scholar] [CrossRef]
- Lundberg, L.; Pinkham, C.; de la Fuente, C.; Brahms, A.; Shafagati, N.; Wagstaff, K.M.; Jans, D.A.; Tamir, S.; Kehn-Hall, K. Selective Inhibitor of Nuclear Export (SINE) Compounds Alter New World Alphavirus Capsid Localization and Reduce Viral Replication in Mammalian Cells. PLoS Negl. Trop. Dis. 2016, 10, e0005122. [Google Scholar] [CrossRef] [PubMed]
- Gauci, P.J.; Wu, J.Q.; Rayner, G.A.; Barabé, N.D.; Nagata, L.P.; Proll, D.F. Identification of Western equine encephalitis virus structural proteins that confer protection after DNA vaccination. Clin. Vaccine Immunol. 2010, 17, 176–179. [Google Scholar] [CrossRef]
- Ma, B.; Cao, Z.; Ding, W.; Zhang, X.; Xiang, Y.; Cao, D. Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus. Cell Rep. 2025, 44, 115724. [Google Scholar] [CrossRef]
- Peng, W.; Peltier, D.C.; Larsen, M.J.; Kirchhoff, P.D.; Larsen, S.D.; Neubig, R.R.; Miller, D.J. Identification of thieno[3,2-b]pyrrole derivatives as novel small molecule inhibitors of neurotropic alphaviruses. J. Infect. Dis. 2009, 199, 950–957. [Google Scholar] [CrossRef]
- Delekta, P.C.; Dobry, C.J.; Sindac, J.A.; Barraza, S.J.; Blakely, P.K.; Xiang, J.; Kirchhoff, P.D.; Keep, R.F.; Irani, D.N.; Larsen, S.D.; et al. Novel Indole-2-Carboxamide Compounds Are Potent Broad-Spectrum Antivirals Active against Western Equine Encephalitis Virus. J. Virol. 2014, 88, 11199–11214. [Google Scholar] [CrossRef]
- Chakravorty, S.; Afzali, B.; Kazemian, M. EBV-associated diseases: Current therapeutics and emerging technologies. Front. Immunol. 2022, 13, 1059133. [Google Scholar] [CrossRef]
- Turk, S.M.; Jiang, R.; Chesnokova, L.S.; Hutt-Fletcher, L.M. Antibodies to gp350/220 enhance the ability of Epstein-Barr virus to infect epithelial cells. J. Virol. 2006, 80, 9628–9633. [Google Scholar] [CrossRef]
- Henson Brandon, W.; Perkins Edward, M.; Cothran Jonathan, E.; Desai, P. Self-Assembly of Epstein-Barr Virus Capsids. J. Virol. 2009, 83, 3877–3890. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Thompson, J.; Swaminathan, S. Spironolactone blocks Epstein–Barr virus production by inhibiting EBV SM protein function. Proc. Natl. Acad. Sci. USA 2016, 113, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Kiflu, A.B. The Immune Escape Strategy of Rabies Virus and Its Pathogenicity Mechanisms. Viruses 2024, 16, 1774. [Google Scholar] [CrossRef]
- Schoehn, G.; Iseni, F.; Mavrakis, M.; Blondel, D.; Ruigrok Rob, W.H. Structure of Recombinant Rabies Virus Nucleoprotein-RNA Complex and Identification of the Phosphoprotein Binding site. J. Virol. 2001, 75, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Nevers, Q.; Scrima, N.; Glon, D.; Le Bars, R.; Decombe, A.; Garnier, N.; Ouldali, M.; Lagaudrière-Gesbert, C.; Blondel, D.; Albertini, A.; et al. Properties of rabies virus phosphoprotein and nucleoprotein biocondensates formed in vitro and in cellulo. PLoS Pathog. 2022, 18, e1011022. [Google Scholar] [CrossRef]
- Zheng, W.; Zhu, X.; Zhu, T.; Luo, Q.; Zhao, Y.; Xu, T. A Novel Protein NLRP12-119aa that Prevents Rhabdovirus Replication by Disrupting the RNP Complex Formation. Adv. Sci. 2025, 12, e2409953. [Google Scholar] [CrossRef]
- Dobritsa, S.V.; Kuok, I.T.; Nguyen, H.; Webster, J.C.; Spragg, A.M.; Morley, T.; Carr, G.J. Development of a high-throughput cell-based assay for identification of IL-17 inhibitors. J. Biomol. Screen. 2013, 18, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Hassen-Khodja, C.; Georget, V.; Rose, T.; Jacob, Y.; Janin, Y.L.; Nisole, S.; Vidalain, P.O.; Arhel, N.J. Measuring the subcellular compartmentalization of viral infections by protein complementation assay. Proc. Natl. Acad. Sci. USA 2021, 118, e2010524118. [Google Scholar] [CrossRef]
- Orkin, C.; Cahn, P.; Castagna, A.; Emu, B.; Harrigan, P.R.; Kuritzkes, D.R.; Nelson, M.; Schapiro, J. Opening the door on entry inhibitors in HIV: Redefining the use of entry inhibitors in heavily treatment experienced and treatment-limited individuals living with HIV. HIV Med. 2022, 23, 936–946. [Google Scholar] [CrossRef]
- Uchida, A.; Isobe, Y.; Asano, J.; Uemura, Y.; Hoshikawa, M.; Takagi, M.; Miura, I. Targeting BCL2 with venetoclax is a promising therapeutic strategy for “double-proteinexpression” lymphoma with MYC and BCL2 rearrangements. Haematologica 2019, 104, 1417–1421. [Google Scholar] [CrossRef] [PubMed]
- Rebolledo-Bustillo, M.; Garcia-Gomez, D.; Dávila, E.M.; Castro, M.E.; Caballero, N.A.; Melendez, F.J.; Baizabal-Aguirre, V.M.; Sanchez-Gaytan, B.L.; Perez-Aguilar, J.M. Structural Basis of the Binding Mode of the Antineoplastic Compound Motixafortide (BL-8040) in the CXCR4 Chemokine Receptor. Int. J. Mol. Sci. 2023, 24, 4393. [Google Scholar] [CrossRef] [PubMed]
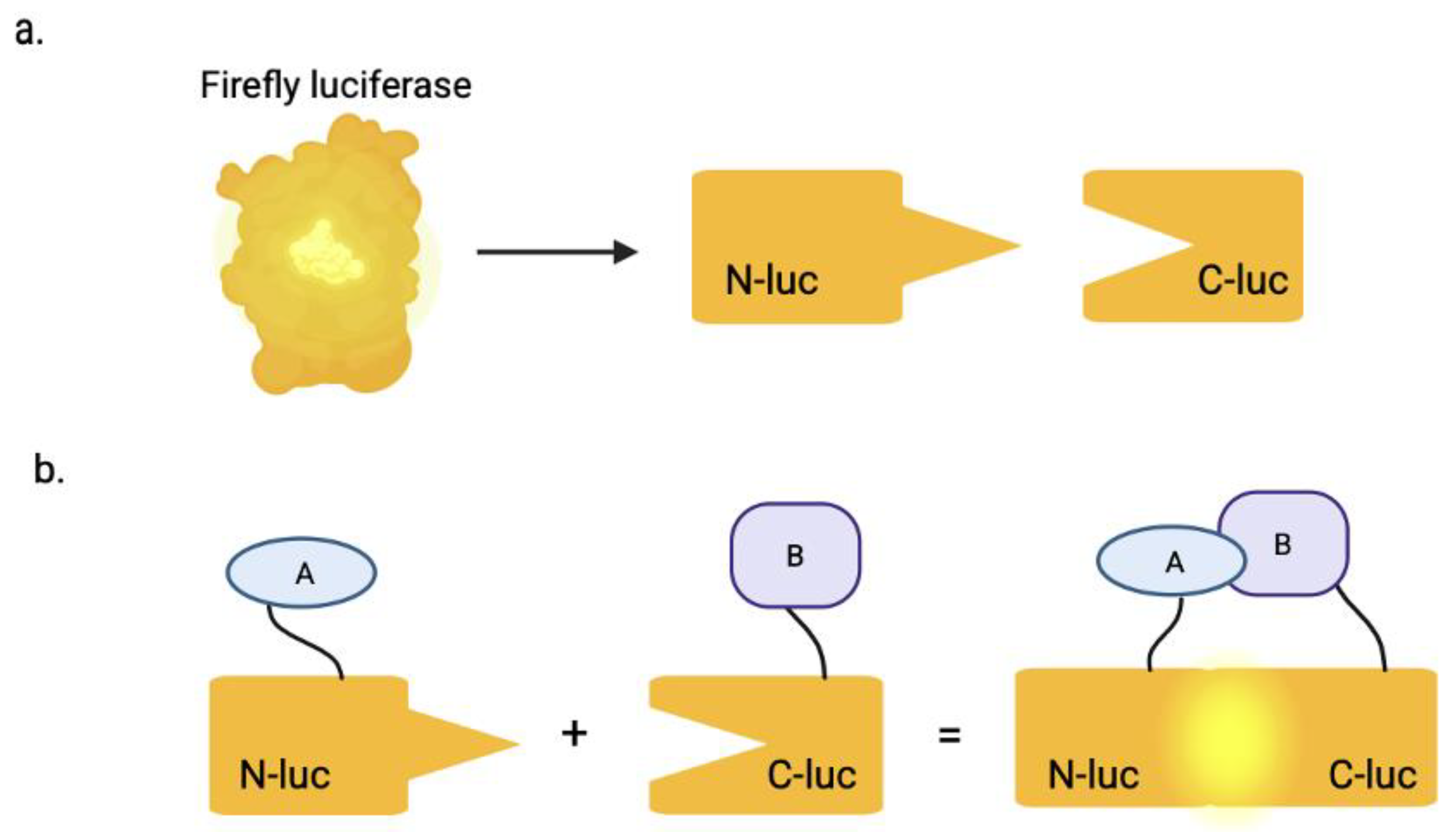
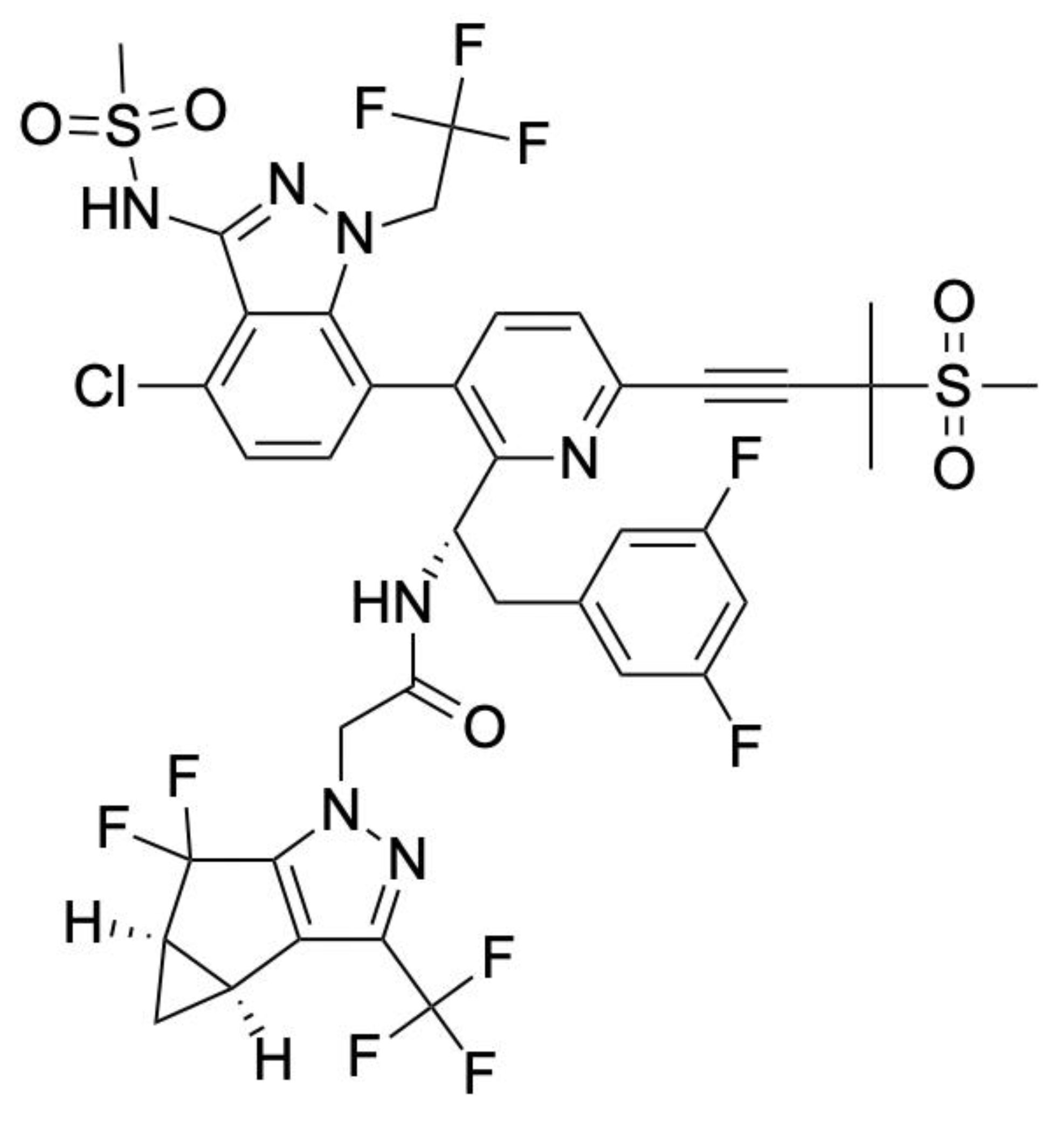
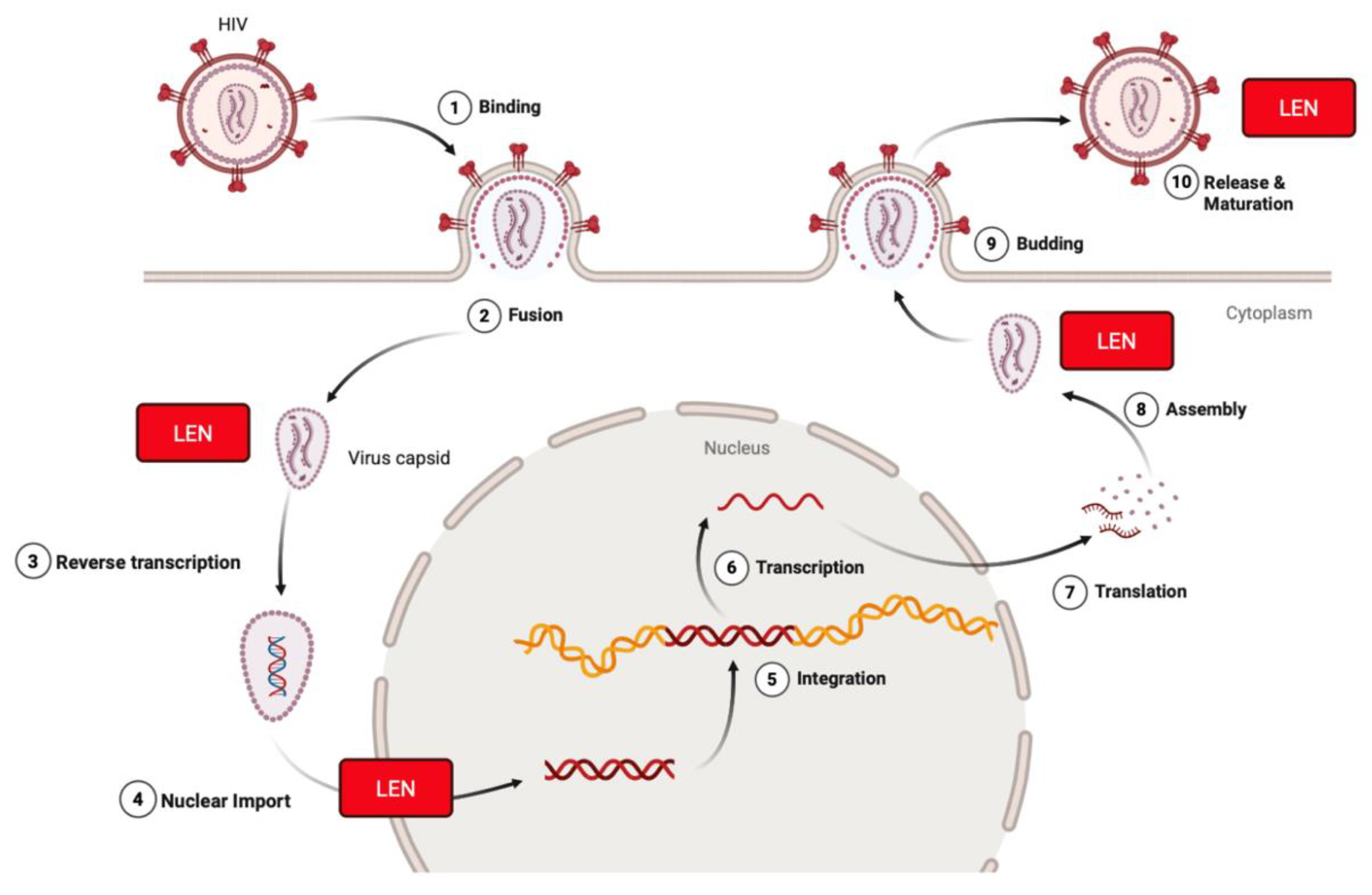
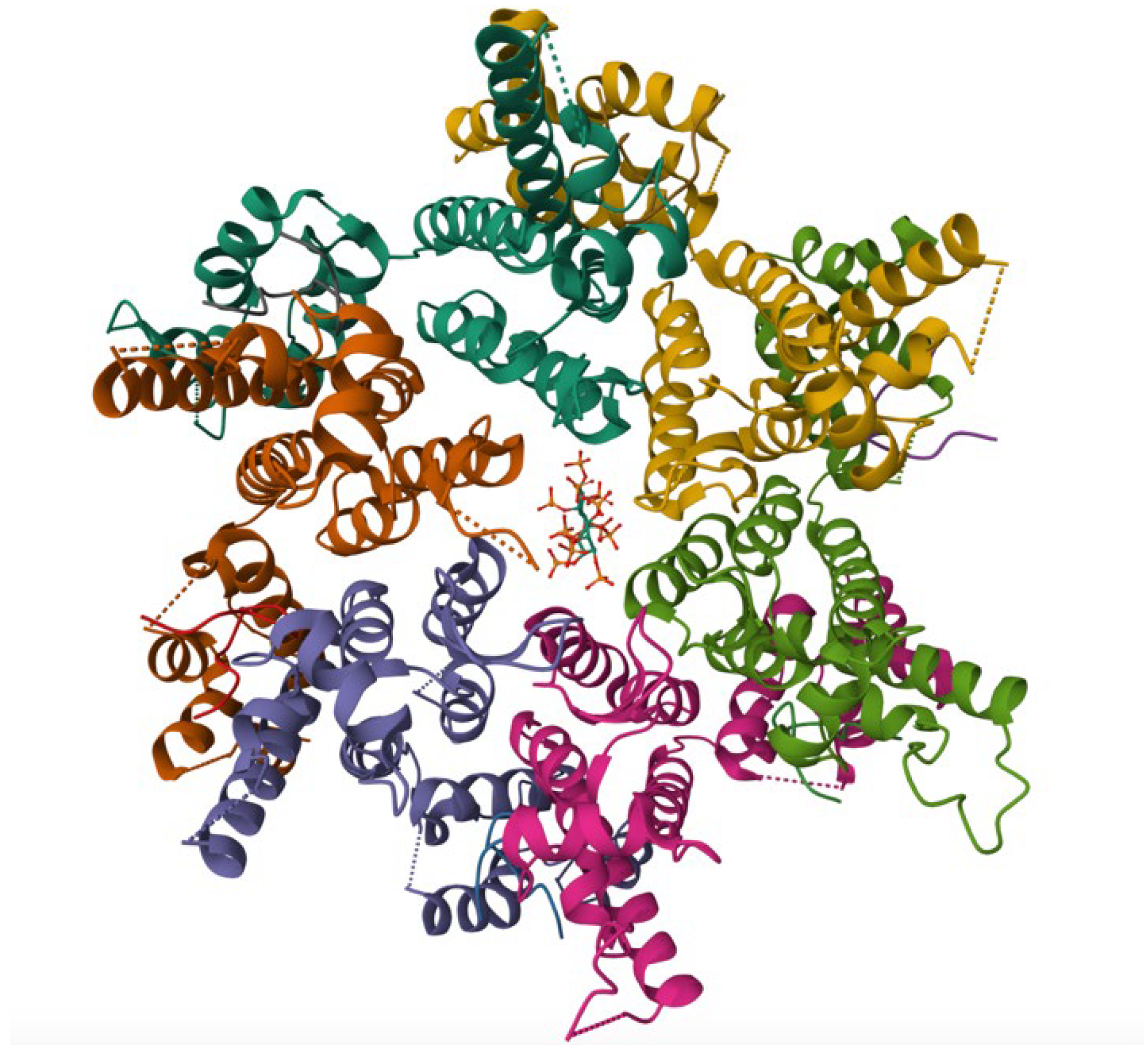
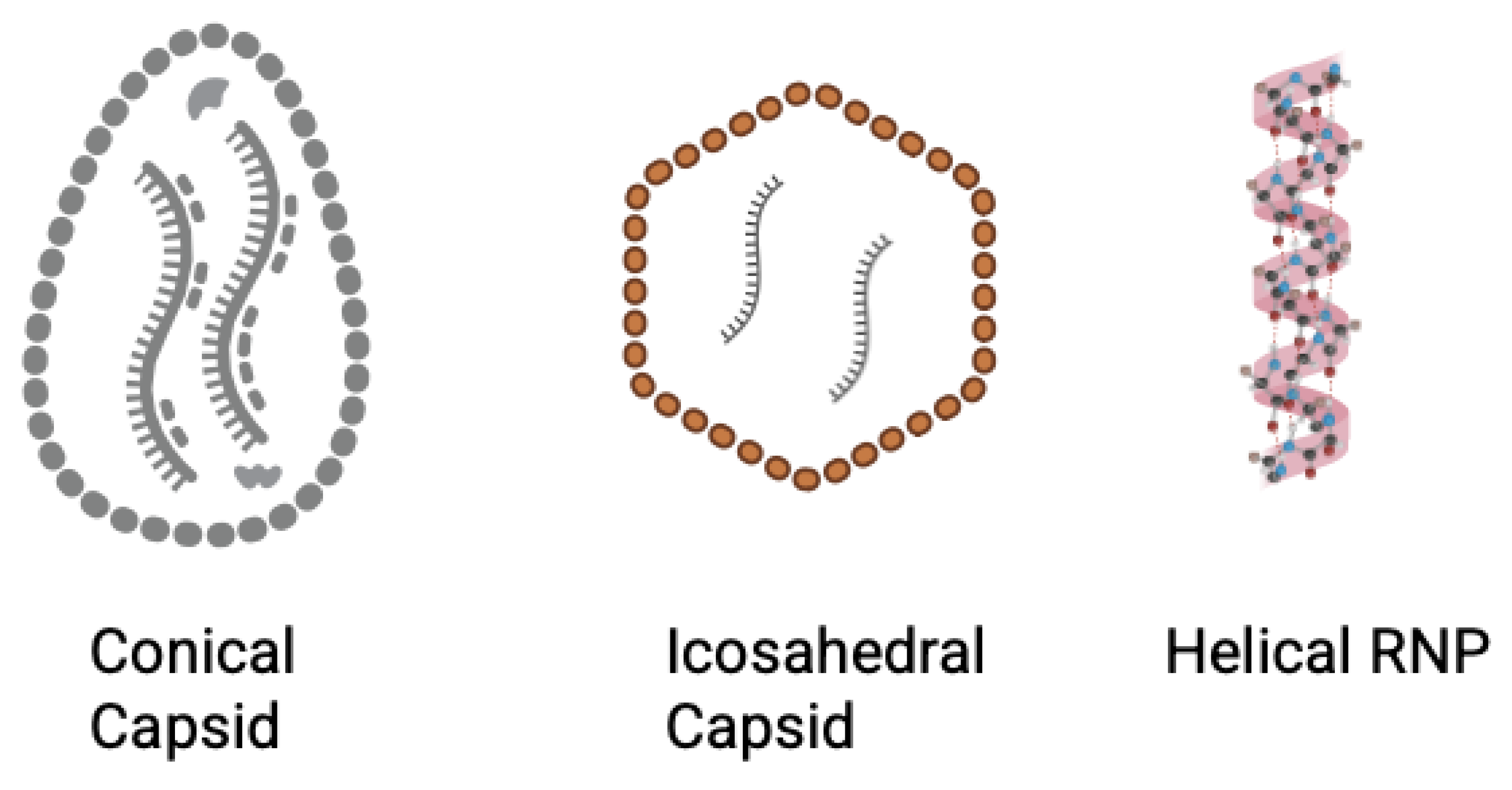
| Compound | Target | Binding Interaction | Disease or Virus | Mechanism of Action |
|---|---|---|---|---|
| Fostemsavir | Viral gp120 | Viral gp120-CD4 | HIV-1 | Blocks viral gp120 from binding to human CD4 receptor, preventing viral entry |
| Maraviroc | CCR5 co-receptor | Viral gp120-CCR5 | HIV-1 | Inhibits R5-tropic viral gp120 from binding to CCR5 co-receptor, blocking viral entry |
| Lenacapavir | Capsid | Capsid–Capsid | HIV-1 | Disrupts capsid assembly and disassembly at multiple stages within the viral cycle |
| Venetoclax | BCL2 | BCL2-BIM | Chronic lymphocytic leukemia, Small lymphocytic leukemia, Acute Myeloid Leukemia | Disrupts anti-apoptotic protein BCL2 and pro-apoptotic protein BIM to induce apoptosis |
| Motixafortide | CXCR4 | CXCL12-CXCR4 | Multiple myeloma | Blocks CXCR4 receptor from binding to its ligand CXCL12, mobilizing hematopoietic stem cells from the bone marrow |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, T.; Sutton, R.E. Use of the Split Luciferase Complementation Assay to Identify Novel Small Molecules That Disrupt Essential Protein–Protein Interactions of Viruses. Biomolecules 2025, 15, 1712. https://doi.org/10.3390/biom15121712
Biswas T, Sutton RE. Use of the Split Luciferase Complementation Assay to Identify Novel Small Molecules That Disrupt Essential Protein–Protein Interactions of Viruses. Biomolecules. 2025; 15(12):1712. https://doi.org/10.3390/biom15121712
Chicago/Turabian StyleBiswas, Tisa, and Richard E. Sutton. 2025. "Use of the Split Luciferase Complementation Assay to Identify Novel Small Molecules That Disrupt Essential Protein–Protein Interactions of Viruses" Biomolecules 15, no. 12: 1712. https://doi.org/10.3390/biom15121712
APA StyleBiswas, T., & Sutton, R. E. (2025). Use of the Split Luciferase Complementation Assay to Identify Novel Small Molecules That Disrupt Essential Protein–Protein Interactions of Viruses. Biomolecules, 15(12), 1712. https://doi.org/10.3390/biom15121712





