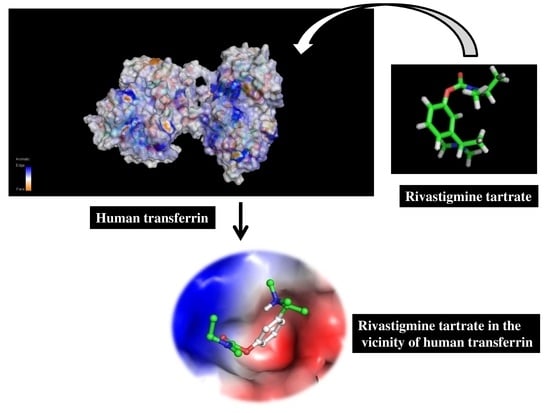
Unraveling Binding Mechanism of Alzheimer’s Drug Rivastigmine Tartrate with Human Transferrin: Molecular Docking and Multi-Spectroscopic Approach towards Neurodegenerative Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Stock Solution of hTf and RT
2.3. Steady State Fluorescence
2.4. Circular Dichroism Spectroscopy (CD)
2.5. Isothermal Titration Calorimetry
2.6. Molecular Docking Analysis
3. Results and Discussion
3.1. Steady State Fluorescence Studies
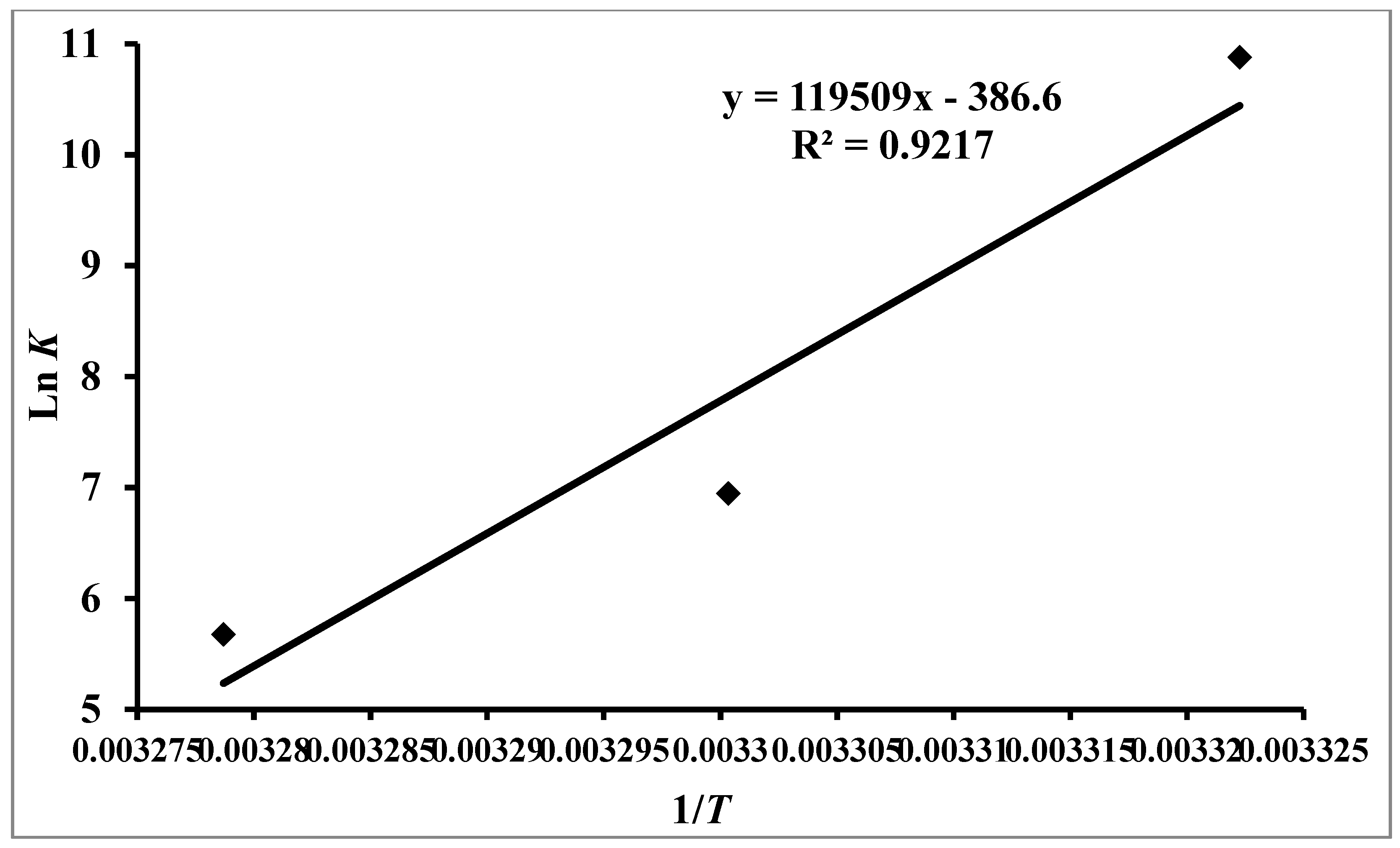
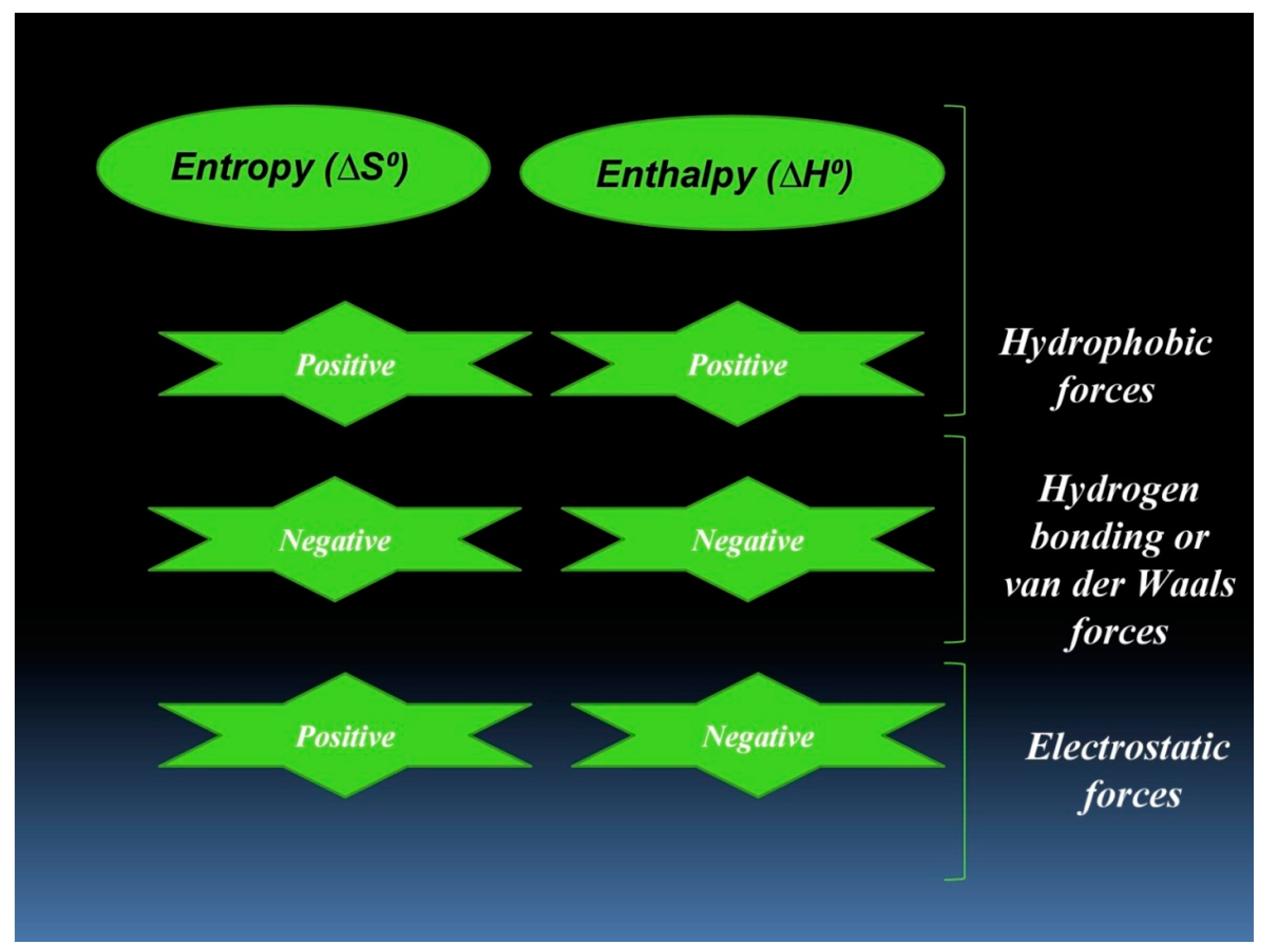
3.2. Thermodynamic Features of hTf–RT Interaction
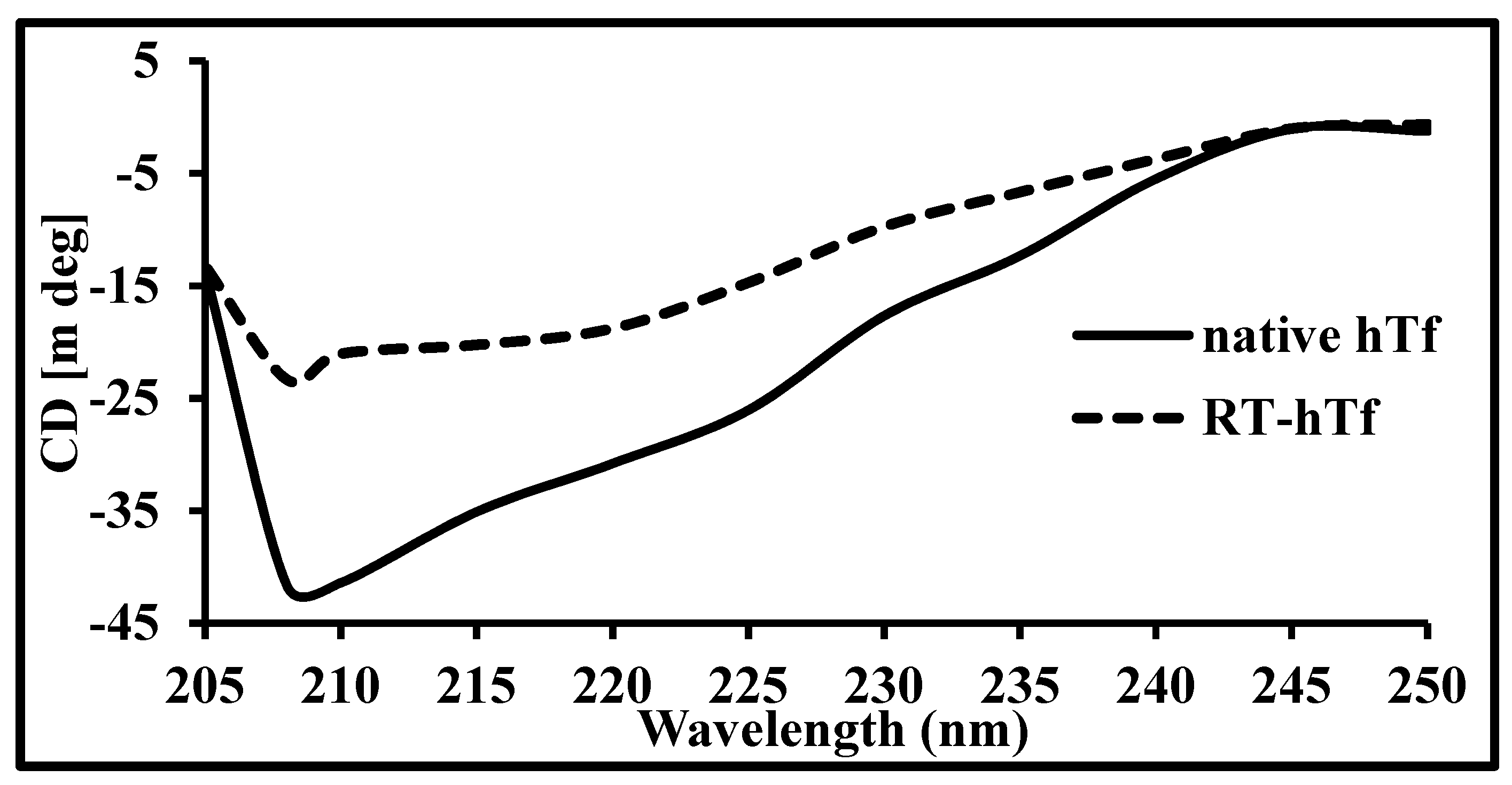
3.3. Circular Dichroism Spectroscopy
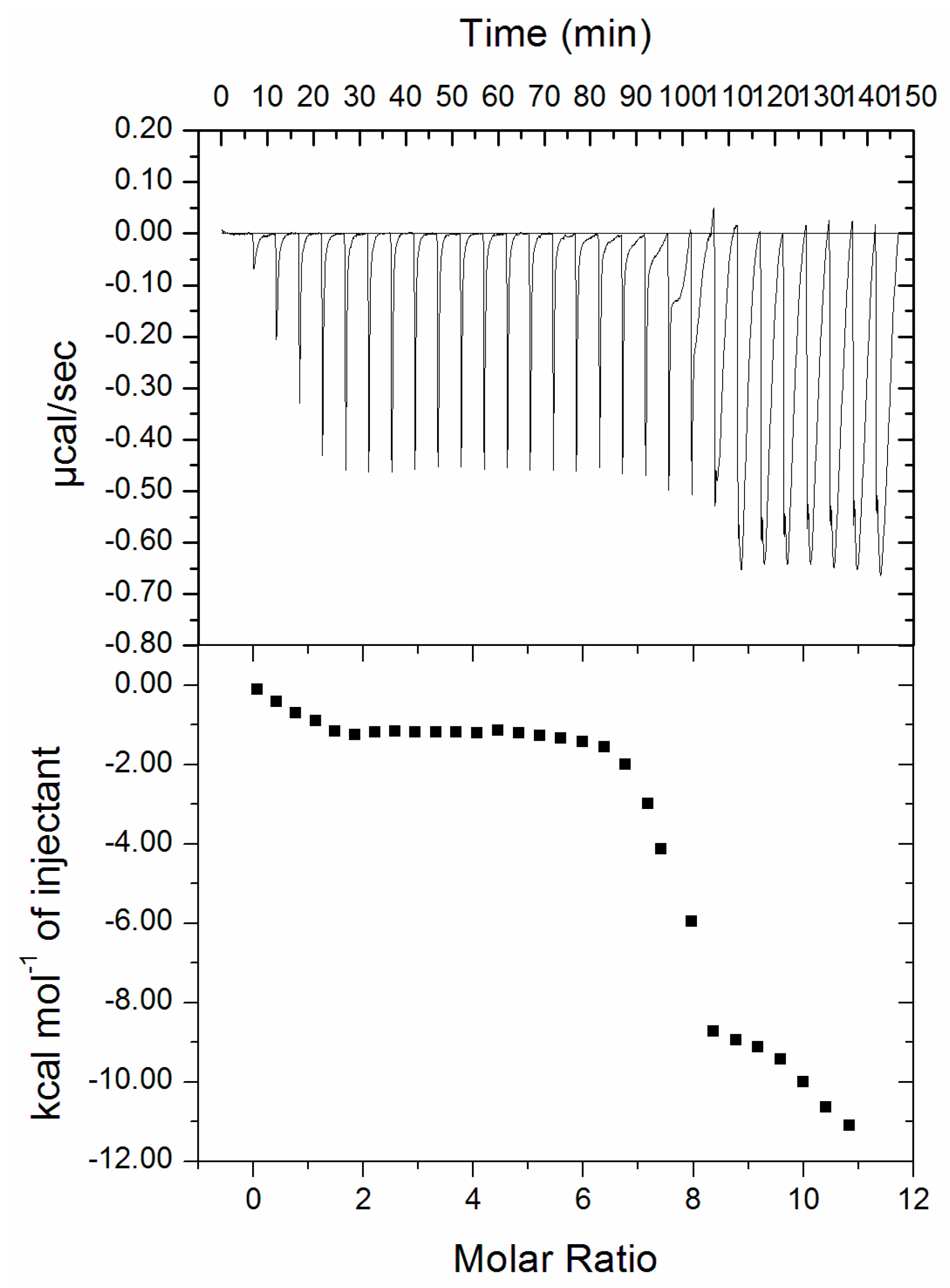
3.4. Isothermal Titration Calorimetry (ITC)
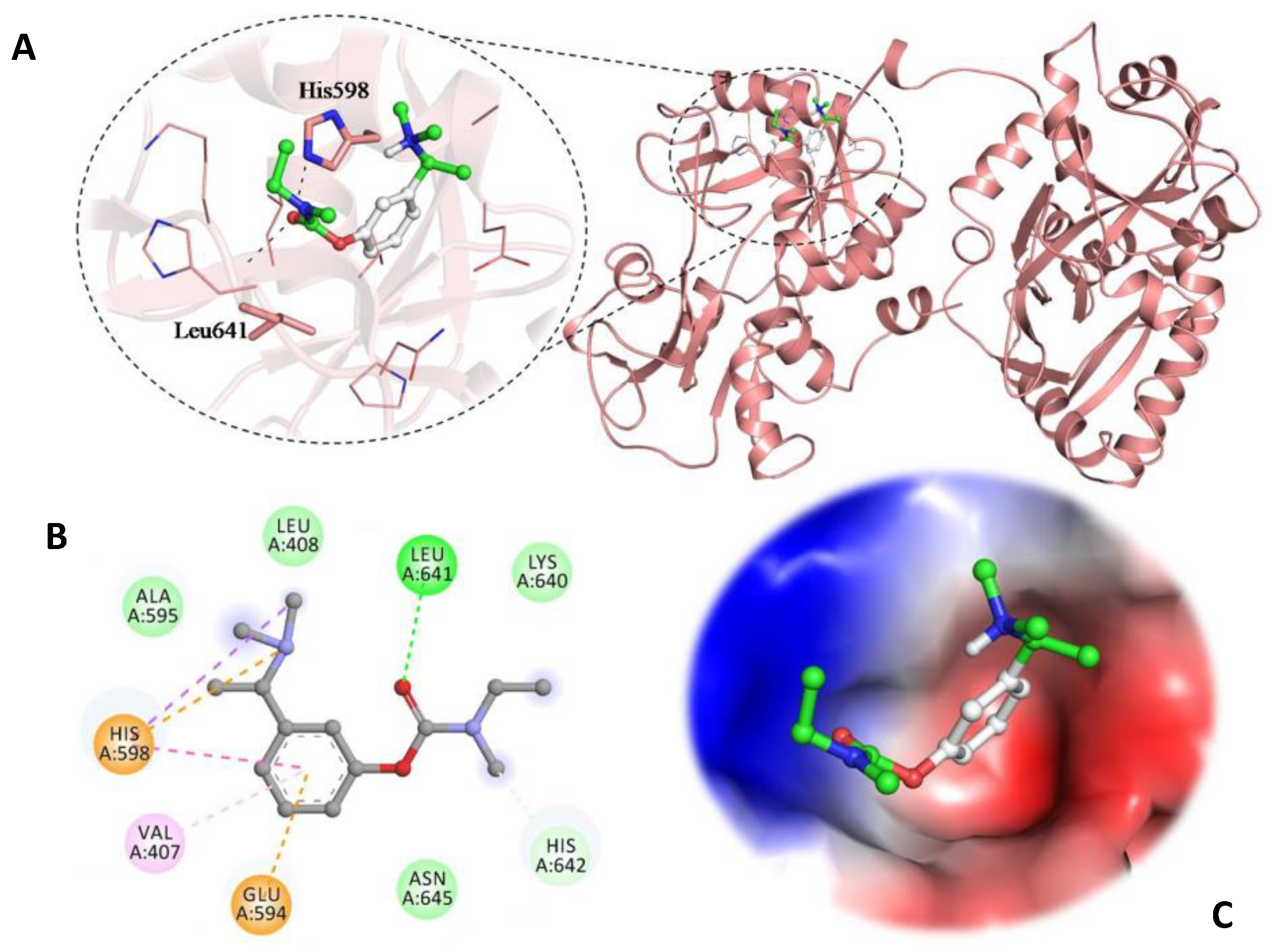
3.5. Molecular Docking Studies
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| hTf | Human transferrin |
| RT | Rivastigmine tartrate |
| ITC | Isotheral titration calorimetry |
| CD | Circular dichroism |
References
- Mills, E.; Dong, X.-P.; Wang, F.; Xu, H. Mechanisms of brain iron transport: Insight into neurodegeneration and CNS disorders. Future Med. Chem. 2010, 2, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Connor, J.R.; Benkovic, S.A. Iron regulation in the brain: Histochemical, biochemical, and molecular considerations. Ann. Neurol. 1992, 32, S51–S61. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Huang, W.; Moir, R.D.; Vanderburg, C.R.; Lai, B.; Peng, Z.; Tanzi, R.E.; Rogers, J.T.; Huang, X. Metal exposure and Alzheimer’s pathogenesis. J. Struct. Biol. 2006, 155, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Youdim, M.B.; Fridkin, M.; Zheng, H. Bifunctional drug derivatives of MAO-B inhibitor rasagiline and iron chelator VK-28 as a more effective approach to treatment of brain ageing and ageing neurodegenerative diseases. Mech. Ageing Dev. 2005, 126, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B. Towards a unifying, systems biology understanding of large-scale cellular death and destruction caused by poorly liganded iron: Parkinson’s, Huntington’s, Alzheimer’s, prions, bactericides, chemical toxicology and others as examples. Arch. Toxicol. 2010, 84, 825–889. [Google Scholar] [CrossRef] [PubMed]
- Roohani, N.; Hurrell, R.; Kelishadi, R.; Schulin, R. Zinc and its importance for human health: An integrative review. J. Res. Med. Sci. 2013, 18, 144. [Google Scholar] [PubMed]
- Kozlowski, H.; Luczkowski, M.; Remelli, M.; Valensin, D. Copper, zinc and iron in neurodegenerative diseases (Alzheimer’s, Parkinson’s and prion diseases). Coord. Chem. Rev. 2012, 256, 2129–2141. [Google Scholar] [CrossRef]
- Wallace, D.F. The regulation of iron absorption and homeostasis. Clin. Biochem. Rev. 2016, 37, 51. [Google Scholar] [PubMed]
- Sian-Hülsmann, J.; Mandel, S.; Youdim, M.B.; Riederer, P. The relevance of iron in the pathogenesis of Parkinson’s disease. J. Neurochem. 2011, 118, 939–957. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-H.; Cahill, C.M.; Vanderburg, C.R.; Scherzer, C.R.; Wang, B.; Huang, X.; Rogers, J.T. Selective translational control of the Alzheimer amyloid precursor protein transcript by iron regulatory protein-1. J. Biol. Chem. 2010, 285, 31217–31232. [Google Scholar] [CrossRef]
- Frackowiak, J.; Potempska, A.; Mazur-Kolecka, B. Formation of amyloid-β oligomers in brain vascular smooth muscle cells transiently exposed to iron-induced oxidative stress. Acta Neuropathol. 2009, 117, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.; Youdim, M.B. Role of iron in neurodegenerative disorders. Top. Magn. Reson. Imaging 2006, 17, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Dexter, D.T.; Jenner, P.; Schapira, A.H.; Marsden, C.D. Alterations in levels of iron, ferritin, and other trace metals in neurodegenerative diseases affecting the basal ganglia. Ann. Neurol. 1992, 32, S94–S100. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.; Menzies, S.; Muckenthaler, M.; Torti, F.M.; Wood, T.; Torti, S.V.; Hentze, M.W.; Beard, J.; Connor, J. Mouse brains deficient in H-ferritin have normal iron concentration but a protein profile of iron deficiency and increased evidence of oxidative stress. J. Neurosci. Res. 2003, 71, 46–63. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Hurd, M.D.; Martorell, P.; Delavande, A.; Mullen, K.J.; Langa, K.M. Monetary costs of dementia in the United States. N. Engl. J. Med. 2013, 368, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Barr, A.J. The biochemical basis of disease. Essays Biochem. 2018, 62, 619–642. [Google Scholar] [CrossRef]
- Abdelhameed, A.S.; Nusrat, S.; Ajmal, M.R.; Zakariya, S.M.; Zaman, M.; Khan, R.H. A biophysical and computational study unraveling the molecular interaction mechanism of a new Janus kinase inhibitor Tofacitinib with bovine serum albumin. J. Mol. Recognit. 2017, 30, e2601. [Google Scholar] [CrossRef]
- Shamsi, A.; Ahmed, A.; Bano, B. Probing the interaction of anticancer drug temsirolimus with human serum albumin: Molecular docking and spectroscopic insight. J. Biomol. Struct. Dyn. 2018, 36, 1479–1489. [Google Scholar] [CrossRef]
- Pan, Y.; Xu, X.; Wang, X. Rivastigmine blocks voltage-activated K+ currents in dissociated rat hippocampal neurons. Br. J. Pharmacol. 2003, 140, 907–912. [Google Scholar] [CrossRef]
- Joshi, S.A.; Chavhan, S.S.; Sawant, K.K. Rivastigmine-loaded PLGA and PBCA nanoparticles: Preparation, optimization, characterization, in vitro and pharmacodynamic studies. Eur. J. Pharm. Biopharm. 2010, 76, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.; Samanta, M.K.; Santhi, K.; Kumar, K.P.S.; Paramakrishnan, N.; Suresh, B. Poly(n-butylcyanoacrylate) nanoparticles coated with polysorbate 80 for the targeted delivery of rivastigmine into the brain to treat Alzheimer’s disease. Brain Res. 2008, 1200, 159–168. [Google Scholar] [CrossRef]
- Williams, B.R.; Nazarians, A.; Gill, M.A. A review of rivastigmine: A reversible cholinesterase inhibitor. Clin. Ther. 2003, 25, 1634–1653. [Google Scholar] [CrossRef]
- Ishtikhar, M.; Khan, S.; Badr, G.; Mohamed, A.O.; Khan, R.H. Interaction of the 5-fluorouracil analog 5-fluoro-2′-deoxyuridine with ‘N’and ‘B’isoforms of human serum albumin: A spectroscopic and calorimetric study. Mol. Biosyst. 2014, 10, 2954–2964. [Google Scholar] [CrossRef] [PubMed]
- Ajmal, M.R.; Nusrat, S.; Alam, P.; Zaidi, N.; Khan, M.V.; Zaman, M.; Shahein, Y.E.; Mahmoud, M.H.; Badr, G.; Khan, R.H. Interaction of anticancer drug clofarabine with human serum albumin and human α-1 acid glycoprotein. Spectroscopic and molecular docking approach. J. Pharm. Biomed. Anal. 2017, 135, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, A.; Ahmed, A.; Khan, M.S.; Husain, F.M.; Amani, S.; Bano, B. Investigating the interaction of anticancer drug temsirolimus with human transferrin: Molecular docking and spectroscopic approach. J. Mol. Recognit. 2018, 31, e2728. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Bourne, P.E.; Westbrook, J.; Zardecki, C. The protein data bank. In Protein Structure; CRC Press: Boca Raton, FL, USA, 2003; pp. 394–410. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2018, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. Discovery Studio Modeling Environment; Accelrys: San Diego, CA, USA, 2017. [Google Scholar]
- Schrodinger, L. The PyMOL Molecular Graphics System; Version 1.3r; Schrödinger, LLC: New York, NY, USA, 2010. [Google Scholar]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Mariam, J.; Dongre, P.; Kothari, D. Study of interaction of silver nanoparticles with bovine serum albumin using fluorescence spectroscopy. J. Fluoresc. 2011, 21, 2193. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Alam, P.; Naseem, F.; Abdelhameed, A.S.; Khan, R.H. Effect of galactose on acid induced molten globule state of soybean agglutinin: Biophysical approach. J. Mol. Struct. 2015, 1099, 149–153. [Google Scholar] [CrossRef]
- Alanazi, A.M.; Abdelhameed, A.S.; Bakheit, A.H.; Almutairi, F.M.; Alkhider, A.; Herqash, R.N.; Darwish, I.A. Unraveling the binding characteristics of the anti-HIV agents abacavir, efavirenz and emtricitabine to bovine serum albumin using spectroscopic and molecular simulation approaches. J. Mol. Liq. 2018, 251, 345–357. [Google Scholar] [CrossRef]
- Abdelhameed, A.S.; Bakheit, A.H.; Almutairi, F.M.; AlRabiah, H.; Kadi, A.A. Biophysical and in silico studies of the interaction between the anti-viral agents acyclovir and penciclovir, and human serum albumin. Molecules 2017, 22, 1906. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.-Q.; Tian, F.-F.; Li, Q.; Li, L.-L.; Xiang, C.; Liu, Y.; Dai, J.; Jiang, F.-L. Probing the adverse temperature dependence in the static fluorescence quenching of BSA induced by a novel anticancer hydrazone. Photochem. Photobiol. Sci. 2012, 11, 1868–1879. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Kelly, S.M.; Price, N.C. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef]
- Bannister, W.; Bannister, J. Circular dichroism and protein structure. Int. J. Biochem. 1974, 5, 673–677. [Google Scholar] [CrossRef]
- Alanazi, A.M.; Abdelhameed, A.S.; Bakheit, A.H.; Darwish, I.A. Exploring the interaction forces involved in the binding of the multiple myeloma drug lenalidomide to bovine serum albumin. J. Mol. Liq. 2017, 238, 3–10. [Google Scholar] [CrossRef]
- Greenfield, N.J.; Fasman, G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 1969, 8, 4108–4116. [Google Scholar] [CrossRef]
- Alam, P.; Chaturvedi, S.K.; Siddiqi, M.K.; Rajpoot, R.K.; Ajmal, M.R.; Zaman, M.; Khan, R.H. Vitamin k3 inhibits protein aggregation: Implication in the treatment of amyloid diseases. Sci. Rep. 2016, 6, 26759. [Google Scholar] [CrossRef]
- Grüner, S.; Neeb, M.; Barandun, L.J.; Sielaff, F.; Hohn, C.; Kojima, S.; Steinmetzer, T.; Diederich, F.; Klebe, G. Impact of protein and ligand impurities on ITC-derived protein–ligand thermodynamics. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2843–2850. [Google Scholar] [CrossRef]
- Rehman, M.T.; Shamsi, H.; Khan, A.U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharm. 2014, 11, 1785–1797. [Google Scholar] [CrossRef]
- Watanabe, H.; Tanase, S.; Nakajou, K.; Maruyama, T.; Kragh-Hansen, U.; Otagiri, M. Role of Arg-410 and Tyr-411 in human serum albumin for ligand binding and esterase-like activity. Biochem. J. 2000, 349, 813–819. [Google Scholar] [CrossRef]
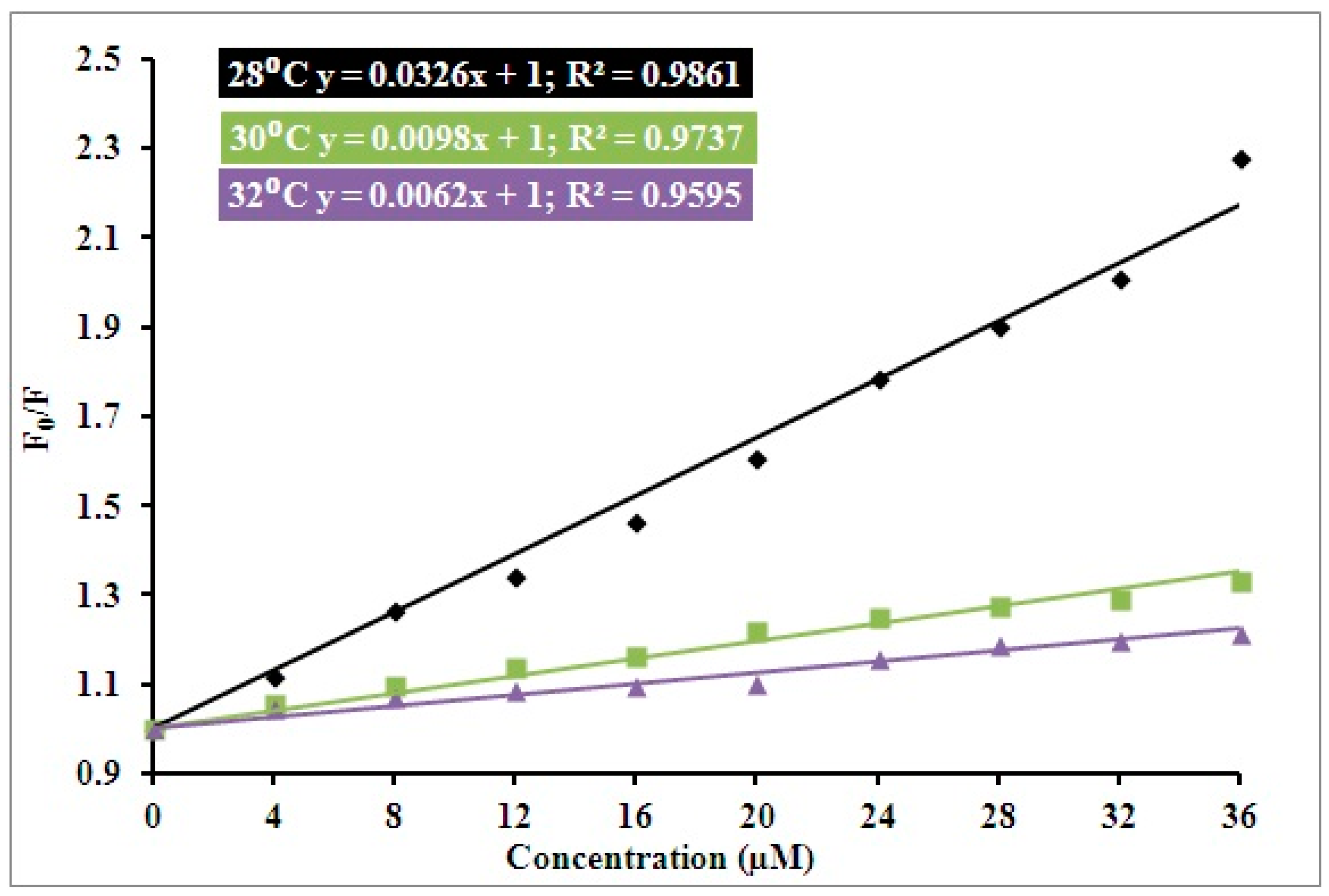
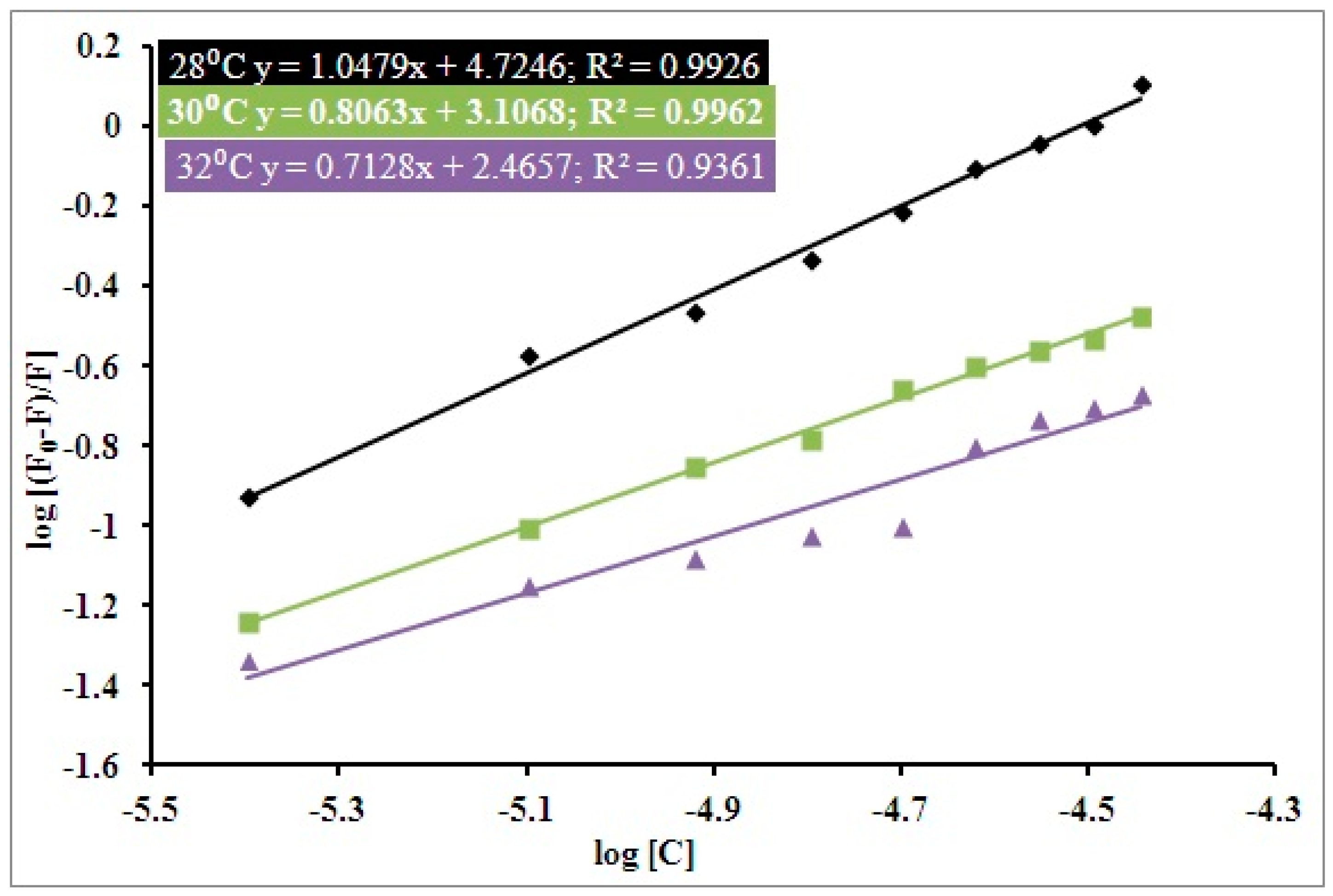
| pH | Temperature (Kelvin) | Ksv (104 M−1) | Kq (1012 M−1 s−1) | R2 |
|---|---|---|---|---|
| 7.4 | 301 | 1.1 | 1.90 | 0.88 |
| 303 | 0.9 | 1.55 | 0.97 | |
| 305 | 0.6 | 1.03 | 0.95 |
| pH | Temperature (K) | K (104 M−1) | N | ∆G0 (kcal mol−1) | ∆S0 (cal mol−1 K−1) | ∆H0 (kcal mol−1) | TΔS0 (kcal mol−1) |
|---|---|---|---|---|---|---|---|
| 7.4 | 301 | 5.3 | 0.99 | −6.24451 | −768.169 | −237.465 | −231.21 |
| 303 | 0.10 | 0.99 | −4.70917 | −232.75 | |||
| 305 | 0.02 | 0.93 | −3.17283 | −234.29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shamsi, A.; Mohammad, T.; Khan, M.S.; Shahwan, M.; Husain, F.M.; Rehman, M.T.; Hassan, M.I.; Ahmad, F.; Islam, A. Unraveling Binding Mechanism of Alzheimer’s Drug Rivastigmine Tartrate with Human Transferrin: Molecular Docking and Multi-Spectroscopic Approach towards Neurodegenerative Diseases. Biomolecules 2019, 9, 495. https://doi.org/10.3390/biom9090495
Shamsi A, Mohammad T, Khan MS, Shahwan M, Husain FM, Rehman MT, Hassan MI, Ahmad F, Islam A. Unraveling Binding Mechanism of Alzheimer’s Drug Rivastigmine Tartrate with Human Transferrin: Molecular Docking and Multi-Spectroscopic Approach towards Neurodegenerative Diseases. Biomolecules. 2019; 9(9):495. https://doi.org/10.3390/biom9090495
Chicago/Turabian StyleShamsi, Anas, Taj Mohammad, Mohd Shahnawaz Khan, Moyad Shahwan, Fohad Mabood Husain, Md. Tabish Rehman, Md. Imtaiyaz Hassan, Faizan Ahmad, and Asimul Islam. 2019. "Unraveling Binding Mechanism of Alzheimer’s Drug Rivastigmine Tartrate with Human Transferrin: Molecular Docking and Multi-Spectroscopic Approach towards Neurodegenerative Diseases" Biomolecules 9, no. 9: 495. https://doi.org/10.3390/biom9090495
APA StyleShamsi, A., Mohammad, T., Khan, M. S., Shahwan, M., Husain, F. M., Rehman, M. T., Hassan, M. I., Ahmad, F., & Islam, A. (2019). Unraveling Binding Mechanism of Alzheimer’s Drug Rivastigmine Tartrate with Human Transferrin: Molecular Docking and Multi-Spectroscopic Approach towards Neurodegenerative Diseases. Biomolecules, 9(9), 495. https://doi.org/10.3390/biom9090495