From Correlation to Causation: Defining Gene and RNA Function in Poultry Muscle Biology Using In Vivo Genetic Tools
Abstract
1. Introduction
2. Poultry-Specific Myogenesis: In Vivo Regulation and Development
2.1. Distinct Developmental Stages Govern Muscle Formation
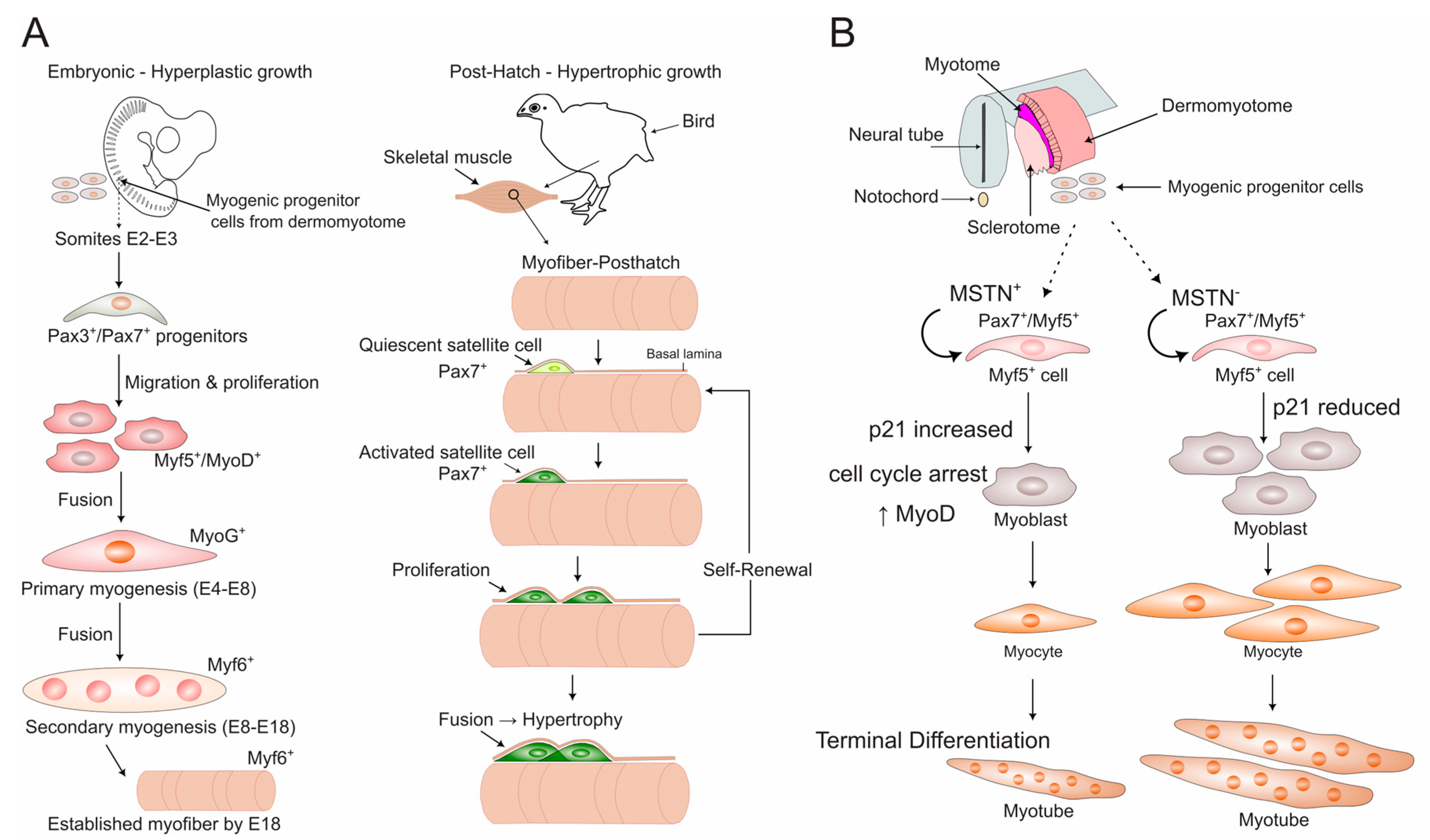
2.2. The Transcriptional Hierarchy of Avian Myogenesis
3. Transcriptional Landscapes: RNA Dynamics in Poultry Muscle
3.1. Developmental Time Courses Reveal Stage-Specific RNA Signatures
3.2. Breed-Specific Signatures Underlie Divergent Growth Phenotypes
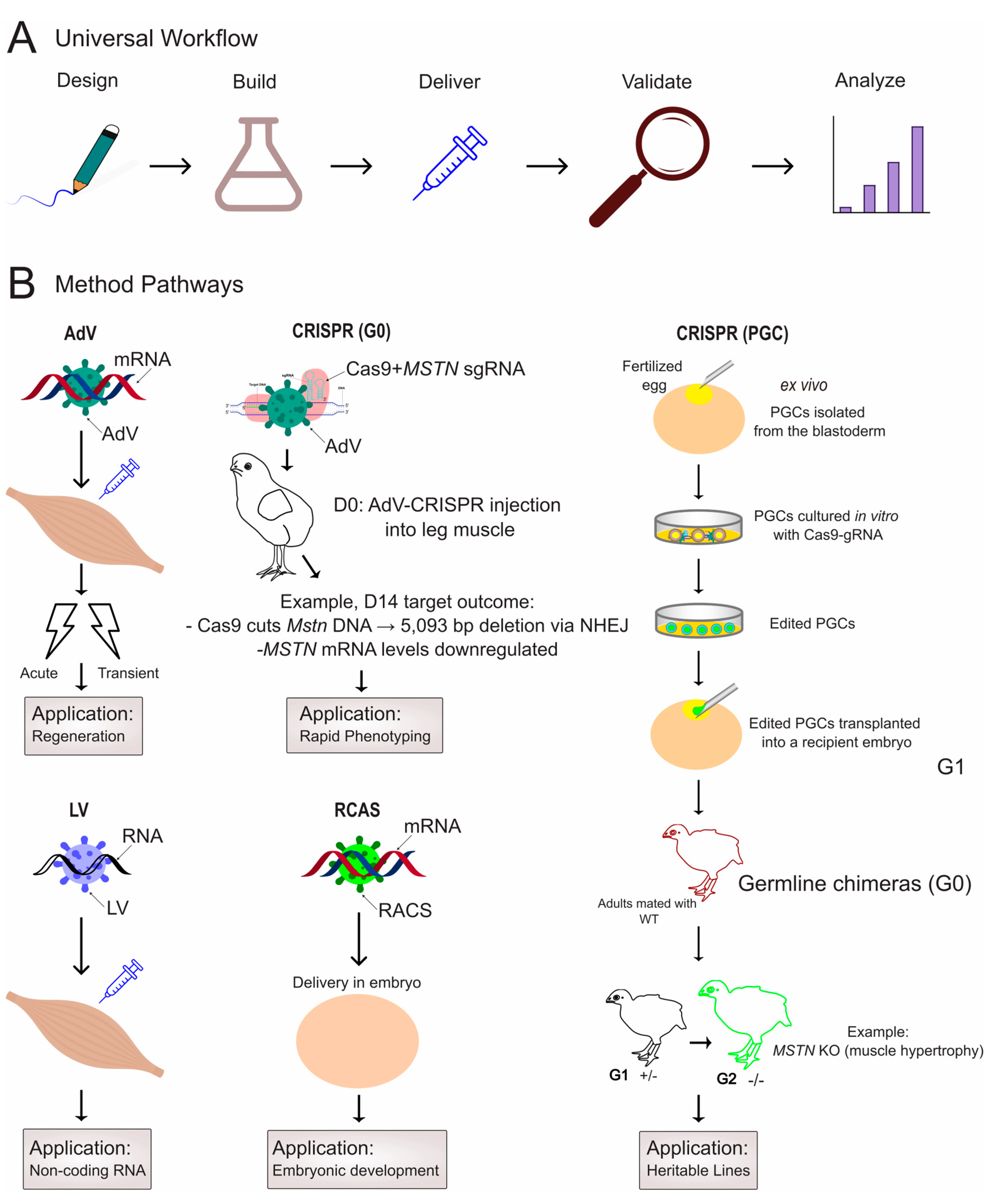
4. In Vivo Methodologies for Functional Validation in Poultry Muscle
4.1. Viral Vector Delivery
4.1.1. Adenovirus: Efficient Transient Expression
4.1.2. Lentivirus: Versatility for Postnatal Manipulation
4.1.3. RCAS Retrovirus: A Key Tool for Embryonic Studies
| Poultry Type (Age) | Method (Dose) | Target RNA | Target Tissue | Duration | Key Finding/Outcome | Ref. |
|---|---|---|---|---|---|---|
| AdV | ||||||
| Chicken (D1) | AdV OV (6 × 108 PFU) | mRNA; VGLL2 | Lateral GAS | Injected once, analyzed 7D post-injection | ↑ muscle fiber diameter, ↑ daily weight gain, ↑ MyoD, MyoG, Myomaker | [66] |
| Chicken (D21) | AdV OV (6 × 106 PFU) | mRNA; CHAC1 | GAS | Injected once, analyzed 1-7 D post-injury | ↑ muscle fiber diameter, ↑ CSA, ↑ regeneration markers (aMyHC, eMyHC, Desmin) | [65] |
| LV | ||||||
| Chicken (D1) | LV OE (106 titers) | mRNA; PPARGC1A | GAS | Injected at 1, 7, 14 D; phenotypic assessment at D21 | ↑ mitochondria, ↑ fatty acid oxidation, fast → slow shift, ↑ muscle mass | [70] |
| Chicken (D1) | LV OV (1 × 106 IU) | mRNA; c-Myc-Δ269–277 | Breast muscle | Two injections over 14D | miRNAs/lincRNAs dysregulation → Hypertrophy | [4] |
| Quail (Fertilized eggs; 4-h) | LV OV (2–3 uL) | mRNA; MSTN-B | Subgerminal space | 42D | ↑ muscle fiber hyperplasia (leg) | [78] |
| Chicken (D1) | LV OV (106 TU) | lncRNA; LncRNA-TBP | Bilateral GAS | 2 doses; analyzed 13D after the initial injection | ↑ slow-twitch fibers, ↓ fat deposition, ↑ muscle hypertrophy via TBP recruitment | [79] |
| Chicken (D1) | LV OV (1 × 107 TU per injection) | circRNA; circMEF2A1/2 | Breast muscle | 2 doses at D1 and D8; Tissue harvest (14D post-first injection) | ↑ breast muscle mass, ↑ muscle/body weight ratio, ↑ myofiber CSA | [80] |
| RCAS | ||||||
| Chick embryo (E4) | RCAS retrovirus (2 × 104 CFU) | mRNA; IGF1 | Hindlimb mesoderm | Analyzed D3-D7 post-injection | ↑ myoblast number, ↑ myofiber hyperplasia, ↑ Muscle mass | [81] |
4.2. CRISPR/Cas Systems: Precision Genome Editing for Functional Genomics
4.2.1. PGC-Mediated Editing for Heritable Modifications
4.2.2. Somatic Editing for Direct Phenotypic Analysis
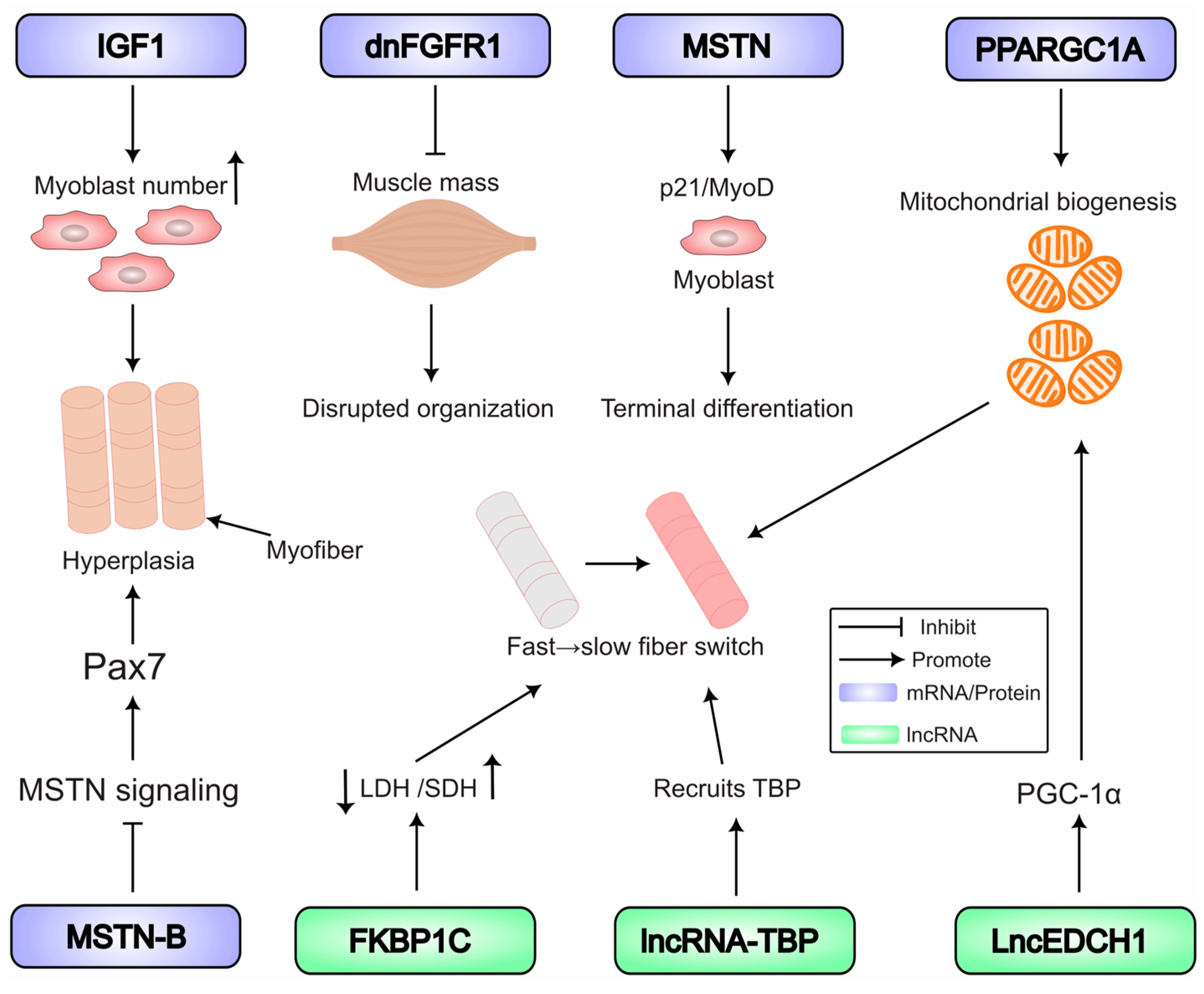
5. Key RNA-Regulated Pathways in Muscle Development
5.1. Coding RNAs: Masters of Myogenic Regulation
5.1.1. The Core Myogenic Regulatory Network
5.1.2. External Growth Factor Signals
5.1.3. The TGF-β Superfamily in Development
5.1.4. Metabolic Regulator
5.2. ncRNAs: The Sophisticated Regulators of Myogenesis
5.2.1. miRNAs: Master Post-Transcriptional Repressors in Development
5.2.2. lncRNAs: Versatile Architects of Developmental Regulation
5.2.3. circRNAs: Stable Regulators in Postnatal Growth
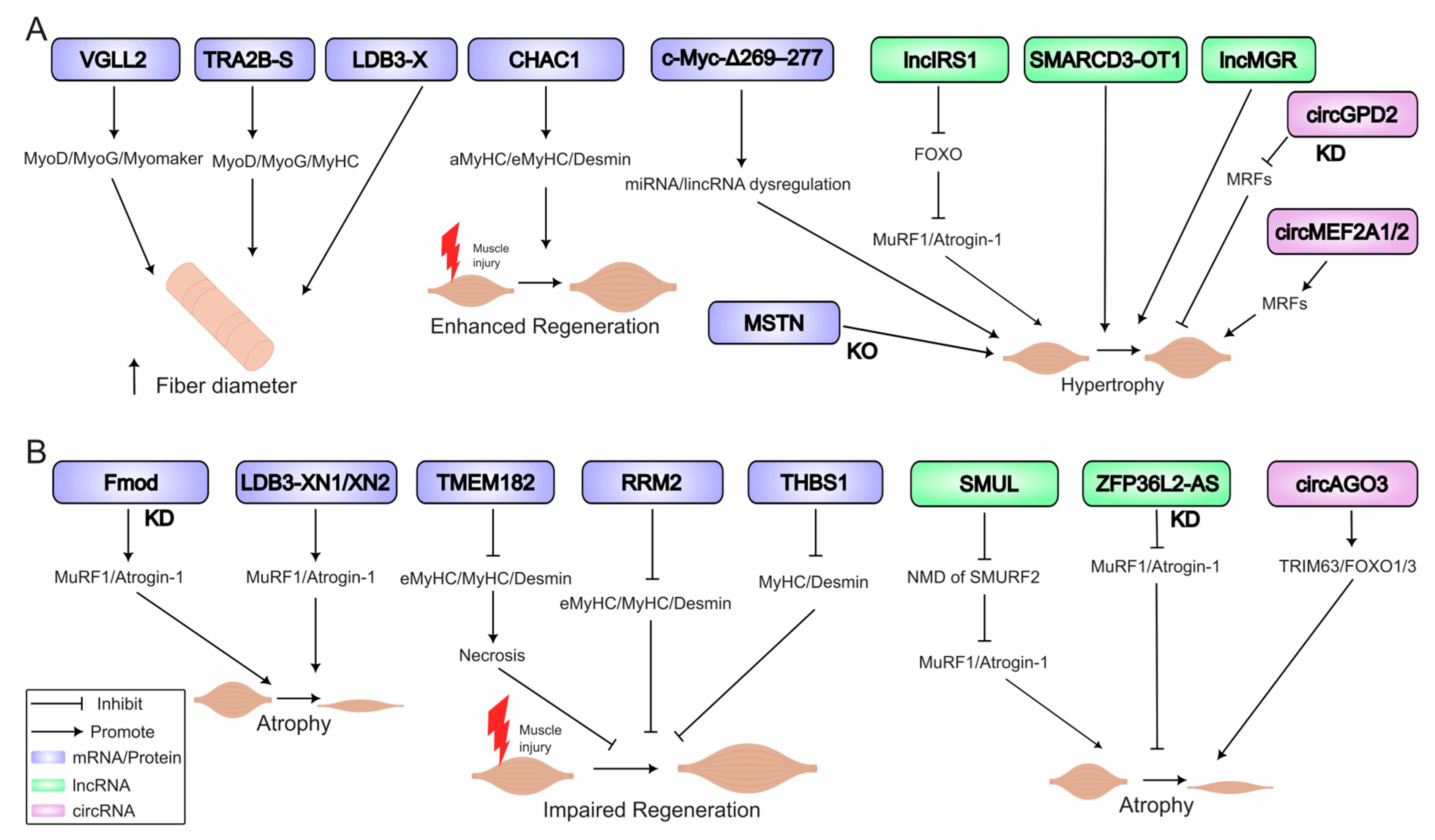
6. Atrophy and Hypertrophy: RNA Networks in Muscle Mass Regulation
6.1. Molecular Triggers of Atrophy
6.2. Drivers of Hypertrophy
6.3. Therapeutic Perspectives and Future Directions
7. Bridging the Functional Gap: Applying Genetic Tools to Unravel Poultry Myopathies
7.1. Conserved Molecular Signatures as a Basis for Functional Inquiry
7.2. A Functional Genomics Roadmap for Myopathy Research
7.3. Future Directions: Integrating Functional Data into a Systems-Level Understanding
8. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BMP | Bone morphogenetic protein |
| Cas | CRISPR-associated protein |
| ceRNA | competing endogenous RNA |
| CHAC1 | Glutathione-specific gamma-glutamylcyclotransferase 1 |
| circRNAs | Circular RNAs |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| DEcircRNAs | Differentially expressed circular RNAs |
| DEGs | Differentially expressed genes |
| DEmiRNAs | Differentially expressed microRNAs |
| DEmRNAs | Differentially expressed mRNAs |
| DElncRNAs | Differentially expressed long non-coding RNAs |
| dnFGFR1 | dominant-negative fibroblast growth factor receptor 1 |
| ECM | Extracellular matrix |
| E | Embryonic day (e.g., E10) |
| GAS | gastrocnemius muscle |
| IGF1 | Insulin-like growth factor 1 |
| lncRNAs | Long non-coding RNAs |
| miRNAs | microRNAs |
| MRF | Muscle regulatory factor (e.g., MyoD, myogenin, Myf5) |
| MSTN | Myostatin |
| MyHC | Myosin heavy chain |
| PGCs | Primordial germ cells |
| PPARGC1A | Peroxisome proliferator-activated receptor gamma-coactivator 1-alpha |
| RCAS | Replication-competent avian sarcoma-leukosis virus (Subgroup A) |
| RNA-seq | RNA sequencing |
| TGF-β | Transforming growth factor beta |
| TMEM182 | Transmembrane protein 182 |
| TRA2B | Transformer-2 beta homolog |
| WB | Wooden breast (myopathy) |
| WGCNA | Weighted gene co-expression network analysis |
| WS | White striping (myopathy) |
References
- Tarpey, P.S.; Smith, R.; Pleasance, E.; Whibley, A.; Edkins, S.; Hardy, C.; O’Meara, S.; Latimer, C.; Dicks, E.; Menzies, A.; et al. A Systematic, Large-Scale Resequencing Screen of X-Chromosome Coding Exons in Mental Retardation. Nat. Genet. 2009, 41, 535–543. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin Signature Reveals over a Thousand Highly Conserved Large Non-Coding RNAs in Mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- OECD; Food and Agriculture Organization of the United Nations. OECD-FAO Agricultural Outlook 2025–2034; OECD-FAO Agricultural Outlook: Rome, Italy, 2025. [Google Scholar] [CrossRef]
- Luo, W.; Chen, J.; Li, L.; Ren, X.; Cheng, T.; Lu, S.; Lawal, R.A.; Nie, Q.; Zhang, X.; Hanotte, O. C-Myc Inhibits Myoblast Differentiation and Promotes Myoblast Proliferation and Muscle Fibre Hypertrophy by Regulating the Expression of Its Target Genes, miRNAs and lincRNAs. Cell Death Differ. 2019, 26, 426–442. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, J.; Wu, P.; Ling, X.; Wang, Q.; Zhou, K.; Li, P.; Zhang, L.; Ye, H.; Zhang, Q.; et al. Transcriptome Sequencing Analysis of circRNA in Skeletal Muscle between Fast- and Slow-Growing Chickens at Embryonic Stages. Animals 2022, 12, 3166. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, Y.; Tian, W.; Wang, Y.; Liu, Y.; Ji, H.; Cai, H.; Han, R.; Tian, Y.; Liu, X.; et al. The Long Noncoding RNA lncMPD2 Inhibits Myogenesis by Targeting the miR-34a-5p/THBS1 Axis. Int. J. Biol. Macromol. 2024, 275, 133688. [Google Scholar] [CrossRef] [PubMed]
- Gibril, B.A.A.; Xiong, X.; Chai, X.; Xu, Q.; Gong, J.; Xu, J. Unlocking the Nexus of Sirtuins: A Comprehensive Review of Their Role in Skeletal Muscle Metabolism, Development, and Disorders. Int. J. Biol. Sci. 2024, 20, 3219–3235. [Google Scholar] [CrossRef]
- Petracci, M.; Soglia, F.; Madruga, M.; Carvalho, L.; Ida, E.; Estévez, M. Wooden-Breast, White Striping, and Spaghetti Meat: Causes, Consequences and Consumer Perception of Emerging Broiler Meat Abnormalities. Compr. Rev. Food Sci. Food Saf. 2019, 18, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G.; Clark, D.L. Histopathologic and Myogenic Gene Expression Changes Associated with Wooden Breast in Broiler Breast Muscles. Avian Dis. 2015, 59, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Abasht, B.; Papah, M.B.; Qiu, J. Evidence of Vascular Endothelial Dysfunction in Wooden Breast Disorder in Chickens: Insights through Gene Expression Analysis, Ultra-Structural Evaluation and Supervised Machine Learning Methods. PLoS ONE 2021, 16, e0243983. [Google Scholar] [CrossRef]
- Xing, T.; Luo, D.; Zhao, X.; Xu, X.; Li, J.; Zhang, L.; Gao, F. Enhanced Cytokine Expression and Upregulation of Inflammatory Signaling Pathways in Broiler Chickens Affected by Wooden Breast Myopathy. J. Sci. Food Agric. 2021, 101, 279–286. [Google Scholar] [CrossRef]
- Bordini, M.; Soglia, F.; Davoli, R.; Zappaterra, M.; Petracci, M.; Meluzzi, A. Molecular Pathways and Key Genes Associated With Breast Width and Protein Content in White Striping and Wooden Breast Chicken Pectoral Muscle. Front. Physiol. 2022, 13, 936768. [Google Scholar] [CrossRef]
- Mutryn, M.F.; Brannick, E.M.; Fu, W.; Lee, W.R.; Abasht, B. Characterization of a Novel Chicken Muscle Disorder through Differential Gene Expression and Pathway Analysis Using RNA-Sequencing. BMC Genom. 2015, 16, 399. [Google Scholar] [CrossRef]
- Zambonelli, P.; Zappaterra, M.; Soglia, F.; Petracci, M.; Sirri, F.; Cavani, C.; Davoli, R. Detection of Differentially Expressed Genes in Broiler Pectoralis Major Muscle Affected by White Striping—Wooden Breast Myopathies. Poult. Sci. 2016, 95, 2771–2785. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cai, B.; Abdalla, B.A.; Zhu, X.; Zheng, M.; Han, P.; Nie, Q.; Zhang, X. LncIRS1 Controls Muscle Atrophy via Sponging miR-15 Family to Activate IGF1-PI3K/AKT Pathway. J. Cachexia Sarcopenia Muscle 2019, 10, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, D.-H.; Lee, K. Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide. Int. J. Mol. Sci. 2020, 21, 1504. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, M.; Zhang, J.; Wang, Z.; Kong, S.; Zhou, Z.; Lian, L.; Zhang, J.; Li, J.; Wang, Y.; et al. LncEDCH1 Improves Mitochondrial Function to Reduce Muscle Atrophy by Interacting with SERCA2. Mol. Ther. Nucleic Acids 2022, 27, 319–334. [Google Scholar] [CrossRef]
- Papah, M.B.; Brannick, E.M.; Schmidt, C.J.; Abasht, B. Gene Expression Profiling of the Early Pathogenesis of Wooden Breast Disease in Commercial Broiler Chickens Using RNA-Sequencing. PLoS ONE 2018, 13, e0207346. [Google Scholar] [CrossRef]
- Li, D.; Hou, T.; Du, X.; Zhao, L.; Zhang, L.; Gao, F.; Xing, T. Integrated Analysis of miRNA and mRNA Expression Profiles Associated with Wooden Breast Myopathy in Broiler Chickens. Int. J. Biol. Macromol. 2025, 284, 137990. [Google Scholar] [CrossRef]
- Marchesi, J.A.P.; Ibelli, A.M.G.; Peixoto, J.O.; Cantão, M.E.; Pandolfi, J.R.C.; Marciano, C.M.M.; Zanella, R.; Settles, M.L.; Coutinho, L.L.; Ledur, M.C. Whole Transcriptome Analysis of the Pectoralis Major Muscle Reveals Molecular Mechanisms Involved with White Striping in Broiler Chickens. Poult. Sci. 2019, 98, 590–601. [Google Scholar] [CrossRef]
- Phillips, C.A.; Reading, B.J.; Livingston, M.; Livingston, K.; Ashwell, C.M. Evaluation via Supervised Machine Learning of the Broiler Pectoralis Major and Liver Transcriptome in Association With the Muscle Myopathy Wooden Breast. Front. Physiol. 2020, 11, 101. [Google Scholar] [CrossRef]
- Velleman, S.G.; McFarland, D.C. Chapter 16—Skeletal Muscle. In Sturkie’s Avian Physiology, 6th ed.; Scanes, C.G., Ed.; Academic Press: San Diego, CA, USA, 2015; pp. 379–402. ISBN 978-0-12-407160-5. [Google Scholar]
- Ricklefs, R.E. Modification of Growth and Development of Muscles of Poultry. Poult. Sci. 1985, 64, 1563–1576. [Google Scholar] [CrossRef]
- Stockdale, F.E. Myogenic Cell Lineages. Dev. Biol. 1992, 154, 284–298. [Google Scholar] [CrossRef]
- Smith, J.H. Relation of Body Size to Muscle Cell Size and Number in the Chicken1,2. Poult. Sci. 1963, 42, 283–290. [Google Scholar] [CrossRef]
- Xu, J.; Velleman, S.G. Critical Role of the mTOR Pathway in Poultry Skeletal Muscle Physiology and Meat Quality: An Opinion Paper. Front. Physiol. 2023, 14, 1228318. [Google Scholar] [CrossRef] [PubMed]
- Theobald, J.; DiMario, J.X. Lineage-Based Primary Muscle Fiber Type Diversification Independent of MEF2 and NFAT in Chick Embryos. J Muscle Res. Cell Motil. 2011, 31, 369–381. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, H.; He, L.; Niu, Y.; Zhang, Y.; Liu, Y.; Tian, Y.; Liu, X.; Li, H.; Kang, X.; et al. Establishment and Analysis of Immortalized Chicken Skeletal Muscle Satellite Cell Lines1. J. Integr. Agric. 2024, 24, 4370–4378. [Google Scholar] [CrossRef]
- Kocamis, H.; McFarland, D.C.; Killefer, J. Temporal Expression of Growth Factor Genes during Myogenesis of Satellite Cells Derived from the Biceps Femoris and Pectoralis Major Muscles of the Chicken. J. Cell. Physiol. 2001, 186, 146–152. [Google Scholar] [CrossRef]
- Manceau, M.; Gros, J.; Savage, K.; Thomé, V.; McPherron, A.; Paterson, B.; Marcelle, C. Myostatin Promotes the Terminal Differentiation of Embryonic Muscle Progenitors. Genes Dev. 2008, 22, 668–681. [Google Scholar] [CrossRef]
- Sweetman, D.; Goljanek, K.; Rathjen, T.; Oustanina, S.; Braun, T.; Dalmay, T.; Münsterberg, A. Specific Requirements of MRFs for the Expression of Muscle Specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol. 2008, 321, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Berti, F.; Nogueira, J.M.; Wöhrle, S.; Sobreira, D.R.; Hawrot, K.; Dietrich, S. Time Course and Side-by-Side Analysis of Mesodermal, Pre-Myogenic, Myogenic and Differentiated Cell Markers in the Chicken Model for Skeletal Muscle Formation. J. Anat. 2015, 227, 361–382. [Google Scholar] [CrossRef]
- Luo, W.; Li, E.; Nie, Q.; Zhang, X. Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken Myoblast Fusion. Int. J. Mol. Sci. 2015, 16, 26186–26201. [Google Scholar] [CrossRef]
- Gu, S.; Huang, Q.; Sun, C.; Wen, C.; Yang, N. Transcriptomic and Epigenomic Insights into Pectoral Muscle Fiber Formation at the Late Embryonic Development in Pure Chicken Lines. Poult. Sci. 2024, 103, 103882. [Google Scholar] [CrossRef]
- Gu, S.; Wen, C.; Li, J.; Liu, H.; Huang, Q.; Zheng, J.; Sun, C.; Yang, N. Temporal Expression of Myogenic Regulatory Genes in Different Chicken Breeds during Embryonic Development. Int. J. Mol. Sci. 2022, 23, 10115. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wu, Y.; Zhang, S.; Zhang, T.; Zhang, G.; Wang, J. Transcriptomic Profile of Leg Muscle during Early Growth and Development in Haiyang Yellow Chicken. Arch. Anim. Breed. 2021, 64, 405–416. [Google Scholar] [CrossRef]
- Xue, Q.; Zhang, G.; Li, T.; Ling, J.; Zhang, X.; Wang, J. Transcriptomic Profile of Leg Muscle during Early Growth in Chicken. PLoS ONE 2017, 12, e0173824. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Fang, W.; Yuan, M.; Sun, H.; Wang, J. Transcriptome Analysis of Leg Muscles and the Effects of ALOX5 on Proliferation and Differentiation of Myoblasts in Haiyang Yellow Chickens. Genes 2023, 14, 1213. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Cao, J.; Zhang, J.; Ge, L.; Zhang, H.; Liu, X. Skeletal Muscle Transcriptome Analysis of Hanzhong Ma Duck at Different Growth Stages Using RNA-Seq. Biomolecules 2021, 11, 315. [Google Scholar] [CrossRef]
- Cao, C.; Cai, Y.; Li, Y.; Li, T.; Zhang, J.; Hu, Z.; Zhang, J. Characterization and Comparative Transcriptomic Analysis of Skeletal Muscle in Female Pekin Duck and Hanzhong Ma Duck during Different Growth Stages Using RNA-Seq. Poult. Sci. 2023, 102, 103122. [Google Scholar] [CrossRef]
- Gu, S.; Huang, Q.; Jie, Y.; Sun, C.; Wen, C.; Yang, N. Transcriptomic and Epigenomic Landscapes of Muscle Growth during the Postnatal Period of Broilers. J. Anim. Sci. Biotechnol. 2024, 15, 91. [Google Scholar] [CrossRef]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.H.; Yatskievych, T.A.; Antin, P.B. MicroRNA Expression during Chick Embryo Development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef]
- Li, C.; Xiong, T.; Zhou, M.; Wan, L.; Xi, S.; Liu, Q.; Chen, Y.; Mao, H.; Liu, S.; Chen, B. Characterization of microRNAs during Embryonic Skeletal Muscle Development in the Shan Ma Duck. Animals 2020, 10, 1417. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA Transcriptome of Breast Muscle from the Embryonic to Post-Hatching Periods in Chickens. BMC Genom. 2021, 22, 64. [Google Scholar] [CrossRef]
- Zeng, B.; Tang, M.; Chen, T.; Jiang, Y.; Tang, W.; Yu, G. Characterization and Analysis of MicroRNA during Leg Muscle Development in Embryonic Stage of Daozhou Grey Goose. Poult. Sci. 2025, 104, 105354. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Y.; Hu, X.; Yang, J.; Lei, Q.; Liu, W.; Han, H.; Li, F.; Cao, D. Transcriptome Analysis Reveals the Profile of Long Non-Coding RNAs During Chicken Muscle Development. Front. Physiol. 2021, 12, 660370. [Google Scholar] [CrossRef]
- Ouyang, H.; Chen, X.; Li, W.; Li, Z.; Nie, Q.; Zhang, X. Circular RNA circSVIL Promotes Myoblast Proliferation and Differentiation by Sponging miR-203 in Chicken. Front. Genet. 2018, 9, 172. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wu, J.; Jiang, H.; Zhou, Y.; Huang, X.; Wang, Y.; Xie, Z.; Liao, Z.; Ding, Z.; Liu, J.; et al. CircFBLN2 Regulates Duck Myoblast Proliferation and Differentiation through miR-22-5p and MEF2C Interaction. Poult. Sci. 2025, 104, 105063. [Google Scholar] [CrossRef]
- Ouyang, H.; Chen, X.; Wang, Z.; Yu, J.; Jia, X.; Li, Z.; Luo, W.; Abdalla, B.A.; Jebessa, E.; Nie, Q.; et al. Circular RNAs Are Abundant and Dynamically Expressed during Embryonic Muscle Development in Chickens. DNA Res. 2018, 25, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhang, Y.; Chen, Y.; Yang, N.; Wang, X.-J.; Zhu, D. Systematic Identification of Genes Involved in Divergent Skeletal Muscle Growth Rates of Broiler and Layer Chickens. BMC Genom. 2009, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Al-Musawi, S.L.; Lock, F.; Simbi, B.H.; Bayol, S.A.M.; Stickland, N.C. Muscle Specific Differences in the Regulation of Myogenic Differentiation in Chickens Genetically Selected for Divergent Growth Rates. Differentiation 2011, 82, 127–135. [Google Scholar] [CrossRef]
- Shin, J.; Velleman, S.G.; Latshaw, J.D.; Wick, M.P.; Suh, Y.; Lee, K. The Ontogeny of Delta-like Protein 1 Messenger Ribonucleic Acid Expression during Muscle Development and Regeneration: Comparison of Broiler and Leghorn Chickens. Poult. Sci. 2009, 88, 1427–1437. [Google Scholar] [CrossRef]
- San, J.; Du, Y.; Wu, G.; Xu, R.; Yang, J.; Hu, J. Transcriptome Analysis Identifies Signaling Pathways Related to Meat Quality in Broiler Chickens—The Extracellular Matrix (ECM) Receptor Interaction Signaling Pathway. Poult. Sci. 2021, 100, 101135. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Choi, Y.M.; Lee, J.; Shin, S.; Kim, S.; Suh, Y.; Lee, K. Differential Expression of MSTN Isoforms in Muscle between Broiler and Layer Chickens. Animals 2022, 12, 539. [Google Scholar] [CrossRef]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The Transient Expression of miR-203 and Its Inhibiting Effects on Skeletal Muscle Cell Proliferation and Differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef]
- Lin, S.; Luo, W.; Ye, Y.; Bekele, E.J.; Nie, Q.; Li, Y.; Zhang, X. Let-7b Regulates Myoblast Proliferation by Inhibiting IGF2BP3 Expression in Dwarf and Normal Chicken. Front. Physiol. 2017, 8, 477. [Google Scholar] [CrossRef]
- Shen, X.; Wei, Y.; Liu, W.; You, G.; Tang, S.; Su, Z.; Du, M.; He, J.; Zhao, J.; Tian, Y.; et al. A Novel Circular RNA circITSN2 Targets the miR-218-5p/LMO7 Axis to Promote Chicken Embryonic Myoblast Proliferation and Differentiation. Front. Cell Dev. Biol. 2021, 9, 748844. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, J.; Zhao, X.; He, H.; Cui, C.; Zhang, Y.; Zhu, Q.; Yin, H.; Han, S. CircLRRFIP1 Promotes the Proliferation and Differentiation of Chicken Skeletal Muscle Satellite Cells by Sponging the miR-15 Family via Activating AKT3-mTOR/p70S6K Signaling Pathway. Poult. Sci. 2023, 102, 103050. [Google Scholar] [CrossRef]
- Ren, T.; Li, Z.; Zhou, Y.; Liu, X.; Han, R.; Wang, Y.; Yan, F.; Sun, G.; Li, H.; Kang, X. Sequencing and Characterization of lncRNAs in the Breast Muscle of Gushi and Arbor Acres Chickens. Genome 2018, 61, 337–347. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiu, M.; Du, H.; Li, Q.; Gan, W.; Xiong, X.; Yu, C.; Peng, H.; Xia, B.; Song, X.; et al. Small RNA Sequencing of Pectoral Muscle Tissue Reveals microRNA-Mediated Gene Modulation in Chicken Muscle Growth. J. Anim. Physiol. Anim. Nutr. 2020, 104, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zhou, K.; Zhang, L.; Li, P.; He, M.; Zhang, X.; Ye, H.; Zhang, Q.; Wei, Q.; Zhang, G. High-Throughput Sequencing Reveals Crucial miRNAs in Skeletal Muscle Development of Bian Chicken. Br. Poult. Sci. 2021, 62, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Scaal, M.; Gros, J.; Lesbros, C.; Marcelle, C. In Ovo Electroporation of Avian Somites. Dev. Dyn. 2004, 229, 643–650. [Google Scholar] [CrossRef]
- Wei, C.; Niu, Y.; Chen, B.; Wang, Y.; Cai, H.; Han, R.; Tian, Y.; Liu, X.; Guo, W.; Kang, X.; et al. Divergent Regulatory Roles of Transcriptional Variants of the Chicken LDB3 Gene in Muscle Shaping. J. Agric. Food Chem. 2024, 72, 12240–12250. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, Y.; Niu, Y.; Wang, Y.; Liu, Y.; Ji, H.; Han, R.; Tian, Y.; Liu, X.; Kang, X.; et al. RRM2 Promotes the Proliferation of Chicken Myoblasts, Inhibits Their Differentiation and Muscle Regeneration. Poult. Sci. 2024, 103, 103407. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Cai, H.; Niu, Y.; Zhang, Y.; Wang, Y.; Liu, Y.; Han, R.; Liu, X.; Kang, X.; Li, Z. Whole Transcriptome Profiling Reveals a lncMDP1 That Regulates Myogenesis by Adsorbing miR-301a-5p Targeting CHAC1. Commun. Biol. 2024, 7, 518. [Google Scholar] [CrossRef]
- Li, W.; Ma, H.; Wang, Y.; Zhang, Y.; Liu, Y.; Han, R.; Li, H.; Cai, H.; Liu, X.; Kang, X.; et al. The VGLL2 Gene Participates in Muscle Development in Gushi Chickens. J. Integr. Agric. 2025, 24, 246–260. [Google Scholar] [CrossRef]
- Guo, Y.; Geng, W.; Chen, Z.; Zhi, Y.; Zhang, K.; Li, Z.; Li, G.; Kang, X.; Tian, W.; Li, H.; et al. LncRNA lncMGR Regulates Skeletal Muscle Development and Regeneration by Recruiting CDK9 and Sponging miRNAs. Int. J. Biol. Macromol. 2024, 266, 131049. [Google Scholar] [CrossRef]
- Ouyang, J.; Alway, S.E. Transgene Expression in Hypertrophied and Aged Skeletal Muscle in Vivo by Lentivirus Delivery. J. Gene Med. 2004, 6, 278–287. [Google Scholar] [CrossRef]
- Chen, G.; Chen, J.; Qi, L.; Yin, Y.; Lin, Z.; Wen, H.; Zhang, S.; Xiao, C.; Bello, S.F.; Zhang, X.; et al. Bulk and Single-Cell Alternative Splicing Analyses Reveal Roles of TRA2B in Myogenic Differentiation. Cell Prolif. 2024, 57, e13545. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Cai, B.; Kong, S.; Zhou, Z.; Zhang, J.; Zhang, X.; Nie, Q. PPARGC1A Is a Moderator of Skeletal Muscle Development Regulated by miR-193b-3p. Int. J. Mol. Sci. 2022, 23, 9575. [Google Scholar] [CrossRef]
- Shen, X.; Cui, C.; Tang, S.; Han, S.; Zhang, Y.; Xia, L.; Tan, B.; Ma, M.; Kang, H.; Yu, J.; et al. MyoG-Enhanced circGPD2 Regulates Chicken Skeletal Muscle Development by Targeting miR-203a. Int. J. Biol. Macromol. 2022, 222, 2212–2224. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, S.; Lei, Z.; Shen, X.; Zhang, Y.; Han, S.; Yin, H.; Cui, C. circAGO3 Facilitates NF-κB Pathway-Mediated Inflammatory Atrophy in Chicken Skeletal Muscle via the miR-34b-5p/TRAF3 Axis. Int. J. Biol. Macromol. 2024, 283, 137614. [Google Scholar] [CrossRef]
- Yin, H.; Cui, C.; Han, S.; Chen, Y.; Zhao, J.; He, H.; Li, D.; Zhu, Q. Fibromodulin Modulates Chicken Skeletal Muscle Development via the Transforming Growth Factor-β Signaling Pathway. Animals 2020, 10, 1477. [Google Scholar] [CrossRef]
- Marcelle, C.; Stark, M.R.; Bronner-Fraser, M. Coordinate Actions of BMPs, Wnts, Shh and Noggin Mediate Patterning of the Dorsal Somite. Development 1997, 124, 3955–3963. [Google Scholar] [CrossRef]
- Capdevila, J.; Johnson, R.L. Endogenous and Ectopic Expression of Noggin Suggests a Conserved Mechanism for Regulation of BMP Function during Limb and Somite Patterning. Dev. Biol. 1998, 197, 205–217. [Google Scholar] [CrossRef]
- Flanagan-Steet, H.; Hannon, K.; McAvoy, M.J.; Hullinger, R.; Olwin, B.B. Loss of FGF Receptor 1 Signaling Reduces Skeletal Muscle Mass and Disrupts Myofiber Organization in the Developing Limb. Dev. Biol. 2000, 218, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Clase, K.L.; Mitchell, P.J.; Ward, P.J.; Dorman, C.M.; Johnson, S.E.; Hannon, K. FGF5 Stimulates Expansion of Connective Tissue Fibroblasts and Inhibits Skeletal Muscle Development in the Limb. Dev. Dyn. 2000, 219, 368–380. [Google Scholar] [CrossRef]
- Chen, P.R.; Suh, Y.; Shin, S.; Woodfint, R.M.; Hwang, S.; Lee, K. Exogenous Expression of an Alternative Splicing Variant of Myostatin Prompts Leg Muscle Fiber Hyperplasia in Japanese Quail. Int. J. Mol. Sci. 2019, 20, 4617. [Google Scholar] [CrossRef]
- Ma, M.; Cai, B.; Zhou, Z.; Kong, S.; Zhang, J.; Xu, H.; Zhang, X.; Nie, Q. LncRNA-TBP Mediates TATA-Binding Protein Recruitment to Regulate Myogenesis and Induce Slow-Twitch Myofibers. Cell Commun. Signal. 2023, 21, 7. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, X.; He, H.; Zhao, J.; Wei, Y.; Chen, Y.; Han, S.; Zhu, Y.; Zhang, Y.; Zhu, Q.; et al. Evolutionary Conserved Circular MEF2A RNAs Regulate Myogenic Differentiation and Skeletal Muscle Development. PLoS Genet. 2023, 19, e1010923. [Google Scholar] [CrossRef]
- Mitchell, P.J.; Johnson, S.E.; Hannon, K. Insulin-like Growth Factor I Stimulates Myoblast Expansion and Myofiber Development in the Limb. Dev. Dyn. 2002, 223, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-D.; Lee, J.H.; Song, S.; Kim, S.W.; Han, J.S.; Shin, S.P.; Park, B.-C.; Park, T.S. Generation of Myostatin-Knockout Chickens Mediated by D10A-Cas9 Nickase. FASEB J. 2020, 34, 5688–5696. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, J.; Huang, Z.; Tang, G.; Lv, G.; Li, D.; Yang, C. Functional Prediction of AMP Deaminase 1 in Jingyuan Chicken and Evaluation of the Biological Activities of Its Expression Vectors. Int. J. Biol. Macromol. 2024, 271, 132546. [Google Scholar] [CrossRef]
- Xu, K.; Han, C.X.; Zhou, H.; Ding, J.M.; Xu, Z.; Yang, L.Y.; He, C.; Akinyemi, F.; Zheng, Y.M.; Qin, C.; et al. Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle. Int. J. Mol. Sci. 2020, 21, 2584. [Google Scholar] [CrossRef]
- Delfini, M.-C.; Duprez, D. Ectopic Myf5 or MyoD Prevents the Neuronal Differentiation Program in Addition to Inducing Skeletal Muscle Differentiation, in the Chick Neural Tube. Development 2004, 131, 713–723. [Google Scholar] [CrossRef]
- Lowe, D.A.; Alway, S.E. Stretch-Induced Myogenin, MyoD, and MRF4 Expression and Acute Hypertrophy in Quail Slow-Tonic Muscle Are Not Dependent upon Satellite Cell Proliferation. Cell Tissue Res. 1999, 296, 531–539. [Google Scholar] [CrossRef]
- Delfini, M.C.; Hirsinger, E.; Pourquié, O.; Duprez, D. Delta 1-Activated Notch Inhibits Muscle Differentiation without Affecting Myf5 and Pax3 Expression in Chick Limb Myogenesis. Development 2000, 127, 5213–5224. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Chu, H.; Li, F.; Han, G.; Cai, Y.; Yi, J.; Lu, M.; Xiang, H.; Kang, H.; Ye, F.; et al. Genome-Wide Identification, Evolution, and miRNA-22 Regulation of Kruppel-Like Factor (KLF) Gene Family in Chicken (Gallus Gallus). Animals 2024, 14, 2594. [Google Scholar] [CrossRef]
- Yu, J.-A.; Wang, Z.; Yang, X.; Ma, M.; Li, Z.; Nie, Q. LncRNA-FKBP1C Regulates Muscle Fiber Type Switching by Affecting the Stability of MYH1B. Cell Death Discov. 2021, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, O.; Fujisawa-Sehara, A.; Nabeshima, Y.; Periasamy, M. Expression of Myogenic Factors in Denervated Chicken Breast Muscle: Isolation of the Chicken Myf5 Gene. Nucleic Acids Res. 1993, 21, 2503–2509. [Google Scholar] [CrossRef]
- Alway, S.E.; Martyn, J.K.; Ouyang, J.; Chaudhrai, A.; Murlasits, Z.S. Id2 Expression during Apoptosis and Satellite Cell Activation in Unloaded and Loaded Quail Skeletal Muscles. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, R540–R549. [Google Scholar] [CrossRef]
- Luo, W.; Lin, Z.; Chen, J.; Chen, G.; Zhang, S.; Liu, M.; Li, H.; He, D.; Liang, S.; Luo, Q.; et al. TMEM182 Interacts with Integrin Beta 1 and Regulates Myoblast Differentiation and Muscle Regeneration. J. Cachexia Sarcopenia Muscle 2021, 12, 1704–1723. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, Z.; Ma, M.; Zhang, J.; Kong, S.; Abdalla, B.A.; Xu, H.; Jebessa, E.; Zhang, X.; Lawal, R.A.; et al. Long Noncoding RNA SMUL Suppresses SMURF2 Production-Mediated Muscle Atrophy via Nonsense-Mediated mRNA Decay. Mol. Ther. Nucleic Acids 2021, 23, 512–526. [Google Scholar] [CrossRef]
- Cai, B.; Ma, M.; Zhang, J.; Kong, S.; Zhou, Z.; Li, Z.; Abdalla, B.A.; Xu, H.; Zhang, X.; Lawal, R.A.; et al. Long Noncoding RNA ZFP36L2-AS Functions as a Metabolic Modulator to Regulate Muscle Development. Cell Death Dis. 2022, 13, 389. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, B.; Ma, M.; Kong, S.; Zhou, Z.; Zhang, X.; Nie, Q. LncRNA SMARCD3-OT1 Promotes Muscle Hypertrophy and Fast-Twitch Fiber Transformation via Enhancing SMARCD3X4 Expression. Int. J. Mol. Sci. 2022, 23, 4510. [Google Scholar] [CrossRef] [PubMed]
- Pampouille, E.; Hennequet-Antier, C.; Praud, C.; Juanchich, A.; Brionne, A.; Godet, E.; Bordeau, T.; Fagnoul, F.; Le Bihan-Duval, E.; Berri, C. Differential Expression and Co-Expression Gene Network Analyses Reveal Molecular Mechanisms and Candidate Biomarkers Involved in Breast Muscle Myopathies in Chicken. Sci. Rep. 2019, 9, 14905. [Google Scholar] [CrossRef]
- Pejšková, L.; Rønning, S.B.; Kent, M.P.; Solberg, N.T.; Høst, V.; Thu-Hien, T.; Wold, J.P.; Lunde, M.; Mosleth, E.; Pisconti, A.; et al. Characterization of Wooden Breast Myopathy: A Focus on Syndecans and ECM Remodeling. Front. Physiol. 2023, 14, 1301804. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Khondowe, P.; Brannick, E.; Abasht, B. Spatial Transcriptomics Reveals Alterations in Perivascular Macrophage Lipid Metabolism in the Onset of Wooden Breast Myopathy in Broiler Chickens. Sci. Rep. 2024, 14, 3450. [Google Scholar] [CrossRef] [PubMed]
- Malila, Y.; Uengwetwanit, T.; Thanatsang, K.V.; Arayamethakorn, S.; Srimarut, Y.; Petracci, M.; Soglia, F.; Rungrassamee, W.; Visessanguan, W. Insights Into Transcriptome Profiles Associated With Wooden Breast Myopathy in Broilers Slaughtered at the Age of 6 or 7 Weeks. Front. Physiol. 2021, 12, 691194. [Google Scholar] [CrossRef]
- Praud, C.; Jimenez, J.; Pampouille, E.; Couroussé, N.; Godet, E.; Le Bihan-Duval, E.; Berri, C. Molecular Phenotyping of White Striping and Wooden Breast Myopathies in Chicken. Front. Physiol. 2020, 11, 633. [Google Scholar] [CrossRef]
- Malila, Y.; Thanatsang, K.; Arayamethakorn, S.; Uengwetwanit, T.; Srimarut, Y.; Petracci, M.; Strasburg, G.M.; Rungrassamee, W.; Visessanguan, W. Absolute Expressions of Hypoxia-Inducible Factor-1 Alpha (HIF1A) Transcript and the Associated Genes in Chicken Skeletal Muscle with White Striping and Wooden Breast Myopathies. PLoS ONE 2019, 14, e0220904. [Google Scholar] [CrossRef]
- Maharjan, P.; Beitia, A.; Weil, J.; Suesuttajit, N.; Hilton, K.; Caldas, J.; Umberson, C.; Martinez, D.; Kong, B.; Owens, C.M.; et al. Woody Breast Myopathy Broiler Show Age-Dependent Adaptive Differential Gene Expression in Pectoralis Major and Altered in-Vivo Triglyceride Kinetics in Adipogenic Tissues. Poult. Sci. 2021, 100, 101092. [Google Scholar] [CrossRef]
- Bordini, M.; Wang, Z.; Soglia, F.; Petracci, M.; Schmidt, C.J.; Abasht, B. RNA-Sequencing Revisited Data Shed New Light on Wooden Breast Myopathy. Poult. Sci. 2024, 103, 103902. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yuan, H.; Liu, S.; Liu, Y.; Qin, Z.; Han, W.; Zhang, R. Gene Coexpression Network Analysis Reveals the Genes and Pathways in Pectoralis Major Muscle and Liver Associated with Wooden Breast in Broilers. Poult. Sci. 2024, 103, 104056. [Google Scholar] [CrossRef]
- Brothers, B.; Zhuo, Z.; Papah, M.B.; Abasht, B. RNA-Seq Analysis Reveals Spatial and Sex Differences in Pectoralis Major Muscle of Broiler Chickens Contributing to Difference in Susceptibility to Wooden Breast Disease. Front. Physiol. 2019, 10, 764. [Google Scholar] [CrossRef] [PubMed]
- Lake, J.A.; Papah, M.B.; Abasht, B. Increased Expression of Lipid Metabolism Genes in Early Stages of Wooden Breast Links Myopathy of Broilers to Metabolic Syndrome in Humans. Genes 2019, 10, 746. [Google Scholar] [CrossRef]
- Velleman, S.G. Relationship of Skeletal Muscle Development and Growth to Breast Muscle Myopathies: A Review. Avian Dis. 2015, 59, 525–531. [Google Scholar] [CrossRef]
- Pampouille, E.; Berri, C.; Boitard, S.; Hennequet-Antier, C.; Beauclercq, S.A.; Godet, E.; Praud, C.; Jégo, Y.; Le Bihan-Duval, E. Mapping QTL for White Striping in Relation to Breast Muscle Yield and Meat Quality Traits in Broiler Chickens. BMC Genom. 2018, 19, 202. [Google Scholar] [CrossRef]
- Li, J.; Yang, D.; Chen, C.; Wang, J.; Wang, Z.; Yang, C.; Yu, C.; Li, Z. Single-Cell RNA Transcriptome Uncovers Distinct Developmental Trajectories in the Embryonic Skeletal Muscle of Daheng Broiler and Tibetan Chicken. BMC Genom. 2025, 26, 187. [Google Scholar] [CrossRef]
- Li, F.; Zhu, C.; Luo, Y.; Li, S.; Wang, Q.; Han, Y.; Wu, Z.; Li, X.; Liang, Y.; Chen, Y.; et al. Transcriptomic Analysis on Pectoral Muscle of European Meat Pigeons and Shiqi Pigeons during Embryonic Development. Animals 2023, 13, 3267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tian, W.; Wang, D.; Guo, Y.; Cheng, Z.; Zhang, Y.; Li, X.; Zhi, Y.; Li, D.; Li, Z.; et al. Comparative Analyses of Dynamic Transcriptome Profiles Highlight Key Response Genes and Dominant Isoforms for Muscle Development and Growth in Chicken. Genet. Sel. Evol. 2023, 55, 73. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Han, R.; Wang, Y.; Li, G.; Liu, X.; Tian, Y.; Kang, X.; Li, Z. Transcriptome Analysis of the Pectoral Muscles of Local Chickens and Commercial Broilers Using Ribo-Zero Ribonucleic Acid Sequencing. PLoS ONE 2017, 12, e0184115. [Google Scholar] [CrossRef]
- Sporer, K.R.B.; Tempelman, R.J.; Ernst, C.W.; Reed, K.M.; Velleman, S.G.; Strasburg, G.M. Transcriptional Profiling Identifies Differentially Expressed Genes in Developing Turkey Skeletal Muscle. BMC Genom. 2011, 12, 143. [Google Scholar] [CrossRef]
- Zhou, K.-Z.; Wu, P.-F.; Zhang, X.-C.; Ling, X.-Z.; Zhang, J.; Zhang, L.; Li, P.-F.; Zhang, T.; Wei, Q.-Y.; Zhang, G.-X. Comparative Analysis of miRNA Expression Profiles in Skeletal Muscle of Bian Chickens at Different Embryonic Ages. Animals 2022, 12, 1003. [Google Scholar] [CrossRef]
- Wu, P.; He, M.; Zhang, X.; Zhou, K.; Zhang, T.; Xie, K.; Dai, G.; Wang, J.; Wang, X.; Zhang, G. miRNA-Seq Analysis in Skeletal Muscle of Chicken and Function Exploration of miR-24-3p. Poult. Sci. 2022, 101, 102120. [Google Scholar] [CrossRef]
- Ju, X.; Liu, Y.; Shan, Y.; Ji, G.; Zhang, M.; Tu, Y.; Zou, J.; Chen, X.; Geng, Z.; Shu, J. Analysis of Potential Regulatory LncRNAs and CircRNAs in the Oxidative Myofiber and Glycolytic Myofiber of Chickens. Sci. Rep. 2021, 11, 20861. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Shan, Y.; Ji, G.; Ju, X.; Tu, Y.; Sheng, Z.; Xie, J.; Zou, J.; Shu, J. miRNA-mRNA Network Regulation in the Skeletal Muscle Fiber Phenotype of Chickens Revealed by Integrated Analysis of miRNAome and Transcriptome. Sci. Rep. 2020, 10, 10619. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y.; Jin, W.; Fu, S.; Li, D.; Zhang, Y.; Sun, G.; Jiang, R.; Han, R.; Li, Z.; et al. Analyses of MicroRNA and mRNA Expression Profiles Reveal the Crucial Interaction Networks and Pathways for Regulation of Chicken Breast Muscle Development. Front. Genet. 2019, 10, 197. [Google Scholar] [CrossRef]
- Jebessa, E.; Ouyang, H.; Abdalla, B.A.; Li, Z.; Abdullahi, A.Y.; Liu, Q.; Nie, Q.; Zhang, X. Characterization of miRNA and Their Target Gene during Chicken Embryo Skeletal Muscle Development. Oncotarget 2018, 9, 17309–17324. [Google Scholar] [CrossRef] [PubMed]
- Khatri, B.; Seo, D.; Shouse, S.; Pan, J.H.; Hudson, N.J.; Kim, J.K.; Bottje, W.; Kong, B.C. MicroRNA Profiling Associated with Muscle Growth in Modern Broilers Compared to an Unselected Chicken Breed. BMC Genom. 2018, 19, 683. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; He, X.; Li, G.; Xu, H.; Jia, X.; Nie, Q.; Zhang, X. Deep Sequencing Analysis of miRNA Expression in Breast Muscle of Fast-Growing and Slow-Growing Broilers. Int. J. Mol. Sci. 2015, 16, 16242–16262. [Google Scholar] [CrossRef]
- Li, T.; Wu, R.; Zhang, Y.; Zhu, D. A Systematic Analysis of the Skeletal Muscle miRNA Transcriptome of Chicken Varieties with Divergent Skeletal Muscle Growth Identifies Novel miRNAs and Differentially Expressed miRNAs. BMC Genom. 2011, 12, 186. [Google Scholar] [CrossRef]
- Rathjen, T.; Pais, H.; Sweetman, D.; Moulton, V.; Munsterberg, A.; Dalmay, T. High Throughput Sequencing of microRNAs in Chicken Somites. FEBS Lett. 2009, 583, 1422–1426. [Google Scholar] [CrossRef]
- Miao, D.; Zhang, C.; Wu, X.; Wang, Y.; Chowdhury, V.S.; Yang, H.; Wang, Z. Transcriptomic Analysis of Long Non-Coding RNA and mRNA in Pigeon Squab Pectoralis Muscle Development. Br. Poult. Sci. 2025, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Xu, D.; Li, W.; Wang, Y.; Cao, N.; Fu, X.; Tian, Y.; Li, Y.; Li, B. Non-Coding RNA Regulation of Magang Geese Skeletal Muscle Maturation via the MAPK Signaling Pathway. Front. Physiol. 2023, 14, 1331974. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Hu, S.; Yan, P.; Wu, J.; Guo, H.; Zhao, L.; Tang, Q.; Ma, J.; Long, K.; Jin, L.; et al. Analysis of mRNA and lncRNA Expression Profiles of Breast Muscle during Pigeon (Columbalivia) Development. Genes 2022, 13, 2314. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated Analysis of Long Non-Coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front. Physiol. 2017, 7, 687. [Google Scholar] [CrossRef]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of Long Non-Protein Coding RNAs in Chicken Skeletal Muscle Using next Generation Sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef]
- Shao, B.; Wang, Z.; Luo, P.; Du, P.; Zhang, X.; Zhang, H.; Si, X.; Ma, S.; Chen, W.; Huang, Y. Identifying Insulin-Responsive circRNAs in Chicken Pectoralis. BMC Genom. 2025, 26, 148. [Google Scholar] [CrossRef]
- Cai, B.; Ma, M.; Zhou, Z.; Kong, S.; Zhang, J.; Zhang, X.; Nie, Q. circPTPN4 Regulates Myogenesis via the miR-499-3p/NAMPT Axis. J. Anim. Sci. Biotechnol. 2022, 13, 2. [Google Scholar] [CrossRef]
- Wu, P.; Zhou, K.; Zhang, J.; Ling, X.; Zhang, X.; Zhang, L.; Li, P.; Wei, Q.; Zhang, T.; Wang, X.; et al. Identification of Crucial circRNAs in Skeletal Muscle during Chicken Embryonic Development. BMC Genom. 2022, 23, 330. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, J.; Zhou, Y.; Liu, S.; Wu, J.; Jiang, H.; Xu, J.; Mao, H.; Liu, S.; Chen, B. Signaling Pathways and Regulatory Networks in Quail Skeletal Muscle Development: Insights from Whole Transcriptome Sequencing. Poult. Sci. 2024, 103, 103603. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, C.; Han, S.; Chen, L.; Ding, H.; Lin, Y.; Zhang, G.; Xie, K.; Wang, J.; Dai, G. Integrated Analysis Reveals a lncRNA-miRNA-mRNA Network Associated with Pigeon Skeletal Muscle Development. Genes 2021, 12, 1787. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, M.; Yuan, R.; Zhou, Z.; Zhang, J.; Kong, S.; Lin, D.; Lian, L.; Li, J.; Zhang, X.; et al. MYH1G-AS Is a Chromatin-Associated lncRNA That Regulates Skeletal Muscle Development in Chicken. Cell. Mol. Biol. Lett. 2024, 29, 9. [Google Scholar] [CrossRef] [PubMed]
- Padilha, S.F.; Ibelli, A.M.G.; Peixoto, J.O.; Cantão, M.E.; Moreira, G.C.M.; Fernandes, L.T.; Tavernari, F.C.; Morés, M.A.Z.; Bastos, A.P.A.; Dias, L.T.; et al. Novel Candidate Genes Involved in an Initial Stage of White Striping Development in Broiler Chickens. Animals 2024, 14, 2379. [Google Scholar] [CrossRef] [PubMed]
- Marciano, C.M.M.; Ibelli, A.M.G.; Marchesi, J.A.P.; de Oliveira Peixoto, J.; Fernandes, L.T.; Savoldi, I.R.; do Carmo, K.B.; Ledur, M.C. Differential Expression of Myogenic and Calcium Signaling-Related Genes in Broilers Affected With White Striping. Front. Physiol. 2021, 12, 712464. [Google Scholar] [CrossRef] [PubMed]
- Malila, Y.; Uengwetwanit, T.; Arayamethakorn, S.; Srimarut, Y.; Thanatsang, K.V.; Soglia, F.; Strasburg, G.M.; Rungrassamee, W.; Visessanguan, W. Transcriptional Profiles of Skeletal Muscle Associated With Increasing Severity of White Striping in Commercial Broilers. Front. Physiol. 2020, 11, 580. [Google Scholar] [CrossRef]
- Pizzol, M.S.D.; Ibelli, A.M.G.; Cantão, M.E.; Campos, F.G.; de Oliveira, H.C.; de Oliveira Peixoto, J.; Fernandes, L.T.; de Castro Tavernari, F.; Morés, M.A.Z.; Bastos, A.P.A.; et al. Differential Expression of miRNAs Associated with Pectoral Myopathies in Young Broilers: Insights from a Comparative Transcriptome Analysis. BMC Genom. 2024, 25, 104. [Google Scholar] [CrossRef]
- Shu, J.; Liu, Y.; Shan, Y.; Ji, G.; Ju, X.; Tu, Y.; Shi, S.; Sheng, Z.; Zhang, M.; Zou, J. Deep Sequencing microRNA Profiles Associated with Wooden Breast in Commercial Broilers. Poult. Sci. 2021, 100, 101496. [Google Scholar] [CrossRef]
- Wang, Z.; Brannick, E.; Abasht, B. Integrative Transcriptomic and Metabolomic Analysis Reveals Alterations in Energy Metabolism and Mitochondrial Functionality in Broiler Chickens with Wooden Breast. Sci. Rep. 2023, 13, 4747. [Google Scholar] [CrossRef]
| Method | Primary Application | Key Advantages | Key Limitations | Ideal Use Case |
|---|---|---|---|---|
| Adenovirus (AdV) | Acute interventions; regeneration models | High-efficiency transduction; high-level transient expression; large payload capacity. | Triggers strong immune response; expression is transient (weeks). | Muscle injury/regeneration models; acute overexpression/knockdown in postnatal birds. |
| CRISPR (G0) Somatic (AdV/Plasmid) | Rapid phenotyping (G0 generation) | Bypasses need for germline transmission; direct functional testing in same generation. | Mosaicism (mix of edited/unedited cells); delivery efficiency to target tissue can be variable. | Rapid validation of gene function in embryonic or postnatal muscle without creating stable lines. |
| CRISPR PGC Editing | Heritable modifications; stable lines | Creates stable, non-mosaic, heritable knockout/knock-in lines; precise edits. | Technically demanding; requires long breeding programs; limited to few avian species. | Generating models for agricultural traits (e.g., MSTN-KO for hypermuscling) and fundamental research. |
| Lentivirus (LV) | Postnatal muscle; ncRNA function | Infects dividing and non-dividing cells (e.g., myofibers); stable, long-term expression; larger payload. | Lower transduction efficiency in some mature tissues; integration risks. | Validating functions of lncRNAs, circRNAs, and mRNAs in postnatal growth, atrophy, and fiber-type specification. |
| RCAS Retrovirus | Embryonic development | Species-specific; infectious spread within embryo for widespread expression. | Only infects dividing cells; limited to avian species; smaller payload capacity. | Studying early myogenesis, somite patterning, and progenitor cell niches. |
| Candidate Gene/RNA | Dysregulation Pattern | Hypothesized Role | Proposed In Vivo Validation Tool | Expected Phenotypic Outcome If Hypothesis Is Correct |
|---|---|---|---|---|
| FABP4 | Embryonic development | Drives early lipid accumulation and metabolic insult. | Adenoviral CRISPR-KO in ovo or postnatal lentiviral shRNA knockdown. | Reduced intramuscular lipidosis; delayed or reduced severity of WB/WS. |
| miR-155 | Postnatal muscle; ncRNA function | Promotes fibrosis and inflammation via target repression. | Lentiviral sponge/decoy vector to sequester miR-155 in vivo. | Attenuated fibrosis (↓ collagen) and reduced inflammatory markers. |
| SDC4 | Acute interventions; regeneration models | Enhances pro-fibrotic signaling and ECM organization. | Lentiviral shRNA knockdown in young broilers. | Improved muscle texture; reduced ECM deposition. |
| PPARGC1A | Rapid phenotyping (G0 generation) | Loss impairs mitochondrial function, promoting metabolic shift. | Lentiviral overexpression in breast muscle pre-onset. | Improved oxidative metabolism; resistance to metabolic stress. |
| COL6A3 | Heritable modifications; stable lines | Key structural component of pathological fibrosis. | Somatic CRISPR-KO in ovo to disrupt fibril formation. | Disorganized collagen deposition; reduced muscle stiffness. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibril, B.A.A.; Chai, X.; Xu, J. From Correlation to Causation: Defining Gene and RNA Function in Poultry Muscle Biology Using In Vivo Genetic Tools. Biomolecules 2025, 15, 1554. https://doi.org/10.3390/biom15111554
Gibril BAA, Chai X, Xu J. From Correlation to Causation: Defining Gene and RNA Function in Poultry Muscle Biology Using In Vivo Genetic Tools. Biomolecules. 2025; 15(11):1554. https://doi.org/10.3390/biom15111554
Chicago/Turabian StyleGibril, Bahareldin Ali Abdalla, Xuewen Chai, and Jiguo Xu. 2025. "From Correlation to Causation: Defining Gene and RNA Function in Poultry Muscle Biology Using In Vivo Genetic Tools" Biomolecules 15, no. 11: 1554. https://doi.org/10.3390/biom15111554
APA StyleGibril, B. A. A., Chai, X., & Xu, J. (2025). From Correlation to Causation: Defining Gene and RNA Function in Poultry Muscle Biology Using In Vivo Genetic Tools. Biomolecules, 15(11), 1554. https://doi.org/10.3390/biom15111554





