Microbial Pyrrolnitrin: Natural Metabolite with Immense Practical Utility
Abstract
:1. Introduction
2. Halometabolites with Potential Functions
3. Pyrrolnitrin (PRN)
3.1. Pyrrolnitrin: Chemical Synthesis
3.2. Microbial Pyrrolnitrin Production and Recovery
3.3. Analytical Characteristics of Pyrrolnitrin
3.4. Biochemistry of Pyrrolnitrin
3.5. Pyrrolnitrin Derivatives
4. Applications of Pyrrolnitrin
4.1. Biological Activity
4.2. Agricultural Applications
4.3. Pharmaceutical Applications
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Challinor, V.L.; Bode, H.B. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 2015, 1354, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Gerth, K.; Steinmetz, H.; Müller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9, 803. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.H.; Stone, M.J.; Hauck, P.R.; Rahman, S.K. Why are secondary metabolites (natural products) biosynthesized? J. Nat. Prod. 1989, 52, 1189–1208. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J. Secondary metabolites and plant/environment interactions: A view through Arabidopsis thaliana tinged glasses. Plant Cell Environ. 2004, 27, 675–684. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.B.; Pelaez, F. Biodiversity, chemical diversity and drug discovery. In Natural Compounds as Drugs Volume I; Birkhäuser: Basel, Switzerland, 2008; pp. 141–174. [Google Scholar]
- Hochberg, M.E. An ecosystem framework for understanding and treating disease. Evol. Med. Public Health 2018, 2018, 270–286. [Google Scholar] [CrossRef]
- Hoddle, M.S.; Warner, K.; Steggall, J.; Jetter, K.M. Classical biological control of invasive legacy crop pests: New technologies offer opportunities to revisit old pest problems in perennial tree crops. Insects 2015, 6, 13–37. [Google Scholar] [CrossRef]
- Gibson, D.M.; Donzelli, B.G.G.; Krasnoff, S.B.; Keyhani, N.O. Discovering the secondary metabolite potential encoded within entomopathogenic fungi. Nat. Prod. Rep. 2018, 2014, 1287–1305. [Google Scholar] [CrossRef]
- Schmidt, C. Living in a microbial world. Nat. Biotechnol. 2017, 35, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Choudoir, M.J.; Pepe-Ranney, C.; Buckley, D.H. Diversification of secondary metabolite biosynthetic gene clusters coincides with lineage divergence in Streptomyces. Antibiotics 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Narcross, L.; Martin, V.J. Engineering plant secondary metabolism in microbial systems. Plant Physiol. 2019, 179, 844–861. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. In History of Modern Biotechnology; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–39. [Google Scholar]
- Gelband, H.; Molly Miller, P.; Pant, S.; Gandra, S.; Levinson, J.; Barter, D.; White, A.; Laxminarayan, R. The state of the world’s antibiotics 2015. Wound Heal. South. Afr. 2015, 8, 30–34. [Google Scholar]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Pee, K.H. Microbial biosynthesis of halometabolites. Arch. Microbiol. 2001, 175, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Gribble, G.W. Biological activity of recently discovered halogenated marine natural products. Mar. Drugs 2015, 13, 4044–4136. [Google Scholar] [CrossRef] [PubMed]
- Field, J.A. Natural production of organohalide compounds in the environment. In Organohalide-Respiring Bacteria; Springer: Berlin/Heidelberg, Germany, 2016; pp. 7–29. [Google Scholar]
- Marumo, S.; Abe, H.; Hattori, H.; Munakata, K. Isolation of a novel auxin, methyl 4-chloroindoleacetate from immature seeds of Pisum sativum. Agric. Biol. Chem. 1968, 32, 117–118. [Google Scholar] [CrossRef]
- Engvild, K.C.; Egsgaard, H.; Larsen, E. Determination of 4-chloroindoleacetic acid methyl ester in Vicieae species by gas chromatography-mass spectrometry. Physiol. Plant. 1981, 53, 79–81. [Google Scholar] [CrossRef]
- Gribble, G.W. The natural production of chlorinated compounds. Environ. Sci. Technol. 1994, 28, 310A–319A. [Google Scholar] [CrossRef]
- Kuo, R.Y.; Chang, F.R.; Chen, C.Y.; Teng, C.M.; Yen, H.F.; Wu, Y.C. Antiplatelet activity of N-methoxycarbonyl aporphines from Rollinia mucosa. Phytochemistry 2001, 57, 421–425. [Google Scholar] [CrossRef]
- Takahashi, Y.; Daitoh, M.; Suzuki, M.; Abe, T.; Masuda, M. Halogenated metabolites from the new Okinawan red alga Laurencia yonaguniensis. J. Nat. Prod. 2002, 65, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Gribble, G.W. Amazing organohalogens: Although best known as synthetic toxicants, thousands of halogen compounds are, in fact, part of our natural enviornment. Am. Sci. 2004, 92, 342–349. [Google Scholar] [CrossRef]
- Hunt, S. Halogenated tyrosine derivatives in invertebrate scleroproteins: Isolation and identification. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1984; Volume 107, pp. 413–438. [Google Scholar] [CrossRef]
- Dème, D.; Fimiani, E.; Pommier, J.; Nunez, J. Free diiodotyrosine effects on protein iodination and thyroid hormone synthesis catalyzed by thyroid peroxidase. Eur. J. Biochem. 1975, 51, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Spande, T.F.; Garraffo, H.M.; Edwards, M.W.; Yeh, H.J.; Pannell, L.; Daly, J.W. Epibatidine: A novel (chloropyridyl) azabicycloheptane with potent analgesic activity from an Ecuadoran poison frog. J. Am. Chem. Soc. 1992, 114, 3475–3478. [Google Scholar] [CrossRef]
- Van Pee, K.H. Biosynthesis of halogenated metabolites by bacteria. Annu. Rev. Microbiol. 1996, 50, 375–399. [Google Scholar] [CrossRef] [PubMed]
- Gribble, G.W. The diversity of naturally produced organohalogens. Chemosphere 2003, 52, 289–297. [Google Scholar] [CrossRef]
- Fitch, R.W.; Spande, T.F.; Garraffo, H.M.; Yeh, H.J.; Daly, J.W. Phantasmidine: An epibatidine congener from the Ecuadorian poison frog Epipedobates anthonyi. J. Nat. Prod. 2010, 73, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, J.; Bartz, Q.R.; Smith, R.M.; Joslyn, D.A. Chloromyeetin, a new antibiotic from a soil actinomycete. Science (Washington) 1947, 417. [Google Scholar] [CrossRef]
- Duggar, B.M. Aureomycin: A product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 1948, 51, 177–181. [Google Scholar] [CrossRef]
- Grove, J.F.; MacMillan, J.; Mulholland, T.P.C.; Rogers, M.T. 762. Griseofulvin. Part IV. Structure. J. Chem. Soc. (Resumed) 1952, 3977–3987. [Google Scholar] [CrossRef]
- Takeda, R. Structure of a new antibiotic, pyoluteorin. J. Am. Chem. Soc. 1958, 80, 4749–4750. [Google Scholar] [CrossRef]
- Oelrichs, P.B.; McEwan, T. Isolation of the toxic principle in Acacia georginae. Nature 1961, 190, 808–809. [Google Scholar] [CrossRef] [PubMed]
- Arima, K.; Imanaka, H.; Kousaka, M.; Fukuta, A.; Tamura, G. Pyrrolnitrin, a new antibiotic substance produced by Pseudomonas. Agric. Biol. Chem. 1964, 28, 575–576. [Google Scholar] [CrossRef]
- Morton, G.O.; Lancaster, J.E.; Van Lear, G.E.; Fulmor, W.; Meyer, W.E. Structure of nucleocidin. III. Revised structure. J. Am. Chem. Soc. 1969, 91, 1535–1537. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.N.; Stone, T.W. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982, 247, 184–187. [Google Scholar] [CrossRef]
- Tunac, J.B.; Underhill, M. 2’-Chloropentostatin: Discovery, fermentation and biological activity. J. Antibiot. 1985, 38, 1344–1349. [Google Scholar] [CrossRef]
- Shiomi, K.; Kakehashi, Y.; Yamanaka, H.; Kikuchi, T. Identification of arsenobetaine and a tetramethylarsonium salt in the clam Meretrix lusoria. Appl. Organomet. Chem. 1987, 1, 177–183. [Google Scholar] [CrossRef]
- Lee, M.D.; Manning, J.K.; Williams, D.R.; Kuck, N.A.; Testa, R.T.; Borders, D.B. Calicheamicins, a novel family of antitumor antibiotics. J. Antibiot. 1989, 42, 1070–1087. [Google Scholar] [CrossRef]
- Zehner, S. Molekulargenetische und biochemische Untersuchungen zur Tryptophan-5-Halogenase aus der Biosynthese von Pyrroindomycin B in Streptomyces rugosporus 2003, Dissertation. Available online: http://www.qucosa.de/fileadmin/data/qucosa/documents/1143/1084445344609-4611.pdf (accessed on 12 July 2019).
- Hanefeld, U.; Floss, H.G.; Laatsch, H. Biosynthesis of the marine antibiotic pentabromopseudilin. Part 1. The benzene ring. J. Org. Chem. 1994, 59, 3604–3608. [Google Scholar] [CrossRef]
- Trimurtulu, G.; Ohtani, I.; Patterson, G.M.; Moore, R.E.; Corbett, T.H.; Valeriote, F.A.; Demchik, L. Total structures of cryptophycins, potent antitumor depsipeptides from the blue-green alga Nostoc sp. strain GSV 224. J. Am. Chem. Soc. 1994, 116, 4729–4737. [Google Scholar] [CrossRef]
- Lang, M.; Spiteller, P.; Hellwig, V.; Steglich, W. Stephanosporin, a “Traceless” Precursor of 2-Chloro-4-nitrophenol in the Gasteromycete stephanospora caroticolor. Angew. Chem. Int. Ed. 2001, 40, 1704–1705. [Google Scholar] [CrossRef]
- Nelson, M.L.; Levy, S.B. The history of the tetracyclines. Ann. N. Y. Acad. Sci. 2011, 1241, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Gkotsi, D.S.; Dhaliwal, J.; McLachlan, M.M.; Mulholand, K.R.; Goss, R.J. Halogenases: Powerful tools for biocatalysis (mechanisms applications and scope). Curr. Opin. Chem. Biol. 2018, 43, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, K.P.; Smith, D.R.; Bogosyan, E.J.; Goss, R.J. Access to high value natural and unnatural products through hyphenating chemical synthesis and biosynthesis. Synthesis 2014, 46, 2122–2132. [Google Scholar]
- Murphy, C.D. New frontiers in biological halogenation. J. Appl. Microbiol. 2003, 94, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Gruschow, S.; Goss, R.J. Scope and potential of halogenases in biosynthetic applications. Curr. Opin. Chem. Biol. 2013, 17, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Lively, D.H.; Gorman, M.; Haney, M.E.; Mabe, J.A. Metabolism of tryptophans by Pseudomonas aureofaciens. I. Biosynthesis of pyrrolnitrin. Antimicrob. Agents Chemother. 1966, 6, 462. [Google Scholar] [PubMed]
- Nakano, H.; Umio, S.; Kariyone, K.; Tanaka, K.; Kishimoto, T.; Noguchi, H.; Ueda, I.; Nakamura, H.; Morimoto, T. Total synthesis of pyrrolnitrin, a new antibiotic. Tetrahedron Lett. 1966, 7, 737–740. [Google Scholar] [CrossRef]
- Hamill, R.; Elander, R.; Mabe, J.; Gorman, M. Metabolism of tryptophans by Pseudomonas aureofaciens. V. Conversion of tryptophan to pyrrolnitrin. Antimicrob. Agents Chemother. 1967, 7, 388–396. [Google Scholar] [PubMed]
- Hamill, R.L.; Elander, R.P.; Mabe, J.A.; Gorman, M. Metabolism of tryptophan by Pseudomonas aureofaciens III. Production of substituted pyrrolnitrins from tryptophan analogues. Appl. Microbiol. 1970, 19, 721–725. [Google Scholar] [PubMed]
- Van Pee, K.H.; Ligon, J.M. Biosynthesis of pyrrolnitrin and other phenylpyrrole derivatives by bacteria. Nat. Prod. Rep. 2000, 17, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Van Pee, K.H.; Salcher, O.; Fischer, P.; Bokel, M.; Lingens, F. The biosynthesis of brominated pyrrolnitrin derivatives by Pseudomonas aureofaciens. J. Antibiot. 1983, 36, 1735–1742. [Google Scholar] [PubMed]
- Burkhead, K.D.; Schisler, D.A.; Slininger, P.J. Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol. 1994, 60, 2031–2039. [Google Scholar] [PubMed]
- Tichenor, M.S.; Boger, D.L. Yatakemycin: Total synthesis, DNA alkylation, and biological properties. Nat. Prod. Rep. 2008, 25, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Gosteli, J. Eine neue Synthese des Antibioticums Pyrrolnitrin. Helv. Chim. Acta 1972, 55, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.D.; Hanthorn, J.J.; Pratt, D.A. Synthesis of pyrrolnitrin and related halogenated phenylpyrroles. Org. Lett. 2009, 11, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.; Guzman, A.; Ruiz, A.; Velarde, E.; Muchowski, J.M. Synthesis of 3-arylpyrroles and 3-pyrrolylacetylenes by palladium-catalyzed coupling reactions. J. Org. Chem. 1992, 57, 1653–1656. [Google Scholar] [CrossRef]
- Elander, R.P.; Mabe, J.A.; Hamill, R.H.; Gorman, M. Metabolism of tryptophans by Pseudomonas aureofaciens VI. Production of pyrrolnitrin by selected Pseudomonas species. Appl. Microbiol. 1968, 16, 753–758. [Google Scholar] [PubMed]
- Cherin, L.; Brandis, A.; Ismailov, Z.; Chet, I. Pyrrolnitrin production by an Enterobacter agglomerans strain with broad-spectrum activity toward fungal and bacterial phytopathogens. Curr. Microbiol. 1996, 32, 208–212. [Google Scholar]
- El-Banna, N.; Winkelmann, G. Pyrrolnitrin from Burkholderia cepacia: Antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 1998, 85, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.P.; McKenna, L.F.; Lakshman, D.K.; Meyer, S.L.F.; Kong, H.; de Souza, J.T.; Lydon, J.; Baker, C.J.; Buyer, J.S.; Chung, S. Suppression of damping-off of cucumber caused by Pythium ultimum with live cells and extracts of Serratia marcescens N4-5. Soil Biol. Biochem. 2007, 39, 2275–2288. [Google Scholar] [CrossRef]
- Parry, R.; Nishino, S.; Spain, J. Naturally-occurring nitro compounds. Nat. Prod. Rep. 2011, 28, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Kamensky, M.; Ovadis, M.; Chet, I.; Chernin, L. Soil-borne strain IC14 of Serratia plymuthica with multiple mechanisms of antifungal activity provides biocontrol of Botrytis cinerea and Sclerotinia sclerotiorum diseases. Soil Biol. Biochem. 2003, 35, 323–331. [Google Scholar] [CrossRef]
- Kalbe, C.; Marten, P.; Berg, G. Strains of the genus Serratia as beneficial rhizobacteria of oilseed rape with antifungal properties. Microbiol. Res. 1996, 151, 433–439. [Google Scholar] [CrossRef]
- Jung, B.K.; Hong, S.J.; Park, G.S.; Kim, M.C.; Shin, J.H. Isolation of Burkholderia cepacia JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl. Biol. Chem. 2018, 61, 173–180. [Google Scholar] [CrossRef]
- Kwak, Y.; Shin, J.H. Complete genome sequence of Burkholderia pyrrocinia 2327T, the first industrial bacterium which produced antifungal antibiotic pyrrolnitrin. J. Biotechnol. 2018, 211, 3–4. [Google Scholar] [CrossRef]
- Ligon, J.M.; Hill, D.S.; Hammer, P.E.; Torkewitz, N.R.; Hofmann, D.; Kempf, H.J.; Pee, K.H.V. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 2000, 56, 688–695. [Google Scholar] [CrossRef]
- Olorunleke, F.E.; Kieu, N.P.; Hofte, M. Recent advances in Pseudomonas biocontrol. In Bacterial-Plant Interactions: Advance Research and Future Trends; Murillo, J., Vinatzer, B.A., Jackson, R.W., Arnold, D.L., Eds.; Caister Academic Press: Cambridgeshire, UK, 2015; pp. 167–198. [Google Scholar]
- Salcher, O.; Lingens, F. Isolation and characterization of a mutant of Pseudomonas aureofaciens ATCC 15926 with an increased capacity for synthesis of pyrrolnitrin. Microbiology 1980, 118, 509–513. [Google Scholar] [CrossRef]
- Elander, R.P.; Mabe, J.A.; Hamill, R.L.; Gorman, M. Biosynthesis of pyrrolnitrins by analogue-resistant mutants of Pseudomonas fluorescens. Folia Microbiol. 1971, 16, 156–165. [Google Scholar] [CrossRef]
- Chang, C.J.; Floss, H.G.; Hook, D.J.; Mabe, J.A.; Manni, P.E.; Martin, L.L.; Schroder, K.; Shieh, T.L. The biosynthesis of the antibiotic pyrrolnitrin by Pseudomonas aureofaciens. J. Antibiot. 1981, 34, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Roitman, J.N.; Mahoney, N.E.; Janisiewicz, W.J.; Benson, M. A new chlorinated phenylpyrrole antibiotic produced by the antifungal bacterium Pseudomonas cepacia. J. Agric. Food Chem. 1990, 38, 538–541. [Google Scholar] [CrossRef]
- Takeda, F.; Janisiewicz, W.J.; Roitman, J.; Mahoney, N.; Abeles, F.B. Pyrrolnitrin delays postharvest fruit rot in strawberries. Hortic. Sci. 1990, 25, 320–322. [Google Scholar]
- Hwang, J.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin production by Burkholderia cepacia and biocontrol of Rhizoctonia stem rot of poinsettia. Biol. Control 2002, 25, 56–63. [Google Scholar] [CrossRef]
- Upadhyay, A.; Srivastava, S. Characterization of a new isolate of Pseudomonas fluorescens strain Psd as a potential biocontrol agent. Lett. Appl. Microbiol. 2008, 47, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Oh, S.A.; Anderson, A.J.; Neiswender, J.; Kim, J.C.; Kim, Y.C. Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Lett. Appl. Microbiol. 2011, 52, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Mujumdar, S.S.; Bashetti, S.P.; Chopade, B.A. Plasmid pUPI126-encoded pyrrolnitrin production by Acinetobacter haemolyticus A19 isolated from the rhizosphere of wheat. World J. Microbiol. Biotechnol. 2014, 30, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Nandi, M.; Selin, C.; Brassinga, A.K.C.; Belmonte, M.F.; Fernando, W.D.; Loewen, P.C.; deKievit, T.R. Pyrrolnitrin and hydrogen cyanide production by Pseudomonas chlororaphis strain PA23 exhibits nematicidal and repellent activity against Caenorhabditis elegans. PLoS ONE 2015, 10, e0123184. [Google Scholar] [CrossRef]
- Purkayastha, G.D.; Mangar, P.; Saha, A.; Saha, D. Evaluation of the biocontrol efficacy of a Serratia marcescens strain indigenous to tea rhizosphere for the management of root rot disease in tea. PLoS ONE 2018, 13, e0191761. [Google Scholar] [CrossRef]
- Martin, L.L.; Chang, C.J.; Floss, H.G.; Mabe, J.A.; Hagaman, E.W.; Wenkert, E. Carbon-13 nuclear magnetic resonance study on the biosynthesis of pyrrolnitrin from tryptophan by Pseudomonas. J. Am. Chem. Soc. 1972, 94, 8942–8944. [Google Scholar] [CrossRef]
- Zhou, P.; Mocek, U.; Siesel, B.; Floss, H.G. Biosynthesis of pyrrolnitrin incorporation of 13C, 15N double-labelled D-and L-tryptophan. J. Basic Microbiol. 1992, 32, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Salcher, O.; Lingens, F.; Fischer, P. Biosynthese von pyrrolnitrin-nachweis von 4-(2′-amino-3′-chlorphenyl) pyrrol-2-carbonsäure. Tetrahedron Lett. 1978, 19, 3097–3100. [Google Scholar] [CrossRef]
- FCtman, J.N.; Mahoney, N.E.; Janisiewicz, W.J. Production and composition of phenylpyrrole metabolites produced by Pseudomonas cepacia. Appl. Microbiol. Biotechnol. 1990, 34, 381–386. [Google Scholar]
- Kirner, S.; Hammer, P.E.; Hill, D.S.; Altmann, A.; Fischer, I.; Weislo, L.J.; Lanahan, M.; van Pee, K.H.; Ligon, J.M. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 1998, 180, 1939–1943. [Google Scholar] [PubMed]
- De Souza, J.T.; Raaijmakers, J.M. Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 2003, 43, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Perneel, M.; Dhondt, L.; De Maeyer, K.; Adiobo, A.; Rabaey, K.; Hofte, M. Phenazines and biosurfactants interact in the biological control of soil-borne diseases caused by Pythium spp. Environ. Microbiol. 2008, 10, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Thongsri, Y.; Aromdee, C.; Yenjai, C.; Kanokmedhakul, S.; Chaiprasert, A.; Hamal, P.; Prariyachatigul, C. Detection of diketopiperazine and pyrrolnitrin, compounds with anti-Pythium insidiosum activity, in a Pseudomonas stutzeri environmental strain. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub 2014, 158, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Gerth, K.; Trowitzsch, W.; Wray, V.; Hofle, G.; Irschik, H.; Reichenbach, H. Pyrrolnitrin from Myxococcus fulvus (myxobacterales). J. Antibiot. 1982, 35, 1101–1103. [Google Scholar] [CrossRef]
- Yoshihisa, H.; Zenji, S.; Fukushi, H.; Katsuhiro, K.; Haruhisa, S.; Takahito, S. Production of antibiotics by Pseudomonas cepacia as an agent for biological control of soilborne plant pathogens. Soil Biol. Biochem. 1989, 21, 723–728. [Google Scholar] [CrossRef]
- Murphy, P.J.; Williams, T.L. Biological inactivation of pyrrolnitrin, Identification and synthesis of pyrrolnitrin metabolites. J. Med. Chem. 1972, 15, 137–139. [Google Scholar] [CrossRef]
- Sultan, M.Z.; Park, K.; Lee, S.Y.; Park, J.K.; Varughese, T.; Moon, S.S. Novel oxidized derivatives of antifungal pyrrolnitrin from the bacterium Burkholderia cepacia K87. J. Antibiot. 2008, 61, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Sako, M.; Kihara, T.; Tanisaki, M.; Maki, Y.; Miyamae, A.; Azuma, T.; Kohda, S.; Masugi, T. Novel photodegradation of the antifungal antibiotic pyrrolnitrin in anhydrous and aqueous aprotic solvents. J. Org. Chem. 2002, 67, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, D.K.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5 B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 1995, 43, 211–216. [Google Scholar] [CrossRef]
- Rosales, A.M.; Thomashow, L.; Cook, R.J.; Mew, T.W. Isolation and identification of antifungal metabolites produced by rice-associated antagonistic Pseudomonas spp. Phytopathology 1995, 85, 1028–1032. [Google Scholar] [CrossRef]
- Mahoney, W.C.; Hermodson, M.A. High-yield cleavage of tryptophanyl peptide bonds by o-iodosobenzoic acid. Biochemistry 1979, 18, 3810–3814. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bimerew, M.; Ma, Y.; Muller, H.; Ovadis, M.; Eberl, L.; Berg, G.; Chernin, L. Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol. Lett. 2007, 270, 299–305. [Google Scholar] [CrossRef] [PubMed]
- van Pee, K.H.; Lingens, F. Detection of a bromoperoxidase in Streptomyces phaeochromogenes. FEBS Lett. 1984, 173, 5–8. [Google Scholar] [CrossRef]
- Hill, D.S.; Stein, J.I.; Torkewitz, N.R.; Morse, A.M.; Howell, C.R.; Pachlatko, J.P.; Becker, J.O.; Ligon, J.M. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 1994, 60, 78–85. [Google Scholar]
- Kirner, S.; Krauss, S.; Sury, G.; Lam, S.T.; Ligon, J.M.; van Pée, K.H. The non-haem chloroperoxidase from Pseudomonas fluorescens and its relationship to pyrrolnitrin biosynthesis. Microbiology 1996, 142, 2129–2135. [Google Scholar] [CrossRef]
- Bonsall, R.F.; Weller, D.M.; Thomashow, L.S. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 1997, 63, 951–955. [Google Scholar]
- Brucker, R.M.; Baylor, C.M.; Walters, R.L.; Lauer, A.; Harris, R.N.; Minbiole, K.P. The identification of 2,4-diacetylphloroglucinol as an antifungal metabolite produced by cutaneous bacteria of the salamander Plethodon cinereus. J. Chem. Ecol. 2008, 34, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; Habibian, R.; Poritsanos, N.; Athukorala, S.N.; Fernando, D.; De Kievit, T.R. Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol. Ecol. 2009, 71, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, Y.; Hashimoto, M.; Hattori, K. The crystal structure of pyrrolnitrin. Tetrahedron Lett. 1968, 9, 209–211. [Google Scholar] [CrossRef]
- Gorman, M.; Lively, D.H. Pyrrolnitrin: A new mode of tryptophan metabolism. In Biosynthesis; Springer: Berlin/Heidelberg, Germany, 1967; pp. 433–438. [Google Scholar]
- Hammer, P.E.; Hill, D.S.; Lam, S.T.; van Pee, K.H.; Ligon, J.M. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 1997, 63, 2147–2154. [Google Scholar] [PubMed]
- Hubbard, B.K.; Walsh, C.T. Vancomycin assembly: nature’s way. Angew. Chem. Int. Ed. 2003, 42, 730–765. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Flecks, S.; Unversucht, S.; Haupt, C.; van Pee, K.H.; Naismith, J.H. Tryptophan 7-halogenase (prnA) structure suggests a mechanism for regioselective chlorination. Science 2005, 309, 2216–2219. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.; Blasiak, L.C.; Koglin, A.; Drennan, C.L.; Walsh, C.T. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry 2007, 46, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- van Pee, K.H.; Zehner, S. Enzymology and molecular genetics of biological halogenation. In Natural Production of Organohalogen Compounds; Springer: Berlin/Heidelberg, Germany, 2003; pp. 171–199. [Google Scholar]
- Lee, J.; Simurdiak, M.; Zhao, H. Reconstitution and characterization of aminopyrrolnitrin oxygenase: A Rieske N-oxygenase that catalyzes unusual arylamine oxidation. J. Biol. Chem. 2005, 280, 36719–36728. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, I.T.; Press, C.M.; Ravel, J.; Kobayashi, D.Y.; Myers, G.S.; Mavrodi, D.V.; DeBoy, R.T.; Seshadri, R.; Ren, Q.; Madupu, R.; et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 2005, 23, 873–878. [Google Scholar] [CrossRef]
- Lee, J.K.; Zhao, H. Identification and characterization of the flavin: NADH reductase (PrnF) involved in a novel two-component arylamine oxygenase. J. Bacteriol. 2007, 189, 8556–8563. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Oscarson, M.; McLellan, R.A. Polymorphic human cytochrome P450 enzymes: An opportunity for individualized drug treatment. Trends Pharmacol. Sci. 1999, 20, 342–349. [Google Scholar] [CrossRef]
- Kendrew, S.G.; Harding, S.E.; Hopwood, D.A.; Marsh, E.N.G. Identification of a flavin: NADH oxidoreductase involved in the biosynthesis of actinorhodin purification and characterization of the recombinant enzyme. J. Biol. Chem. 1995, 270, 17339–17343. [Google Scholar] [CrossRef] [PubMed]
- Thibaut, D.; Ratet, N.; Bisch, D.; Faucher, D.; Debussche, L.; Blanche, F. Purification of the two-enzyme system catalyzing the oxidation of the D-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J. Bacteriol. 1995, 177, 5199–5205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parry, R.J.; Li, W. An NADPH: FAD oxidoreductase from the valanimycin producer, Streptomyces viridifaciens cloning, analysis, and overexpression. J. Biol. Chem. 1997, 272, 23303–23311. [Google Scholar] [CrossRef]
- Duffner, F.M.; Muller, R. A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: Nucleotide sequence and analysis of the genes. FEMS Microbiol. Lett. 1998, 161, 37–45. [Google Scholar] [CrossRef]
- Galan, B.; Diaz, E.; Prieto, M.A.; Garcia, J.L. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: A prototype of a new flavin: NAD(P)H reductase subfamily. J. Bacteriol. 2000, 182, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Xun, L.; Sandvik, E.R. Characterization of 4-hydroxyphenylacetate3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 2000, 66, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, U.; Westphal, A.H.; Muller, R.; van Berkel, W.J. Phenol hydroxylase from Bacillus thermoglucosidasius A7 a two-protein component monooxygenase with a dual role for FAD. J. Biol. Chem. 2003, 278, 47545–47553. [Google Scholar] [CrossRef] [PubMed]
- Umio, S.; Kariyone, K.; Tanaka, K.; Kishimoto, T.; Nakamura, H.; Nishida, M. Structure-activity studies of pyrrolnitrin analogues. Chem. Pharm. Bull. 1970, 18, 1414–1425. [Google Scholar] [CrossRef]
- Warden, J.T.; Edwards, D.L. Electron spin resonance investigations of mitochondrial electron transport in Neurospora crassa: Characterization of paramagnetic intermediates in a standard strain. Eur. J. Biochem. 1976, 71, 411–418. [Google Scholar] [CrossRef]
- Di Santo, R.; Costi, R.; Artico, M.; Massa, S.; Lampis, G.; Deidda, D.; Pompei, R. Pyrrolnitrin and related pyrroles endowed with antibacterial activities against Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 1998, 8, 2931–2936. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Hori, Y.; Abe, Y.; Nakajima, H.; Takase, S.; Fujita, T.; Goto, T.; Okuhara, M.; Kohsaka, M. WB2838 [3-chloro-4-(2-amino-3-chlorophenyl)-pyrrole]: Non-steroidal androgen-receptor antagonist produced by a Pseudomonas. J. Antibiot. 1993, 46, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Fernando, W.D.; Nakkeeran, S.; de Kievit, T.; Poritsanos, N.; Zhang, Y.; Paulit, T.C.; Li, Z.; Ramarathnam, R. Multiple mechanisms of biocontrol by Pseudomonas chlororaphis PA23 affect stem rot of canola caused by Sclerotinia sclerotiorum. In Proceedings of the 12th International Rapeseed Congress, Wuhan, China, 26–30 March 2007. [Google Scholar]
- Lambowitz, A.M.; Slayman, C.W.; Slayman, C.L.; Bonner, W.D. The electron transport components of wild type and poky strains of Neurospora crassa. J. Biol. Chem. 1972, 247, 1536–1545. [Google Scholar] [PubMed]
- Liu, X.; Yu, X.; Yang, Y.; Heeb, S.; Gao, S.; Chan, K.G.; Camara, M.; Gao, K. Functional identification of the prnABCD operon and its regulation in Serratia plymuthica. Appl. Microbiol. Biotechnol. 2018, 102, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef] [PubMed]
- Gokdee, R.; Matthews, T.R. Evaluation of the in vitro and in vivo antifungal activity of pyrrolnitrin. Eval. In Vitro In Vivo Antifung. Act. Pyrrolnitrin 1968, 378–387. [Google Scholar]
- Gordee, R.S.; Matthews, T.R. Systemic antifungal activity of pyrrolnitrin. Appl. Environ. Microbiol. 1969, 17, 690–694. [Google Scholar]
- Nose, M.; Arima, K. On the mode of action of a new antifungal antibiotic, pyrrolnitrin. J. Antibiot. 1969, 22, 135–143. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Gottlieb, D. Mechanism of action of the antifungal antibiotic pyrrolnitrin. J. Bacteriol. 1969, 100, 318. [Google Scholar]
- Howell, C.R.; Stipanovic, R.D. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 1980, 70, 712–715. [Google Scholar] [CrossRef]
- Lambert, B.; Leyns, F.; van Rooyen, L.; Gossele, F.; Papon, Y.; Swings, J. Rhizobacteria of maize and their antifungal activities. Appl. Environ. Microbiol. 1987, 53, 1866–1871. [Google Scholar] [PubMed]
- Pfender, W.F.; Kraus, J.; Loper, J.E. A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici-repentis in wheat straw. Phytopathology 1993, 83, 1223–1228. [Google Scholar] [CrossRef]
- Watts, R.; Dahiya, J.; Chaudhary, K.; Tauro, P. Isolation and characterization of a new antifungal metabolite of Trichoderma reesei. Plant Soil 1988, 107, 81–84. [Google Scholar] [CrossRef]
- Huang, R.; Feng, Z.; Chi, X.; Sun, X.; Lu, Y.; Zhang, B.; Lu, R.; Luo, W.; Wang, Y.; Miao, J.; et al. Pyrrolnitrin is more essential than phenazines for Pseudomonas chlororaphis G05 in its suppression of Fusarium graminearum. Microbiol. Res. 2018, 215, 55–64. [Google Scholar] [CrossRef]
- Wolf, C.R.; Westwood, A.L.; Sleigh, R. Pyrrolnitrin derivatives. Cxr Biosci Ltd., Dundee Technopole James Lindsay Place Dundee DD1 5JJ, GB PCT/GB2012/050937. 2012. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2012146936&recNum=22&docAn=GB2012050937&queryString=((num& (accessed on 12 July 2019).
- Nisr, R.B.; Russell, M.A.; Chrachri, A.; Moody, A.J.; Gilpin, M.L. Effects of the microbial secondary metabolites pyrrolnitrin, phenazine and patulin on INS-1 rat pancreatic β-cells. FEMS Immunol. Med. Microbiol. 2011, 63, 17–227. [Google Scholar] [CrossRef] [PubMed]
- Janisiewicz, W.; Yourman, L.; Roitman, J.; Mahoney, N. Postharvest control of blue mold and gray mold of apples and pears by dip treatment with pyrrolnitrin, a metabolite of Pseudomonas cepacia. Plant Dis. 1991, 75, 490–494. [Google Scholar] [CrossRef]
- Nyfeler, R.; Ackermann, P. Phenylpyrroles, a new class of agricultural fungicides related to the natural antibiotic pyrrolnitrin. In Synthesis and Chemistry of Agrochemicals III: ACS Symposium Series; Americal Chemical Society: Washington, DC, USA, 1992; pp. 395–404. [Google Scholar]
- Kilani, J.; Fillinger, S. Phenylpyrroles: 30 years, two molecules and (nearly) no resistance. Front. Microbiol. 2016, 7, 2014. [Google Scholar] [CrossRef]
- Nishida, M.; Matsubara, T.; Watanabe, N. Pyrrolnitrin, a new antifungal antibiotic, Microbiological and toxicological observations. J. Antibiot. 1965, 18, 211–219. [Google Scholar]
- Tawara, S.; Matsumoto, S.; Hirose, T.; Matsumoto, Y.; Nakamoto, S.; Mitsuno, N.; Kamimura, T.; Yamaguchi, H. In vitro antifungal synergism between pyrrolnitrin and clotrimazole. Jpn. J. Med. Mycol. 1989, 30, 202–210. [Google Scholar] [CrossRef]
- Umio, S.; Kamimura, T.; Kamishita, T.; Mine, Y. Antifungal composition employing pyrrolnitrin in combination with an imodazole compound. US patent US4636520A, 13 January 1987. [Google Scholar]
- Nyfeler, R.; Ehrenfraund, J. Difluorobenzodioxyl cyanopyrrole microbicidal compositions. U.S. Patent 4925840A, 15 May 1990. [Google Scholar]
- Jespers, A.B.; Davidse, L.C.; De Waard, M.A. Biochemical effects of the phenylpyrrole fungicide fenpiclonil in Fusarium sulphureum (Schlecht). Pestic. Biochem. Physiol. 1993, 45, 116–129. [Google Scholar] [CrossRef]
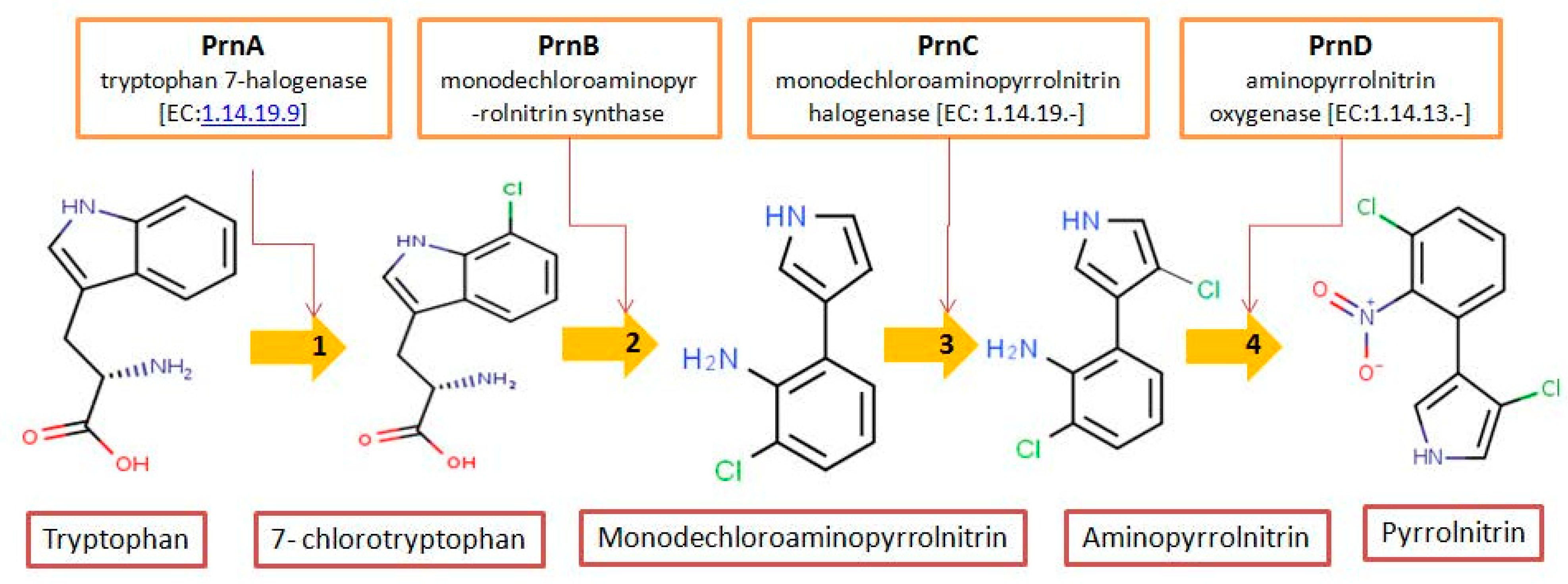

| Organohalogens | Bioactivity | Halogen Type and Number | Source; Habitat | Reference(s) |
|---|---|---|---|---|
| Plants | ||||
| 4-Chloroindole Ester | Plant growth promoting hormone | Cl (01) | Pisum Sativum (Lentil, Sweet Pea, Sea Pea, Vetch); Soil | [21,22] |
| 3-Chloroindole acetate | Plant hormone | Cl (01) | Ptychodero Povo Loysanica; Marine acorn worm | [23] |
| Romucosine B | Plant alkaloids | Cl (01) | Rollinia mucosa; Tropical south America | [24] |
| Neoirietetrao | Diterpene | Br (01) | Laurencia yonaguniensis; Yonaguni island, Japan | [25] |
| Bromomethane | Fumigant; pesticides | Br (01) | Cabbage, Broccoli, Turnips, Rapeseeds (Family: Brassicaceae); Soil | [26] |
| 2-Chloro-4-Nitrophenol | Fungicide | Cl (01) | Stephanospora Caroticolor; Soil | [26] |
| Animals | ||||
| Tyrosine derivative | Improving adhesion between protein fiber, sheets | Cl (01-03) | Marine Sponges, Sea fans, Gorgonians; Sea water | [27] |
| Diiodotyrosine | Precursor in production of thyroid hormone | I (02) | Gorgonia Cavolii, Sea Fan; Western Atlantic Ocean | [28] |
| Ecuadoran | Analgesic activity | Cl (01) | Epipedobotes; Eastern Atlantic Ocean | [29] |
| Tyrian Purple Dye | Dye | Br (02) | Murex Brandaris; Sea snail | [30] |
| Drosophilin A | Antibiotic | Cl (04) | Drosophila Substrata; Ligninolytic Basidiomycetes; overripe or rotting fruit | [31] |
| 2,6 Dichlorophenol | Sex pheromone; growth hormone | Cl (02) | Female; Penicillium Mold; Decaying material | [26] |
| 2,4 Dichlorophenol | Broad spectrum herbicides | Cl (02) | Penicillium Spp.; Agricultural inoculant | [26] |
| Epibatidine | Pain killer | Cl (01) | Epipedobates Anthonyi (Frog); Central; Southern cuador | [32] |
| Microorganisms | ||||
| Chloramphenicol | Antibiotic | Cl (02) | Streptomyces venezuelae; Soil, decaying vegetation | [33] |
| Chlortetracycline | Antibiotic | Cl (01) | Streptomyces aurefaciens; Agricultural soil | [34] |
| Grisiofulvin | Antifungal drug | Cl (01) | Penicillium grisiofulvum; Soil | [35] |
| Pyoluteorin | Antibiotic | Cl (02) | Pseudomonas aeruginosa; Rhizospheric soil | [36] |
| Fluoroacetic Acid | Pesticide | F (01) | Streptomyces cattleya; Soil | [37] |
| Pyrrolnitrin | Antifungal antibiotic | Cl (02) | Burkholderia pyrrocinia, P. fluorescence, Serratia plymuthica; Rhizospheric soil | [38] |
| Nucleocidin | Nucleoside antibiotic | F (01) | Streptomyces calvus; Soil | [39] |
| Vancomycin | Antibiotic | Cl (02) | Amycolatopsis orientalis; Soil | [40] |
| 2′Chloropentostatin | Nucleoside antibiotic | Cl (01) | Actinomadura sp.; Soil | [41] |
| Napyradiomycin | Antibiotic | Cl (02) | Chainia rubra; Soil | [42] |
| Calicheamicin Β1 | Cytotoxin | Br (01) | Micromonospora echinospora; Rhizospheric soil | [43] |
| Pyrroindomycine B | Antibiotic | Cl (01) | Streptomyces rugosporus; Soil | [44] |
| Pentabromopseudilin | Marine antibiotic | Br (05) | Pseudomonas bromoutilis; Coastal area | [45] |
| Cryptophycin A | Anticancer | Cl (01) | Cyanobacterium; Terrestrial, aquatic habitat | [46] |
| 2-Chloro-4-Nitrophenol | Fungicide | Cl (01) | Stephanospora caroticolor; Rotting wood or plant debris | [47] |
| 3,5 Dichloro-Hexanophenone | Inhibit fruiting body formation | Cl (02) | Dictyostelium discoideum; Decaying peach | [31] |
| Rebeccamycin | Weak Topoisomerase I Inhibitor, antitumor | Cl (02) | Streptomyces sp.; Rhizosphere, agricultural soil | [26] |
| Chlortetracycline | Antibiotic | Cl (01) | Streptomyces aureofaciens; Sanborn field | [48] |
| IUPAC Name | Common Name | Structure | Extinction Coefficient ƛmax MeOH (log ε) | Molecular Formula | Molar Mass/ Molecular Weight | |
|---|---|---|---|---|---|---|
| 3-(2-amino-3-chlorophenyl)-pyrrole | Mono-chloro-amino-pyrrolnitrin (MCA) | 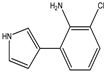 | - | 303 (3.52) | C10H9ClN2 | Exact Mass: 192.05 Mol. Wt.: 192.64 |
| 3-chloro-4(2-amino-3-chlorophenyl)-pyrrole | Di-chloro-amino (DCA) (amino-pyrrolnitrin) | 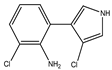 | 212 (4.46) | 302 (3.57) | C10H8Cl2N2 | Exact Mass: 226.01 Mol. Wt.: 227.09 |
| 2, 3 dichloro-4-(2-amino-3-chlorophenyl)-pyrrole | Tri-chloro-amino (TCA) | 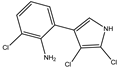 | 212 (4.54) | 302 (3.61) | C10H7Cl3N2 | Exact Mass: 259.97 Mol. Wt.: 261.53 |
| 3-chloro-4-(3-chloro-2nitro-phenyl)-1H pyrrole | Pyrrolnitrin (PRN) | 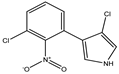 | 212 (4.39) | 252 (3.83) | C10H6Cl2N2O2 | Exact Mass: 255.98 Mol. Wt.: 257.07 |
| 2, 3 dichloro-4-(2-nitro-3-chlorophenyl) pyrrole | 2-chloro-pyrrolnitrin (2-CPRN) |  | 212 (4.47) | 240 280 | C10H5Cl3N2O2 | Exact Mass: 289.94 Mol. Wt.: 291.52 |
| 2-(2-Heptenyl)-3-methyl-4(1H) quinolone | - | 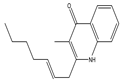 | - | - | C17H21NO | Exact Mass: 255.16 Mol. Wt.: 255.35 |
| 2,3-dichloro-4-(2-nitrophenyl) pyrrole | Iso-pyrrolnitrin | 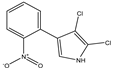 | - | - | C10H6Cl2N2O2 | Exact Mass: 255.98 Mol. Wt.: 257.07 |
| 3-chloro-4-(2-nitro-3-chloro-6-hydroxyphenyl) pyrrole | Oxy-pyrrolnitrin | 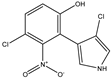 | - | - | C10H6Cl2N2O3 | Exact Mass: 271.98 Mol. Wt.: 273.07 |
| Sr. no. | Producer | Habitat | Medium | Physical Condition | Incubation Period (Days) | Concentration | Significance | Reference |
|---|---|---|---|---|---|---|---|---|
| 1. | Pseudomonas pyrrocinia | - | Bouillon Medium | - | - | ND | Antibiotic, antifungal nature | [26,38] |
| 2. | P. aureofaciens, P. fluorescens, P. multivorans | - | CMM, Synthetic C, E | 27 °C, shaker | 7 | 0.32–126 (µg mL−1) | PRN widespread in groups of Pseudomonas | [64] |
| 3. | P. aureofaciens | - | CMM | 27 °C, shaker | 5 | 9.5 to 50 (µg mL−1) | Production of substituted PRN from Tryptophan analogs | [56] |
| 4. | P. aureofaciens | - | CMM | 30 °C, shaker | 5 | 18.35–19.9 (µm) | Possible pathway discussed | [77] |
| 5. | Pseudomonas cepacia B37w (NRRL B-14858) | Rhizosphere | Sabouraud Maltose Broth | - | 6 | 2.133 (mg L−1) | Efficacy against F. Sambucinum incited potato dry rot disease | [59] |
| 6. | Pseudomonas cepacia LT4-12- W | Apple leaves | Mineral Salt, Nutrient Broth, Kings medium B | 27 °C, 200 rpm | 7 | 1) MS: 51.50 (mg L−1) 2) NB: 7.20 (mg L−1) 3) KMB: 5.50 (mg L−1) | Production of phenylpyrrole metabolites with respect to time | [78] |
| 7. | B. cepacian | - | Mineral Salt | 27 °C, shaker | 7 | ND | Delays postharvest fruit rot in strawberries | [79] |
| 8. | Enterobacter agglomerans IC1270 | Grapes rhizosphere | Potato Dextrose Agar | Incubated on agar plate | 5 | ND | Possible role of a combination of Chitinases and pyrrolnitrin in antagonism | [65] |
| 9. | B. cepacia NB-1 | Ponds in botanical garden of Tubingen, Germany | Minimal medium | 27 °C, aeration rate 0·5 vvm, stirrer speed 150 rev min−1, pH −7.0 | 5 | 0.54 (mg L−1) | PRN block ETS Neurospora crassa 74 A; inhibition of Streptomycine spp. | [66] |
| 10. | Burkholderia cepacia 5.5B (ATCC 55344) Wild Type | Soil sample, North Carolina | Nutrient broth, Mineral salt | 25 °C, at 200 rpm, pH 5.8 | 5 | NB: 35.59; MS: 28.54 (mg 1012 cfu) | Biocontrol of Rhizoctonia stem rot of poinsettia | [80] |
| 11. | Pseudomonas fluorescens psd | Roots of Vigna mungo | Standard succinate medium (SSM) | - | - | ND | Biocontrol property of plants protected from strain | [81] |
| 12. | Pseudomonas chlororaphis O6 | - | Nutrient broth, Mung bean medium | 28 °C 200 rpm | - | 1.7 (µg mL−1) | Regulation by glucose of PRN production influenced biocontrol of tomato leaf blight | [82] |
| 13. | Acinetobacter haemolyticus A19 | Wheat rhizosphere | Luria broth | - | 2 | 15 (mg L−1) | Plasmid-mediated pyrrolnitrin production by A. Haemolyticus A19 | [83] |
| 14. | Pseudomonas chlororaphis strain PA23 | - | M9 medium + 1 mm MgSO4 + 0.2% glucose | - | 5 | ND | Nematicidal and repellent activity against Caenorhabditis elegans | [84] |
| 15. | Serratia marcescens ETR17 | Tea rhizosphere | Semi-solid pigment producing media | 30 °C | 8 | ND | Effective reduction of root-rot disease tea plant on talc-based formulations; Plant growth promoting activity | [85] |
| Matrix | Column | Organic Phase | Detection | Reference |
|---|---|---|---|---|
| Silica gel G | 35 cm × 1.5 cm | Chloroform: methanol (9:1) | - | [83] |
| Silica gel (40 μm) | 35.6 cm × 1.75 cm | Benzene: hexane (2:1); Benzene; Benzene: acetone (1:1); Acetone; methanol | TLC - bioautography | [59] |
| Silica gel (60 μm) | - | Chloroform: hexane (1:1, 1.5:1, 2:1, 5:1) (v/v); chloroform; chloroform-acetone (5:1, 1:1) (v/v); acetone | Bioassay with R. solani | [65] |
| Sephadex LH-20 | - | Methanol | pHPLC | [66] |
| Silica gel 60 (0.015–0.040 mm; Merck) | - | Dichloromethane then methanol | TLC | [93] |
| Silica gel (H60) | - | Dichloromethane | Bioautography | [94] |
| Silica gel | (20 × 170 mm, Wakogel C-200) | Benzene, 10% ethyl acetatobenzene, 20% ethyl acetate benzene and finally ethyl acetate | TLC | [95] |
| Silicic acid | (240 × 22 mm) | Diethyl ether and methanol | - | [96] |
| - | RP C-18 flash | Water and methanol | TLC | [97] |
| - | RP C-18 (MPLC) | 50% to 100% aq methanol | HPLC | [97] |
| Silica gel (60 μm) | - | Toluene | - | [98] |
| Column | Flow Rate (mL/min−1) | Solvent System | Detector | Retention Time (min) | References |
|---|---|---|---|---|---|
| RP 18 | 2 | Methanol: water (70:30; v/v) | - | - | [103] |
| 50 mm × 4.6 mm I.D. guard | 1.0 | Acetonitrile: methanol: water (1:1:1) | UV (254 nm) | 10 | [78] |
| Rainin Dynamax C18 (21.4 × 250 mm) | - | Acetonitrile: water (3:2; v/v) fractions collected at 9.5 to 12.5 min and re-chromatographed on a silica column eluted with chloroform: hexane (1:1; v/v) | - | 13.5 | [79] |
| C-18 column, 5 µm | - | Isocratic acetonitrile: methanol: water (1:1:1) | - | - | [59] |
| Hypersil octyldecyl saline (2.1 mm diameter by 10 cm) | - | Water: methanol from 0%: 100 % and gradually changing up to 100%: 0% | - | between 10-15 | [104] |
| Reverse phase 18 | 0.7 | 0 min 50% methanol in water 15 min 100 % methanol in water 17 min 100% methanol in water 20 min 50% methanol in water 25 min 50% methanol in water | UV (252 nm) | 15.4 | [65] |
| C-18 reverse phase (125 × 4.6 mm) | - | Methanol: water (70:30; v/v) | UV (252 nm) | - | [105] |
| - | 1.0 | 2-min initialization at 10% ACN: 0.1% TFA; 20-min linear gradient to 100% ACN: 0.1% TFA | 990-photodiode array detector | - | [91,106] |
| Nucleosil C-18 | Acetonitrile: water (20 to 100%) | - | 27.5 | [66] | |
| RP C-18 column | 1.0 | Isocratically 45% water: 30% acetonitrile: 25% methanol | - | - | [102] |
| C-18 RP column | 10% acetonitrile: water (v/v) (both acidified with 0.1% amino acid) run for 2min linear gradient 100% acetonitrile (acidified with 0.1% amino acid) | - | 18 | [107] | |
| - | - | 30 ~ 60% aq acetonitrile (for 70 min) | - | 68.9 | [97] |
| Gemini C18 column (100 × 4.6 mm; 5mm particle diameter) | 1.0 | Isocratically 45% acetonitrile: 35% water: 20% methanol | Dionex AD20 (Dionex,Sunnyvale, CA) (225 nm) | - | [84,108] |
| Cosmosil C18 | 0.7 | 18 min linear gradient from 50 to 100% methanol and 0.1% trifluoracetic acid in methanol | - | - | [82] |
| Sr. No. | Name of Test Microorganism | PRN (µg mL−1) | Reference |
|---|---|---|---|
| Bacteria | |||
| 1. | Staphylococcus aureus 209-P | 6.2 | [38] |
| 2. | Escherichia coli | 250 | [38] |
| 3. | M. tuberculosis CIP 103471 | 4.0 | [129] |
| 4. | M. avium CIP 103317 | 8.0 | [129] |
| 5. | M. smegmatis CIP 103599 | 16.0 | [129] |
| 6. | M. gordonae CIP 6427 | >16.0 | [129] |
| 7. | M. marinum CIP 6423 | >16.0 | [129] |
| Yeast | |||
| 8. | Candida albicans | 1.0 | [38] |
| 9. | Saccharomyces cerevisiae | 10.0 | [38] |
| 10. | Cryptococcus neoformans | < 0.78 | [136] |
| 11. | Candida albicans | 12.5 | [137] |
| 12. | Cryptococcus neoformans | 0.78 | [137] |
| 13. | Candida utilis | 10.0 | [138] |
| Fungi | |||
| 14. | Trichophyton asteroids | 0.05 | [38] |
| 15. | Sporotrichum schenckii | < 0.78 | [136] |
| 16. | Penicillium atrovenetwn | 10.0 | [139] |
| 17. | P. oxalicwn | 10.0 | [139] |
| 18. | Sporotrichum schenckii | 3.12 | [137] |
| 19. | Blastomyces dermatitidis | < 0.78 | [137] |
| 20. | Histoplasma capsulatu | 0.156 | [137] |
| 21. | Sclerotinia sclerotiorum | 0.01 | [59] |
| 22. | Rhizoctonia solani | 50 (µg/disc) | [97] |
| Nematode | |||
| 23. | Caenorhabditis elegans | 0.1 | [84] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pawar, S.; Chaudhari, A.; Prabha, R.; Shukla, R.; Singh, D.P. Microbial Pyrrolnitrin: Natural Metabolite with Immense Practical Utility. Biomolecules 2019, 9, 443. https://doi.org/10.3390/biom9090443
Pawar S, Chaudhari A, Prabha R, Shukla R, Singh DP. Microbial Pyrrolnitrin: Natural Metabolite with Immense Practical Utility. Biomolecules. 2019; 9(9):443. https://doi.org/10.3390/biom9090443
Chicago/Turabian StylePawar, Shraddha, Ambalal Chaudhari, Ratna Prabha, Renu Shukla, and Dhananjaya P. Singh. 2019. "Microbial Pyrrolnitrin: Natural Metabolite with Immense Practical Utility" Biomolecules 9, no. 9: 443. https://doi.org/10.3390/biom9090443
APA StylePawar, S., Chaudhari, A., Prabha, R., Shukla, R., & Singh, D. P. (2019). Microbial Pyrrolnitrin: Natural Metabolite with Immense Practical Utility. Biomolecules, 9(9), 443. https://doi.org/10.3390/biom9090443





