Elevated SH3BP5 Correlates with Poor Outcome and Contributes to the Growth of Acute Myeloid Leukemia Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Gene Expression Omnibus (GEO) and Clinical Databases
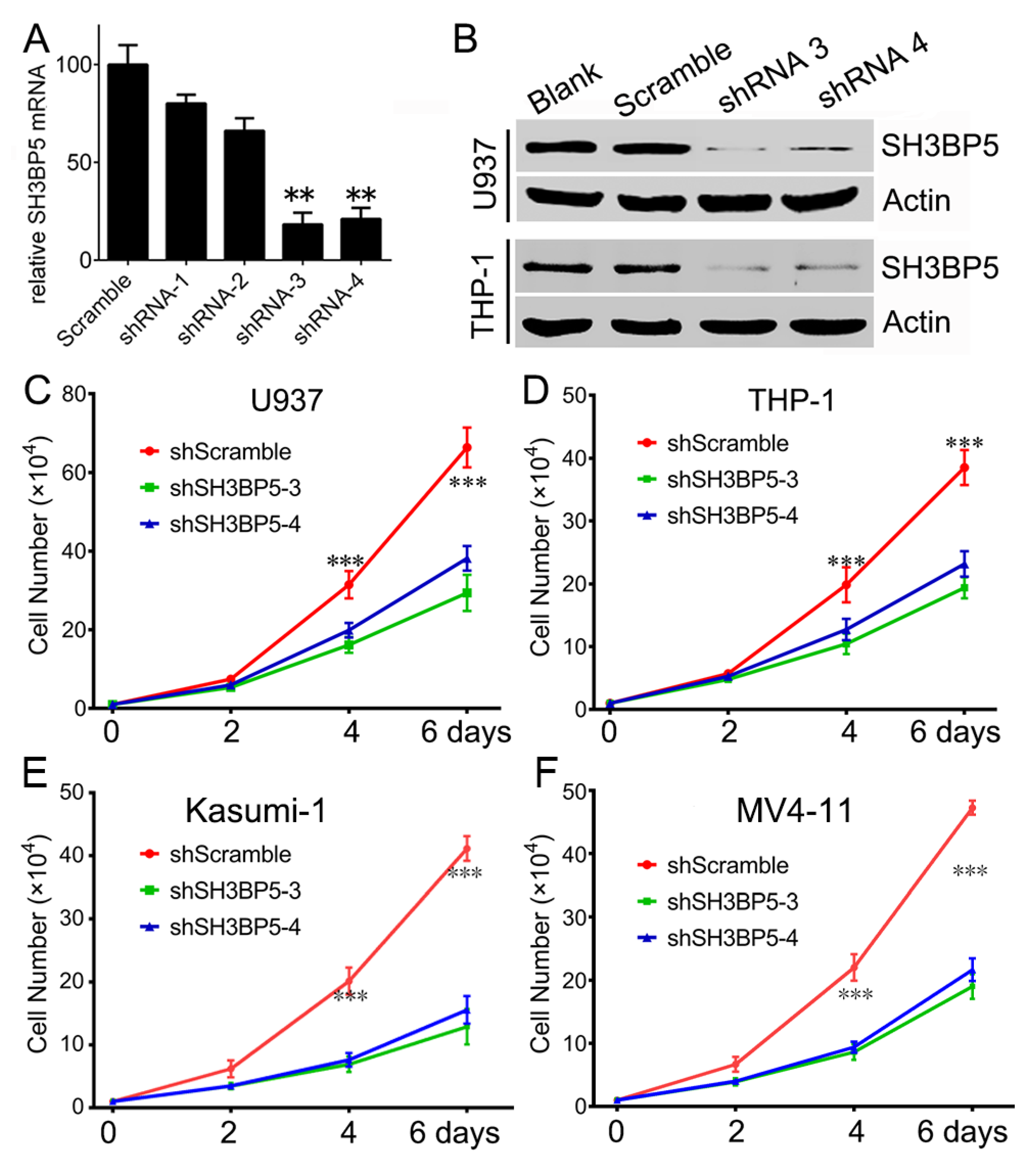
2.4. Lentiviral shRNA Constructs, Production, and Infection
2.5. Quantitative Real-Time RT-PCR
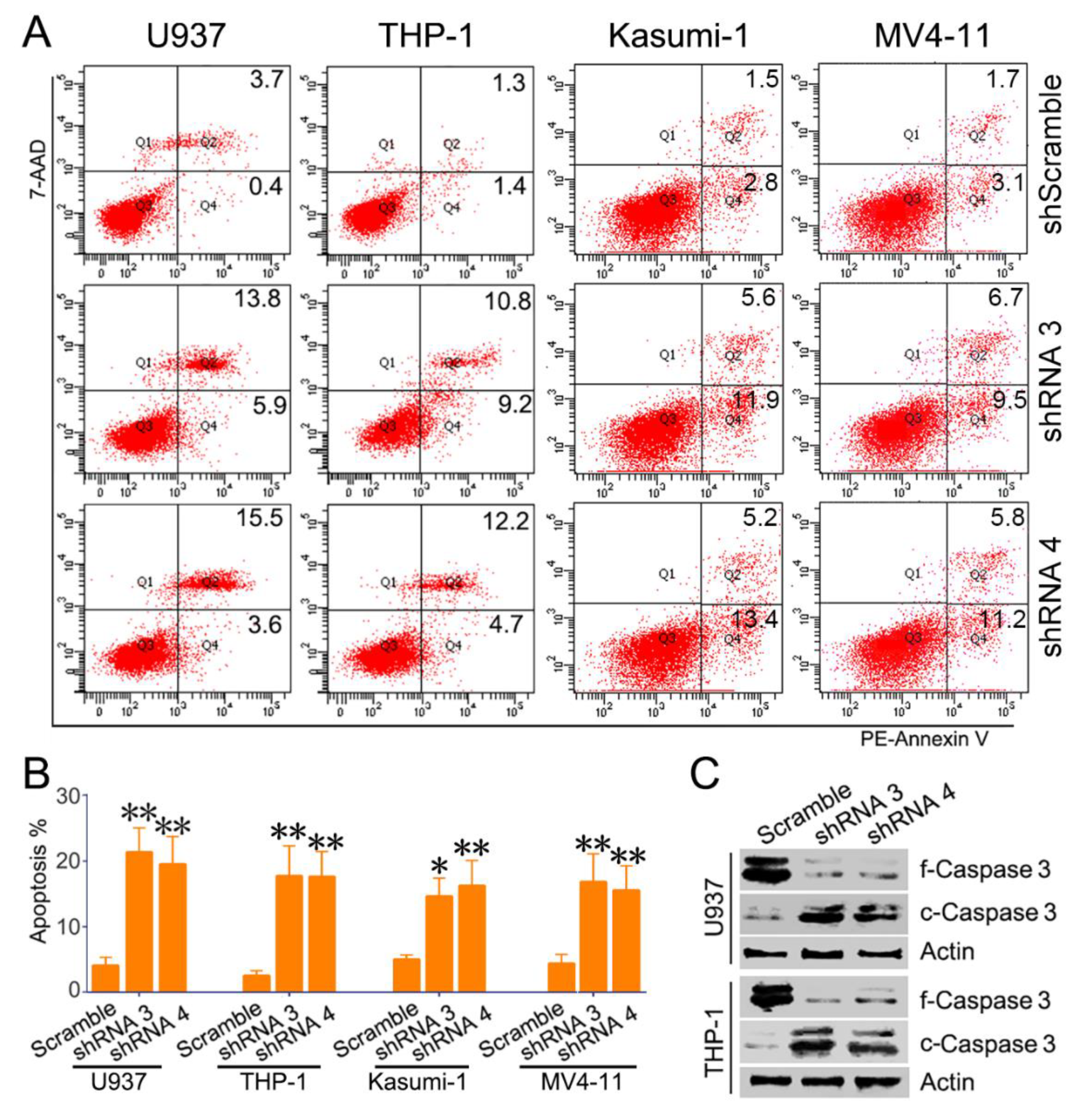
2.6. Cell Growth and Apoptosis Assays
2.7. Western Blot
2.8. Statistical Analyses
3. Results
3.1. Expression of SH3BP5 in AML and Its Connection to AML Pathological Characteristics
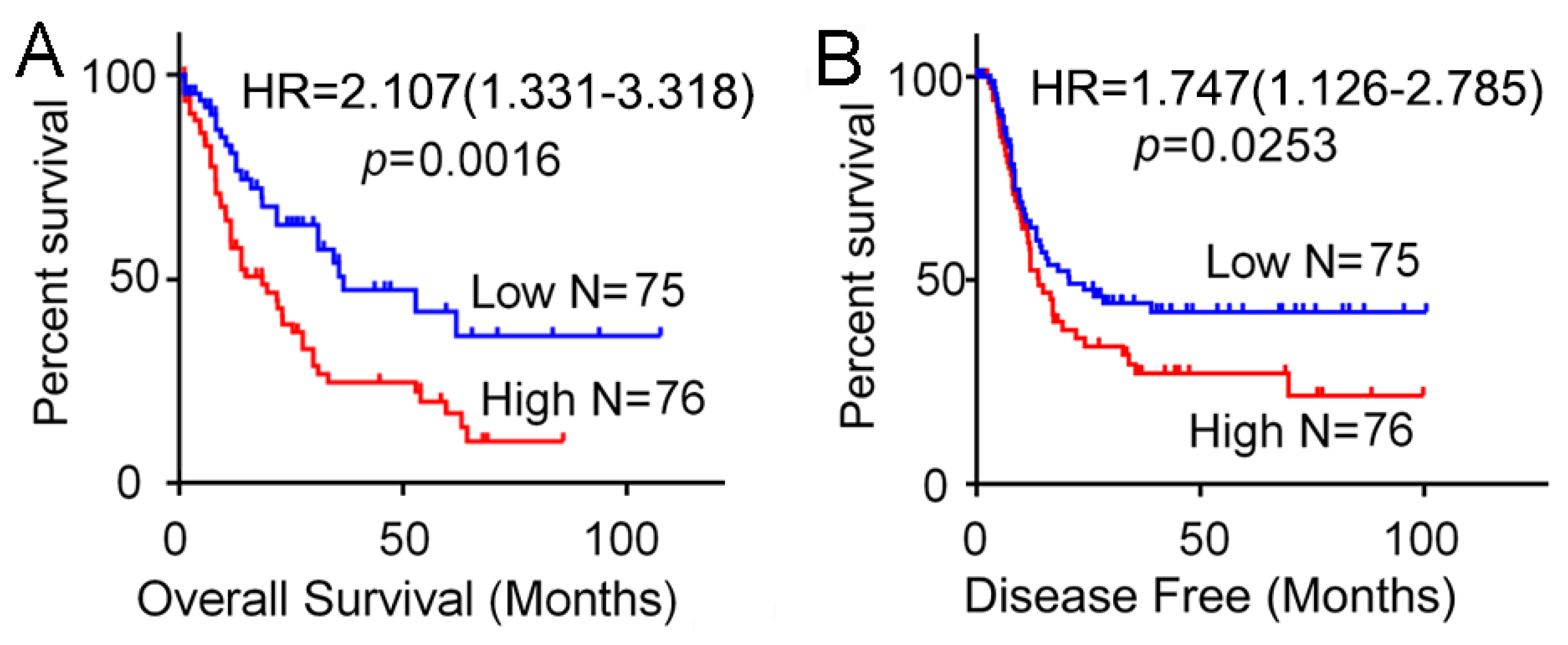
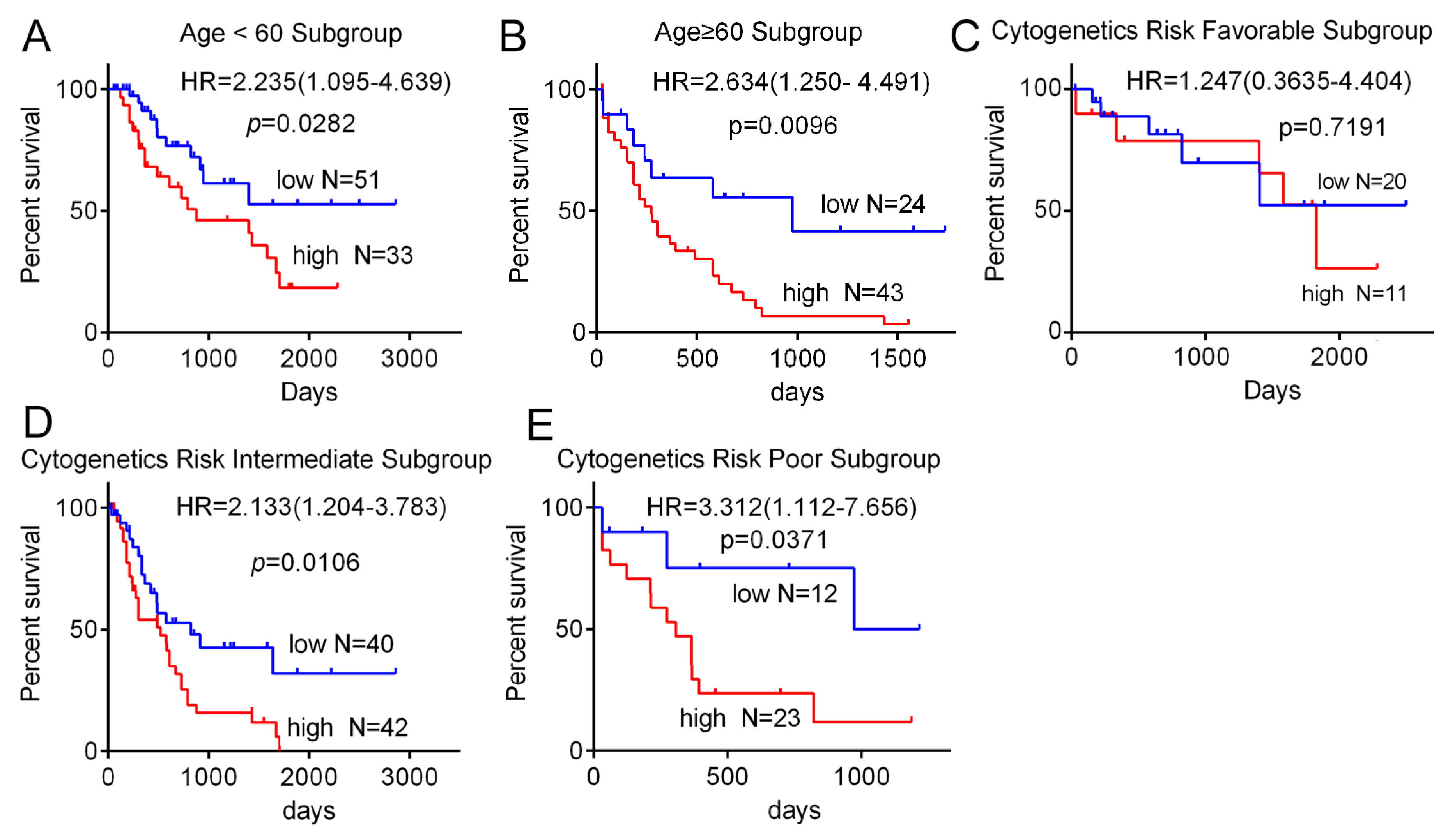
3.2. High Expression of SH3BP5 is Connected to Worse Survival and is a Separate Indicator of Poor Survival in AML Patients
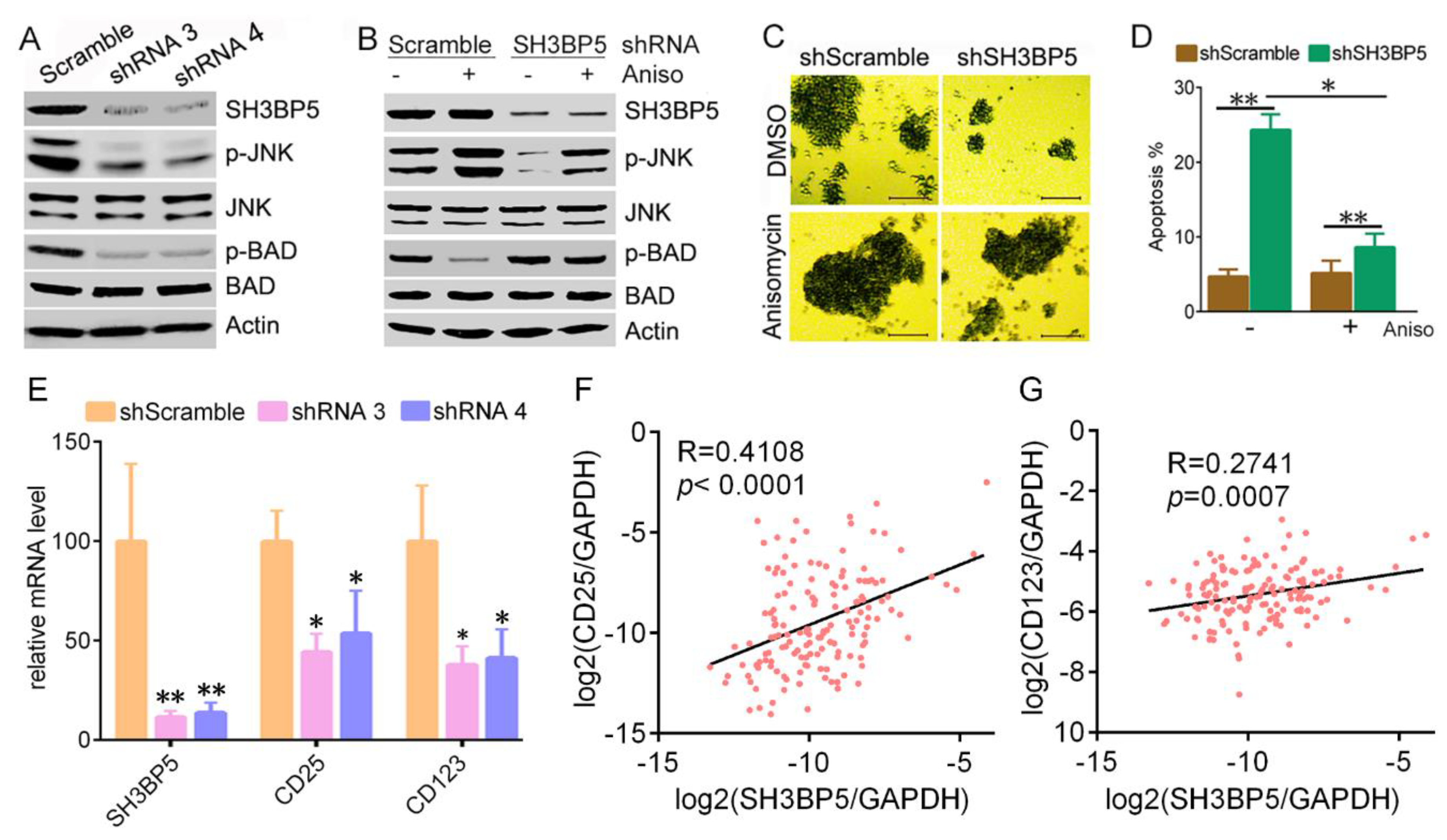
3.3. Knockdown of SH3BP Inhibits Cell Viability and Induces Apoptosis of AML Cells
3.4. SH3BP5/JNK/BAD Signaling Regulates Survival of AML Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xie, J.; Lu, Z.; Xie, J.; Lu, Z.; Li, M.; Wang, Y.; Zhang, C.C. ADCY7 supports development of acute myeloid leukemia. Biochem. Biophys. Res. Commun. 2015, 465, 47–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Yin, Y.; Li, C.; Chen, J.; Xie, J.; Lu, Z.; Li, M.; Wang, Y.; Zhang, C.C. ACER3 supports development of acute myeloid leukemia. Biochem. Biophys. Res. Commun. 2016, 478, 33–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, C.; Zhan, R.; Yin, Y. SPG6 supports development of acute myeloid leukemia by regulating BMPR2-Smad-Bcl-2/Bcl-xl signaling. Biochem. Biophys. Res. Commun. 2018, 501, 220–225. [Google Scholar] [CrossRef]
- Kang, X.; Lu, Z.; Cui, C.; Deng, M.; Fan, Y.; Dong, B.; Han, X.; Xie, F.; Tyner, J.W.; Coligan, J.E.; et al. The ITIM-containing receptor LAIR1 is essential for acute myeloid leukaemia development. Nat. Cell Biol. 2015, 17, 665–677. [Google Scholar] [CrossRef] [Green Version]
- Wiltshire, C.; Matsushita, M.; Tsukada, S.; Gillespie, D.A.; May, G.H. A new c-Jun N-terminal kinase (JNK)-interacting protein, Sab (SH3BP5), associates with mitochondria. Biochem. J. 2002, 367, 577–585. [Google Scholar] [CrossRef]
- Wiltshire, C.; Gillespie, D.A.; May, G.H. Sab (SH3BP5), a novel mitochondria-localized JNK-interacting protein. Biochem. Soc. Trans. 2004, 32, 1075–1077. [Google Scholar] [CrossRef]
- Win, S.; Than, T.A.; Fernandez-Checa, J.C.; Kaplowitz, N. JNK interaction with Sab mediates ER stress induced inhibition of mitochondrial respiration and cell death. Cell Death Dis. 2014, 5, e989. [Google Scholar] [CrossRef]
- Win, S.; Than, T.A.; Le, B.H.; Garcia-Ruiz, C.; Fernandez-Checa, J.C.; Kaplowitz, N. Sab (Sh3bp5) dependence of JNK mediated inhibition of mitochondrial respiration in palmitic acid induced hepatocyte lipotoxicity. J. Hepatol. 2015, 62, 1367–1374. [Google Scholar] [CrossRef] [Green Version]
- Win, S.; Than, T.A.; Kaplowitz, N. The Regulation of JNK Signaling Pathways in Cell Death through the Interplay with Mitochondrial SAB and Upstream Post-Translational Effects. Int. J. Mol. Sci. 2018, 19, 3657. [Google Scholar] [CrossRef]
- Win, S.; Than, T.A.; Zhang, J.; Oo, C.; Min, R.W.M.; Kaplowitz, N. New insights into the role and mechanism of c-Jun-N-terminal kinase signaling in the pathobiology of liver diseases. Hepatology 2018, 67, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jin, C.; Xu, M.; Zhou, L.; Li, D.; Yin, Y. Bifunctional enzyme ATIC promotes propagation of hepatocellular carcinoma by regulating AMPK-mTOR-S6 K1 signaling. Cell Commun. Signal. 2017, 15, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Gao, J.; Li, D.; Yin, Y. CEP55 Promotes Cell Motility via JAK2(-)STAT3(-)MMPs Cascade in Hepatocellular Carcinoma. Cells 2018, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zhou, L.; Zhan, R.; Zhang, Q.; Li, M. Identification of WDR12 as a novel oncogene involved in hepatocellular carcinoma propagation. Cancer Manag. Res. 2018, 10, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ikezoe, T.; Nishioka, C.; Honda, G.; Yokoyama, A. Thrombomodulin-induced differentiation of acute myelomonocytic leukemia cells via JNK signaling. Leuk. Res. 2012, 36, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Volk, A.; Li, J.; Xin, J.; You, D.; Zhang, J.; Liu, X.; Xiao, Y.; Breslin, P.; Li, Z.; Wei, W.; et al. Co-inhibition of NF-kappaB and JNK is synergistic in TNF-expressing human AML. J. Exp. Med. 2014, 211, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Win, S.; Than, T.A.; Min, R.W.; Aghajan, M.; Kaplowitz, N. JNK mediates mouse liver injury through a novel Sab (SH3BP5)-dependent pathway leading to inactivation of intramitochondrial Src. Hepatology 2016, 63, 1987–2003. [Google Scholar] [CrossRef] [PubMed]
- Bode, A.M.; Dong, Z. The functional contrariety of JNK. Mol. Carcinog. 2007, 46, 591–598. [Google Scholar] [CrossRef] [Green Version]
- Chen, F. JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer Res. 2012, 72, 379–386. [Google Scholar] [CrossRef]
- Liu, Z.; Xia, Y.; Zhang, X.; Liu, L.; Tu, S.; Zhu, W.; Yu, L.; Wan, H.; Yu, B.; Wan, F. Roles of the MST1-JNK signaling pathway in apoptosis of colorectal cancer cells induced by Taurine. Libyan J. Med. 2018, 13, 1500346. [Google Scholar] [CrossRef]
- Lin, X.; Fang, Q.; Chen, S.; Liu, L.; Tu, S.; Zhu, W.; Yu, L.; Wan, H.; Yu, B.; Wan, F. Heme oxygenase-1 suppresses the apoptosis of acute myeloid leukemia cells via the JNK/c-JUN signaling pathway. Leuk. Res. 2015, 39, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Minemoto, Y.; Zhang, J.; Liu, J.; Tang, F.; Bui, T.N.; Xiang, J.; Lin, A. JNK suppresses apoptosis via phosphorylation of the proapoptotic Bcl-2 family protein BAD. Mol. Cell 2004, 13, 329–340. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Reddy, E.P. JNK-signaling: A multiplexing hub in programmed cell death. Genes Cancer 2017, 8, 682–694. [Google Scholar] [PubMed] [Green Version]
- Ventura, J.J.; Hubner, A.; Zhang, C.; Liu, J.; Tang, F.; Bui, T.N.; Xiang, J.; Lin, A. Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell 2006, 21, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Papa, S.; Zazzeroni, F.; Bubici, C.; Jayawardena, S.; Alvarez, K.; Matsuda, S.; Nguyen, D.U.; Pham, C.G.; Nelsbach, A.H.; Melis, T.; et al. Gadd45 beta mediates the NF-kappa B suppression of JNK signalling by targeting MKK7/JNKK2. Nat. Cell Biol. 2004, 6, 146–153. [Google Scholar] [CrossRef] [PubMed]

| Patient’s Parameters | SH3BP5 Low, 75 | SH3BP5 High, 76 | p |
|---|---|---|---|
| Sex, male/female | 42/33 | 41/35 | 0.7999 |
| Median age, years (range) | 51 (21–81) | 61(25–88) | 0.0152 * |
| FAB classifications 1 | 0.0801 | ||
| M0 | 5 | 10 | |
| M1 | 20 | 15 | |
| M2 | 25 | 13 | |
| M3 | 6 | 9 | |
| M4 | 14 | 15 | |
| M5 | 4 | 11 | |
| M6 | 0 | 2 | |
| M7 | 0 | 1 | |
| Not Classified | 1 | 0 | |
| Cytogenetics Risk | 0.0278 * | ||
| Favorable | 20 | 11 | |
| Intermediate | 40 | 42 | |
| Poor | 12 | 23 | |
| No data | 3 | 0 | |
| Karyotypes | 0.0117 * | ||
| Normal | 36 | 36 | |
| t (8;21) | 7 | 0 | |
| t (15;17) | 5 | 7 | |
| complex | 4 | 15 | |
| others | 15 | 10 | |
| No data | 8 | 8 | |
| Gene mutations | |||
| FLT3 mutation P/N | 19/56 | 28/48 | 0.1267 |
| RAS expression P/N | 5/70 | 4/72 | 0.7157 |
| NPM1 expression P/N | 13/62 | 21/55 | 0.1298 |
| IDH1 expression P/N | 10/65 | 16/60 | 0.2090 |
| Characteristics | HR | Univariate 95% CI | p | HR | Multivariate 95% CI | p |
|---|---|---|---|---|---|---|
| Sex (female vs. male) | 1.006 | 0.644–1.573 | 0.9781 | |||
| Age (≥60 vs. <60) | 2.615 | 1.906–4.927 | 0.0001 * | 2.645 | 1.655–4.228 | 0.0001 * |
| FAB classifications | /// | /// | 0.0001 * | /// | /// | 0.1316 |
| Cytogenetics risk | /// | /// | 0.1599 | 1.487 | 1.043–2.121 | 0.0285 * |
| Karyotypes | /// | /// | 0.0165 * | /// | /// | 0.7959 |
| FLT3 (mutation vs. normal) | 1.328 | 0.8237–2.234 | 0.2344 | |||
| NPM1 (positive vs. negative) | 0.5966 | 0.3821–1.021 | 0.0647 | |||
| IDH1 (positive vs. negative) | 0.7025 | 0.4116–1.278 | 0.2707 | |||
| SH3BP5 (high vs. low) | 2.107 | 1.331–3.318 | 0.0035 * | 2.020 | 1.271–3.215 | 0.0029 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Hao, S.; Li, C.; Xiao, H.; Sun, L.; Yu, Z.; Zhang, N.; Xiong, Y.; Zhao, D.; Yin, Y. Elevated SH3BP5 Correlates with Poor Outcome and Contributes to the Growth of Acute Myeloid Leukemia Cells. Biomolecules 2019, 9, 505. https://doi.org/10.3390/biom9090505
Li M, Hao S, Li C, Xiao H, Sun L, Yu Z, Zhang N, Xiong Y, Zhao D, Yin Y. Elevated SH3BP5 Correlates with Poor Outcome and Contributes to the Growth of Acute Myeloid Leukemia Cells. Biomolecules. 2019; 9(9):505. https://doi.org/10.3390/biom9090505
Chicago/Turabian StyleLi, Minjing, Shiyu Hao, Chunling Li, Huimin Xiao, Liyuan Sun, Zhenhai Yu, Naili Zhang, Yanlian Xiong, Dongmei Zhao, and Yancun Yin. 2019. "Elevated SH3BP5 Correlates with Poor Outcome and Contributes to the Growth of Acute Myeloid Leukemia Cells" Biomolecules 9, no. 9: 505. https://doi.org/10.3390/biom9090505





