Identification of miRNAs and Their Target Genes Involved in Cucumber Fruit Expansion Using Small RNA and Degradome Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. sRNA Library Construction and Sequencing
2.3. Bioinformatics Analysis of Sequencing Data
2.4. Differential Expression Analysis of miRNAs
2.5. qRT-PCR Analysis
2.6. Degradome Sequencing
3. Results
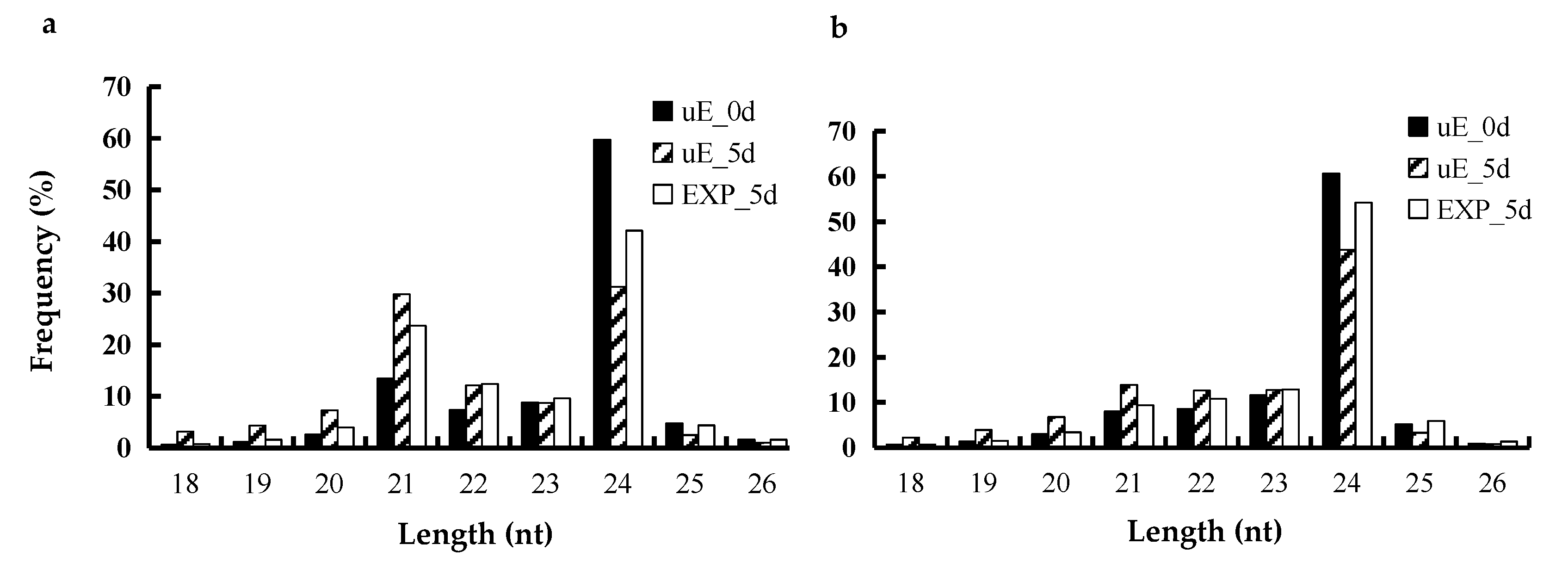
3.1. Overview of miRNAs Sequencing
3.2. Identification of Known and Novel miRNAs
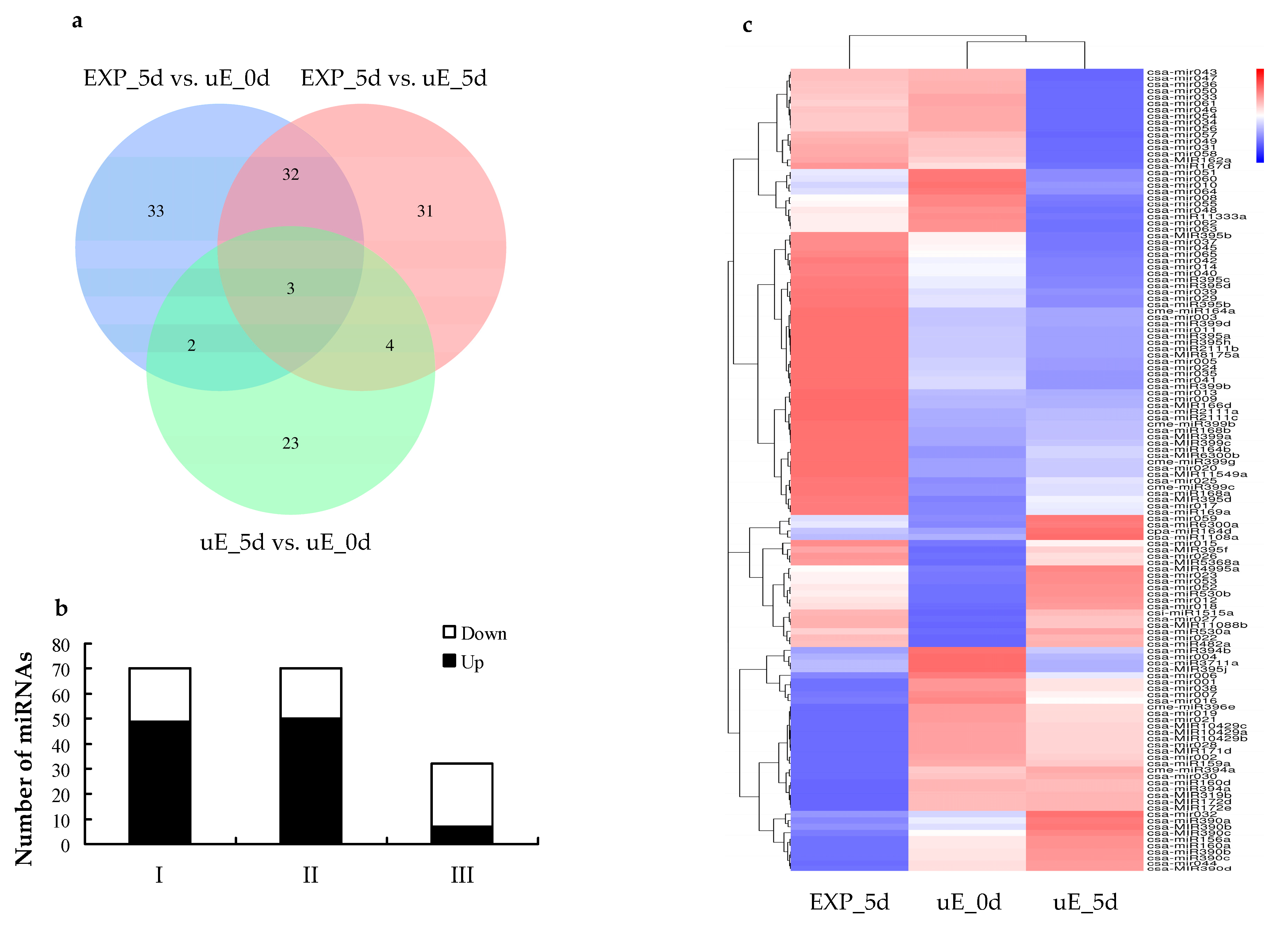
3.3. Differential Expression of miRNAs
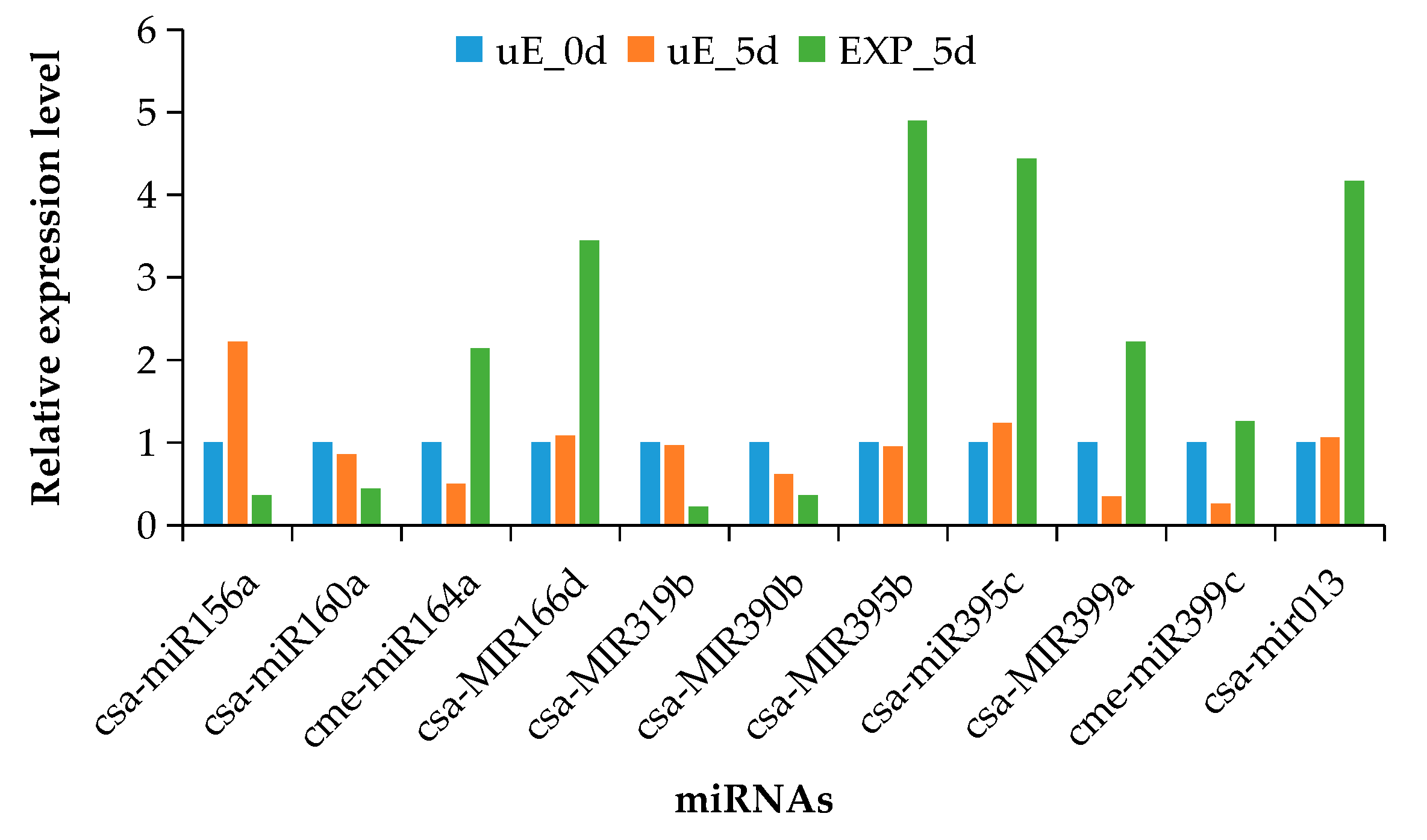
3.4. Validation of miRNA Expression by qRT-PCR
3.5. Identification of Target Genes
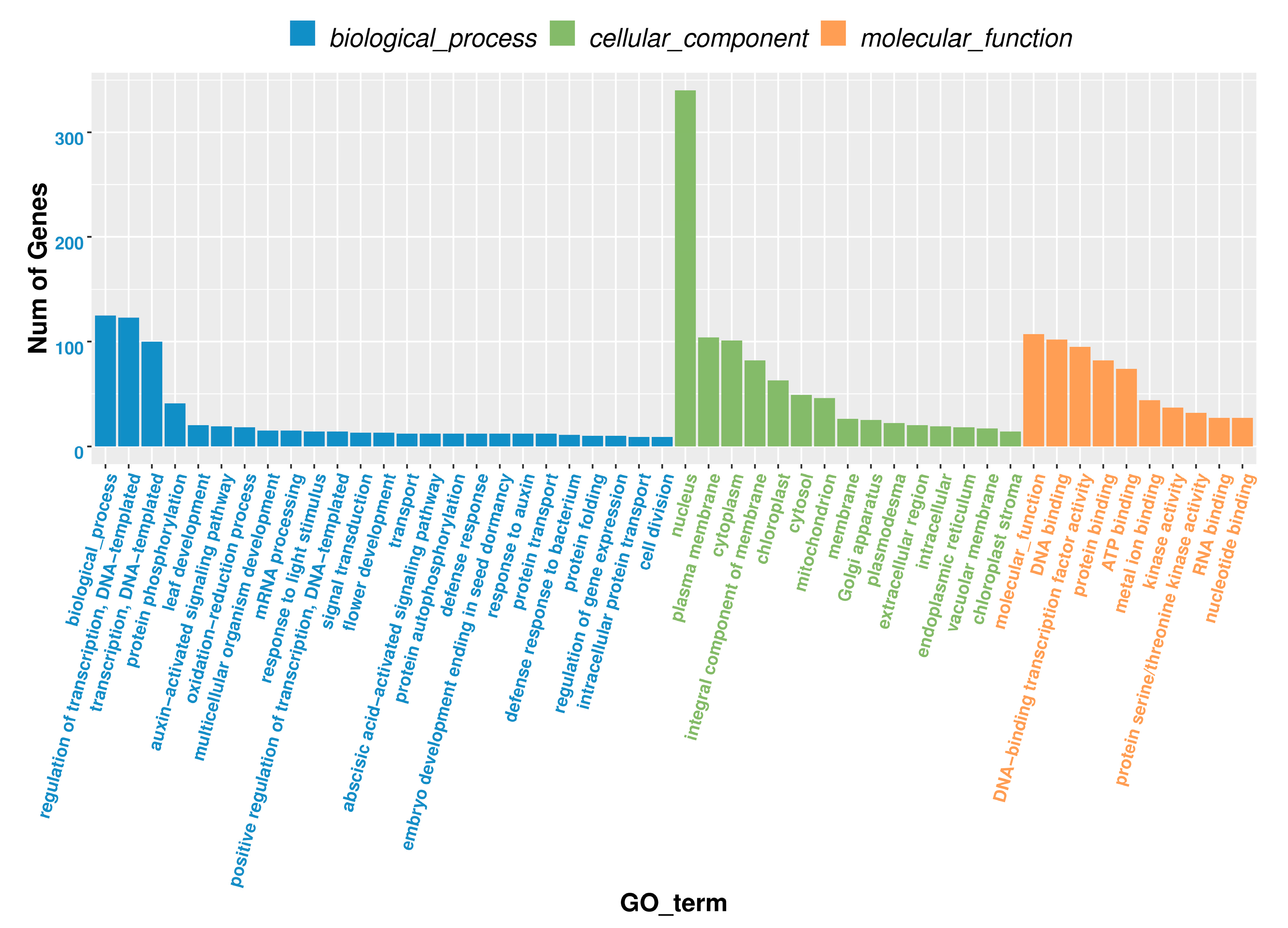
3.6. The Analysis of GO and KEGG
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. 2019. Available online: http://www.fao.org/faostat/ (accessed on 25 August 2019).
- Sjut, V.; Bangerth, F. Effect of pollination or treatment with growth regulators on levels of extractable hormones in tomato ovaries and young fruits. Physiol. Plant 1981, 53, 76–78. [Google Scholar] [CrossRef]
- Kim, I.S.; Okubo, H.; Fujieda, K. Endogenous levels of IAA in relation to parthenocarpy in cucumber (Cucumis sativus L.). Sci. Hortic. 1992, 52, 1–8. [Google Scholar] [CrossRef]
- Yu, J.Q.; Li, Y.; Qian, Y.R.; Zhu, Z.J. Changes of endogenous hormone level in pollinated and N-(2-chloropyridyl)-N0-phenylurea (CPPU)-induced parthenocarpic fruits of Lagenaria leucantha. J. Hortic. Sci. Biotech. 2001, 76, 231–234. [Google Scholar]
- Boonkorkaew, P.; Hikosaka, S.; Sugiyama, N. Effect of pollination on cell division, cell enlargement, and endogenous hormones in fruit development in a gynoecious cucumber. Sci. Hortic. 2008, 116, 1–7. [Google Scholar] [CrossRef]
- Ogawa, Y.; Aoki, S. Prominent promotion on the fruit growth in Cucumis sativus L. by gibberellin A4+7 and benzyladenine. J. Jpn. Soc. Hortic. Sci. 1977, 46, 245–249. [Google Scholar] [CrossRef]
- Takeno, K.; Ise, H.; Minowa, H.; Dounowaki, T. Fruit growth induced by benzyladenine in Cucumis sativus L.: Influence of benzyladenine on cell division, cell enlargement and indole-3-acetic acid content. J. Jpn. Soc. Hortic. Sci. 1992, 60, 915–920. [Google Scholar] [CrossRef]
- Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Asami, T.; Yu, J.Q. A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 2008, 59, 2299–2308. [Google Scholar] [CrossRef]
- Sheng, H.; Qin, Z.W.; Li, W.B.; Zhou, X.Y.; Wu, T.; Xin, M. Genome-wide identification and expression analysis of auxin response factor (ARF) family in cucumber. Sci. Agri. Sin. 2014, 47, 1985–1994. [Google Scholar]
- Sun, Y.D.; Luo, W.R.; Li, Z.X.; Li, X.Z. Endogenous hormones levels and Csexpansin 10 gene expression in the fruit set and early development of cucumber. J. Chem. Soc. Pak. 2017, 39, 59–64. [Google Scholar]
- Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Asami, T.; Yu, J.Q. Spatio-temporal changes in cell division, endoreduplication and expression of cell cycle-related genes in pollinated and plant growth substances-treated ovaries of cucumber. Plant Biol. 2010, 12, 98–107. [Google Scholar] [CrossRef]
- Cui, L.; Li, J.; Zhang, T.; Guo, Q.; Xu, J.; Lou, Q.; Chen, J. Identification and expression analysis of D-type cycling genes in early developing fruit of cucumber (Cucumis sativus L.). Plant Mol. Biol. Rep. 2014, 32, 209–218. [Google Scholar] [CrossRef]
- Cui, L.; Zhang, T.L.; Li, J.; Lou, Q.F.; Chen, J.F. Cloning and expression analysis of Cs-TIR1/AFB2: The fruit development-related genes of cucumber (Cucumis sativus L.). Acta Physiol. Plant 2014, 36, 139–149. [Google Scholar] [CrossRef]
- Yang, X.Y.; Wang, Y.; Jiang, W.J.; Liu, X.L.; Zhang, X.M.; Yu, J.H.; Huang, S.W.; Liu, G.Q. Characterization and expression profiling of cucumber kinesin genes during early fruit development: revealing the roles of kinesins in exponential cell production and enlargement in cucumber fruit. J. Exp. Bot. 2013, 64, 4541–4557. [Google Scholar] [CrossRef] [Green Version]
- Ando, K.; Carr, K.M.; Grumet, R. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genomics 2012, 13, 518–534. [Google Scholar] [CrossRef]
- Tang, G.; Reinhart, B.J.; Bartel, D.P. Abiochemical framework for RNA silencing in plants. Gene. Dev. 2003, 17, 49–63. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Chen, X.A. microRNA as translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Sun, C.; Zhao, Q.; Liu, D.D.; You, C.X.; Hao, Y.J. Ectopic expression of the apple Md-miRNA156h gene regulates flower and fruit development in Arabidopsis. Plant Cell Tiss. Org. 2013, 112, 343–351. [Google Scholar] [CrossRef]
- Tang, F.; Wei, H.R.; Zhao, S.T.; Wang, L.J.; Zheng, H.Q.; Lu, M.Z. Identification of microRNAs involved in regeneration of the secondary vascular system in Populus tomentosa Carr. Front. Plant Sci. 2016, 7, 724–741. [Google Scholar] [CrossRef]
- Chen, Z.H.; Bao, M.L.; Sun, Y.Z.; Yang, Y.J.; Xu, X.H.; Wang, J.H.; Han, N.; Bian, H.W.; Zhu, M.Y. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 2011, 77, 619–629. [Google Scholar] [CrossRef]
- Zhao, Z.; Xue, Y.D.; Yang, H.L.; Li, H.M.; Sun, G.Y.; Zhao, X.F.; Ding, D.; Tang, J.H. Genome-wide identification of miRNAs and their targets involved in the developing internodes under maize ears by responding to hormone signaling. PLoS ONE 2016, 11, e0164026. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, L.F.; Wang, H.; Liu, Z.J.; Zhang, Z.X.; Zheng, Y.L. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Chen, C.J.; Zeng, Z.H.; Liu, Z.R.; Xia, R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018, 63. [Google Scholar] [CrossRef]
- Mohorianu, I.; Schwach, F.; Jing, R.; Lopez-Gomollon, S.; Moxon, S.; Szittya, G.; Sorefan, K.; Moulton, V.; Dalmay, T. Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 2011, 67, 232–246. [Google Scholar] [CrossRef]
- Silva, G.; Silva, E.; Silva, A.M.; Guivin, M.; Ramiro, D.A.; Figueiredo, C.R.; Nogueira, F. microRNA 156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Meyers, B.C. The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 2017, 29, 1232–1247. [Google Scholar] [CrossRef]
- Yao, J.L.; Tomes, S.; Xu, J.; Gleave, A.P. How microRNA172 affects fruit growth in different species is dependent on fruit type. Plant Signal. Behav. 2016, 11, e1156833. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, J.; Cui, L.; Zhang, T.; Wu, Z.; Zhu, P.Y.; Meng, Y.J.; Zhang, K.J.; Yu, X.Q.; Lou, Q.F.; et al. New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biol. 2017, 17, 130–144. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, L.; Wang, H.S.; Wang, G.Z.; Ma, X.L.; Li, M.H.; Wu, H.B.; Fu, Q.S.; Zhang, Y.; Yi, H.P. Genome-wide identification of Hami melon miRNAs with putative roles during fruit development. PLoS ONE 2017, 24, e0180600. [Google Scholar] [CrossRef]
- Ye, X.L.; Song, T.F.; Liu, C.; Feng, H.; Liu, Z.Y. Identification of fruit related microRNAs in cucumber (Cucumis sativus L.) using high-throughput sequencing technology. Hereditas 2014, 151, 220–228. [Google Scholar] [CrossRef]
- Mao, W.H.; Li, Z.Y.; Xia, X.J.; Li, Y.D.; Yu, J.Q. A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS ONE 2012, 7, e33040. [Google Scholar] [CrossRef]
- Wang, L.; Wang, M.B.; Tu, J.X.; Helliwell, C.A.; Waterhouse, P.M.; Dennis, E.S.; Fu, T.D.; Fan, Y.L. Cloning and characterization of microRNAs from Brassica napus. FEBS Lett. 2007, 581, 3848–3856. [Google Scholar] [CrossRef]
- Sunkar, R.; Girke, T.; Jain, P.K.; Zhu, J.K. Cloning and characterization of microRNAs from rice. Plant Cell 2005, 17, 1397–1411. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Li, C.; Zhang, B.H. MicroRNAs in control of plant development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576. [Google Scholar] [CrossRef]
- Mallory, A.C.; Dugas, D.V.; Bartel, D.P.; Bartel, B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef]
- Karlova, R.; van Haarst, J.C.; Maliepaard, C.; van de Geest, H.; Bovy, A.G.; Lammers, M.; Angenent, G.C.; de Maagd, R.A. Identifification of microRNA targets in tomato fruit development using high throughput sequencing and degradome analysis. J. Exp. Bot. 2013, 64, 1863–1878. [Google Scholar] [CrossRef]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baun, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Sakaguchi, J.; Watanabe, Y. miR165/166 and the development of land plants. Dev. Growth Differ. 2012, 54, 93–99. [Google Scholar] [CrossRef]
- Zuo, J.; Zhu, B.; Fu, D.; Zhu, Y.; Ma, Y.; Chi, L.; Ju, Z.; Wang, Y.X.; Zhai, B.Q.; Luo, Y.B. Sculpting the maturation, softening and ethylene pathway: The influences of microRNAs on tomato fruits. BMC Genomics 2012, 13, 7–19. [Google Scholar] [CrossRef]
- Bi, F.; Meng, X.; Ma, C.; Yi, G. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genomics 2015, 16, 776–791. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef]
- Alvarez, J.P.; Pekker, I.; Goldshmidt, A.; Blum, E.; Amsellem, Z.; Eshed, Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 2006, 18, 1134–1151. [Google Scholar] [CrossRef]
- Hunter, C. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 2006, 133, 2973–2981. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Golm, S.P. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.X.; Cui, W.X.; Guan, C.Y.; Mao, W.J.; Zhang, Z.H. Improvement in fruit quality by overexpressing miR399a in woodland strawberry. J. Agric. Food Chem. 2017, 65, 7361–7370. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Primer Sequence (5’–3’) |
|---|---|
| U6-R | GGGGACATCCGATAAAATTGG |
| U6-F | GATTTGTGCGTGTCATCCTT |
| csa-miR156a-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAATGT |
| csa-miR156a-F | CCGGCGTGACAGAAGAGAGT |
| csa-miR160a-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGCAT |
| csa-miR160a-F | CGTGCCTGGCTCCCTGT |
| cme-miR164a-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCACG |
| cme-miR164a-F | ACGGTGGAGAAGCAGGGC |
| csa-MIR166d-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATCTCG |
| csa-MIR166d-F | GCCGAATGTTGTCTGGTGC |
| csa-MIR319b-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGAGCC |
| csa-MIR319b-F | TCGCAGCTGCTGACTCGTT |
| csa-MIR390b-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAACT |
| csa-MIR390b-F | TTCCGGCGCTATCTATCCTG |
| csa-MIR395b-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGATGAA |
| csa-MIR395b-F | TGGCGGAGTTTCCCTGAAT |
| csa-miR395c-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGAGTT |
| csa-miR395c-F | GCGGCTGAAGTGTTTGGG |
| csa-MIR399a-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCTGCC |
| csa-MIR399a-F | GTGGCGGGGCAATTACTCT |
| cme-miR399c-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCGGGC |
| cme-miR399c-F | AGCGGTGCCAAAGGAGATT |
| csa-mir013-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAAA |
| csa-mir013-F | GCGGTGGAGGGTCGAATT |
| Sequence Type | uE_0d | uE_5d | EXP_5d | |||
|---|---|---|---|---|---|---|
| Total sRNA Number | Unique sRNA Number | Total sRNA Number | Unique sRNA Number | Total sRNA Number | Unique sRNA Number | |
| Raw reads | 19,718,088 | 5,363,469 | 20,035,416 | 4,862,572 | 22,463,142 | 4,840,446 |
| 3ADT and length filter | 1,026,683 | 692,365 | 1,664,644 | 1,001,119 | 756,053 | 751,373 |
| Junk reads | 65,726 | 32,144 | 53,301 | 24,553 | 104,331 | 42,517 |
| Rfam | 1,295,187 | 84,578 | 2,077,132 | 92,228 | 1,922,733 | 106,090 |
| mRNA | 633,103 | 188,082 | 738,616 | 273,020 | 695,312 | 191,756 |
| Repeats | 4849 | 1546 | 5029 | 2307 | 3974 | 1505 |
| rRNA | 1,038,125 | 55,560 | 1,746,084 | 61,791 | 1,607,783 | 71,409 |
| tRNA | 121,383 | 17,572 | 90,316 | 14,269 | 142,211 | 21,113 |
| snoRNA | 28,947 | 2787 | 38,706 | 4733 | 46,098 | 3276 |
| snRNA | 35,054 | 3965 | 91,704 | 5469 | 60,823 | 4781 |
| other Rfam RNA | 71,679 | 4694 | 110,322 | 5967 | 65,818 | 5511 |
| valid reads | 16,698,831 | 4,366,701 | 15,521,125 | 3,472,867 | 18,989,300 | 3,749,311 |
| Small RNA | Transcript | Transcript Annotation | log2 (Fold Change) | Degradome Cleavage Site |
|---|---|---|---|---|
| csa-miR160a | XM_011657993.1 | auxin response factor 7-like | −1.30 | 1681 |
| csa-miR160a | XM_011657992.1 | auxin response factor 7-like | −1.30 | 2114 |
| csa-miR160a | XM_004150892.2 | auxin response factor 7-like | −1.14 | 1568 |
| csa-miR160a | XM_004143204.2 | auxin response factor 7-like | −0.87 | 2118 |
| csa-miR160a | NM_001288596.1 | auxin response factor 7 | −0.22 | 1346 |
| csa-miR160a | NM_001281787.1 | auxin response factor 7-like | −1.30 | 1334 |
| cme-miR164a | XM_011661856.1 | NAC domain-containing protein 89-like | 4.02 | 828 |
| cme-miR164a | XM_011656041.1 | exopolygalacturonase-like | - | N |
| cme-miR164a | XM_011653931.1 | NAC domain-containing protein 89-like | −2.10 | 861 |
| cme-miR164a | XM_011653227.1 | NAC domain-containing protein 89-like | −0.85 | 730 |
| cme-miR164a | XM_011652994.1 | NAC domain-containing protein 89-like | - | 803 |
| cme-miR164a | XM_011651827.1 | disease resistance protein RPM1 | - | N |
| cme-miR164a | XM_004150728.2 | NAC domain-containing protein 89-like | −0.85 | 739 |
| cme-miR164a | XM_004146051.2 | NAC domain-containing protein 89-like | - | 960 |
| cme-miR164a | XM_004141527.2 | NAC domain-containing protein 89-like | - | N |
| cme-miR164a | XM_004137540.2 | NAC domain-containing protein 89-like | −0.90 | 970 |
| cme-miR164a | XM_004136650.2 | NAC domain-containing protein 89-like | −2.10 | 864 |
| cme-miR164a | XM_004136327.2 | 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B-like | - | 322 |
| csa-MIR166d | XM_011652521.1 | putative ABC transporter C family member 15 | - | N |
| cme-miR399c | XM_011659907.1 | probable ubiquitin-conjugating enzyme E2 24 | −0.82 | 473 |
| cme-miR399c | XM_004134656.2 | probable ubiquitin-conjugating enzyme E2 24 | −0.82 | 714 |
| csa-mir013 | XM_011661833.1 | calcium-dependent protein kinase 8 | - | N |
| csa-mir013 | XM_011661179.1 | myosin class 11-1 (ISS) | - | N |
| csa-mir013 | XM_011659724.1 | cell division control-like protein | - | N |
| csa-mir013 | XM_011659130.1 | probable L-ascorbate peroxidase 6 | −0.47 | 1272 |
| csa-mir013 | XM_011658857.1 | chitin elicitor receptor kinase 1-like | - | N |
| csa-mir013 | XM_011656847.1 | glutamate decarboxylase 1 | - | 268 |
| csa-mir013 | XM_011652440.1 | mitogen-activated protein kinase homolog D5-like | - | N |
| csa-mir013 | XM_011651425.1 | NADP-dependent malic enzyme | - | N |
| csa-mir013 | XM_011650949.1 | THO complex subunit 3 | - | N |
| csa-mir013 | XM_004150014.2 | THO complex subunit 3 | - | N |
| csa-mir013 | XM_004149751.2 | probable L-ascorbate peroxidase 6 | −0.47 | 1273 |
| csa-mir013 | XM_004148951.2 | omega-hydroxypalmitate O-feruloyl transferase-like | - | N |
| csa-mir013 | XM_004148279.2 | homogentisate phytyltransferase 1 | - | N |
| csa-mir013 | XM_004144564.2 | exopolygalacturonase clone GBGA483 | - | N |
| csa-mir013 | XM_004143155.2 | chitin elicitor receptor kinase 1-like | - | N |
| csa-mir013 | XM_004137239.2 | putative uncharacterized protein At4g01020 | - | N |
| csa-mir013 | XM_004135249.2 | signal recognition particle subunit SRP72-like | - | 1800 |
| csa-mir013 | XM_004133898.2 | ent-kaurenoic acid oxidase 1 | - | N |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Luo, W.; Chang, H.; Li, Z.; Zhou, J.; Li, X.; Zheng, J.; Hao, M. Identification of miRNAs and Their Target Genes Involved in Cucumber Fruit Expansion Using Small RNA and Degradome Sequencing. Biomolecules 2019, 9, 483. https://doi.org/10.3390/biom9090483
Sun Y, Luo W, Chang H, Li Z, Zhou J, Li X, Zheng J, Hao M. Identification of miRNAs and Their Target Genes Involved in Cucumber Fruit Expansion Using Small RNA and Degradome Sequencing. Biomolecules. 2019; 9(9):483. https://doi.org/10.3390/biom9090483
Chicago/Turabian StyleSun, Yongdong, Weirong Luo, Huaicheng Chang, Zhenxia Li, Junguo Zhou, Xinzheng Li, Jinliang Zheng, and Mingxian Hao. 2019. "Identification of miRNAs and Their Target Genes Involved in Cucumber Fruit Expansion Using Small RNA and Degradome Sequencing" Biomolecules 9, no. 9: 483. https://doi.org/10.3390/biom9090483
APA StyleSun, Y., Luo, W., Chang, H., Li, Z., Zhou, J., Li, X., Zheng, J., & Hao, M. (2019). Identification of miRNAs and Their Target Genes Involved in Cucumber Fruit Expansion Using Small RNA and Degradome Sequencing. Biomolecules, 9(9), 483. https://doi.org/10.3390/biom9090483




