The Common Partner of Several Methyltransferases TRMT112 Regulates the Expression of N6AMT1 Isoforms in Mammalian Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cell Culture and Transfections
2.3. RNA Interference
2.4. Treatment with MG132
2.5. Immunoprecipitation and Immunoblotting
2.6. Protein Alignment and Interaction Modeling on Protein Structures
3. Results
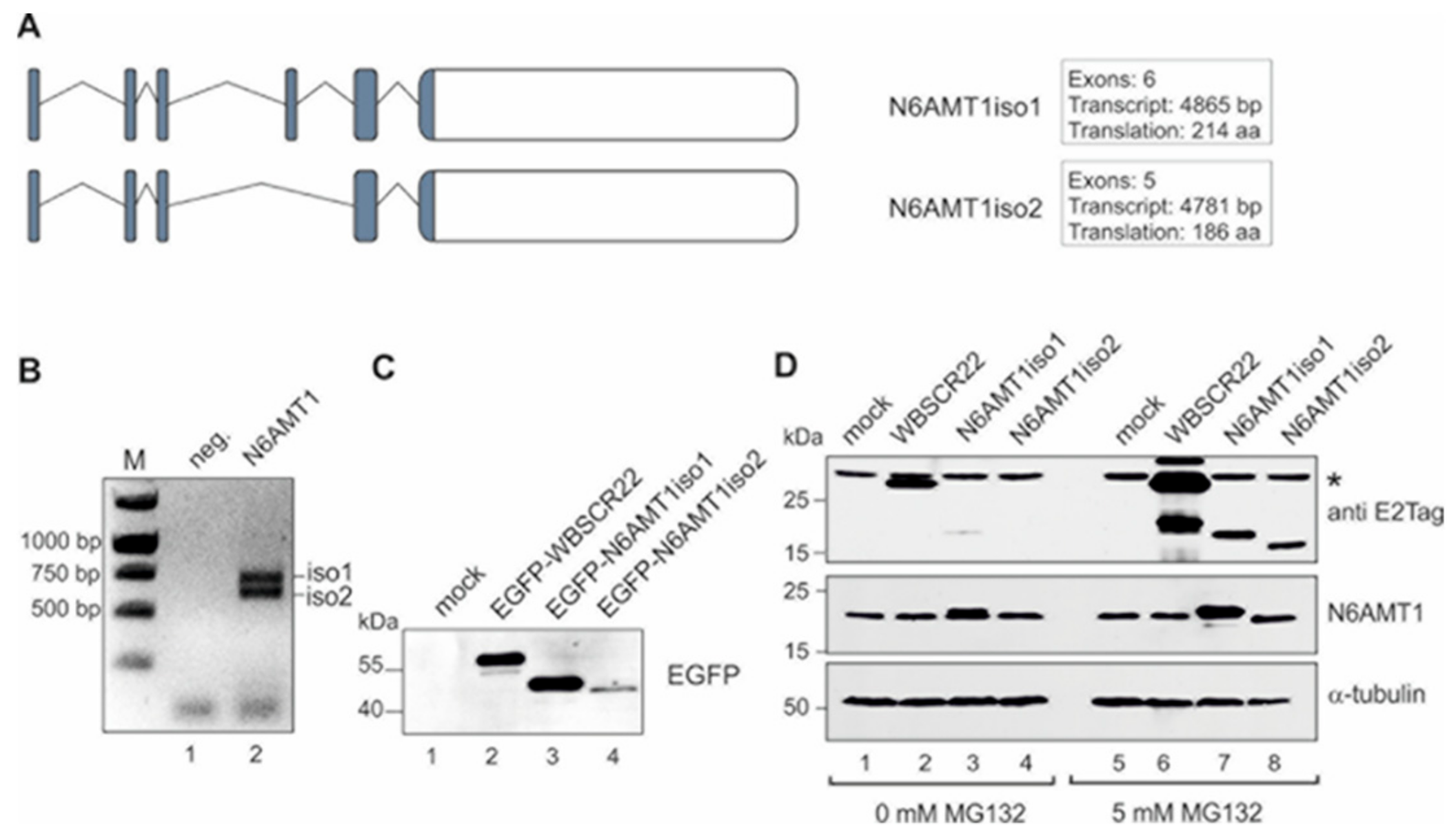
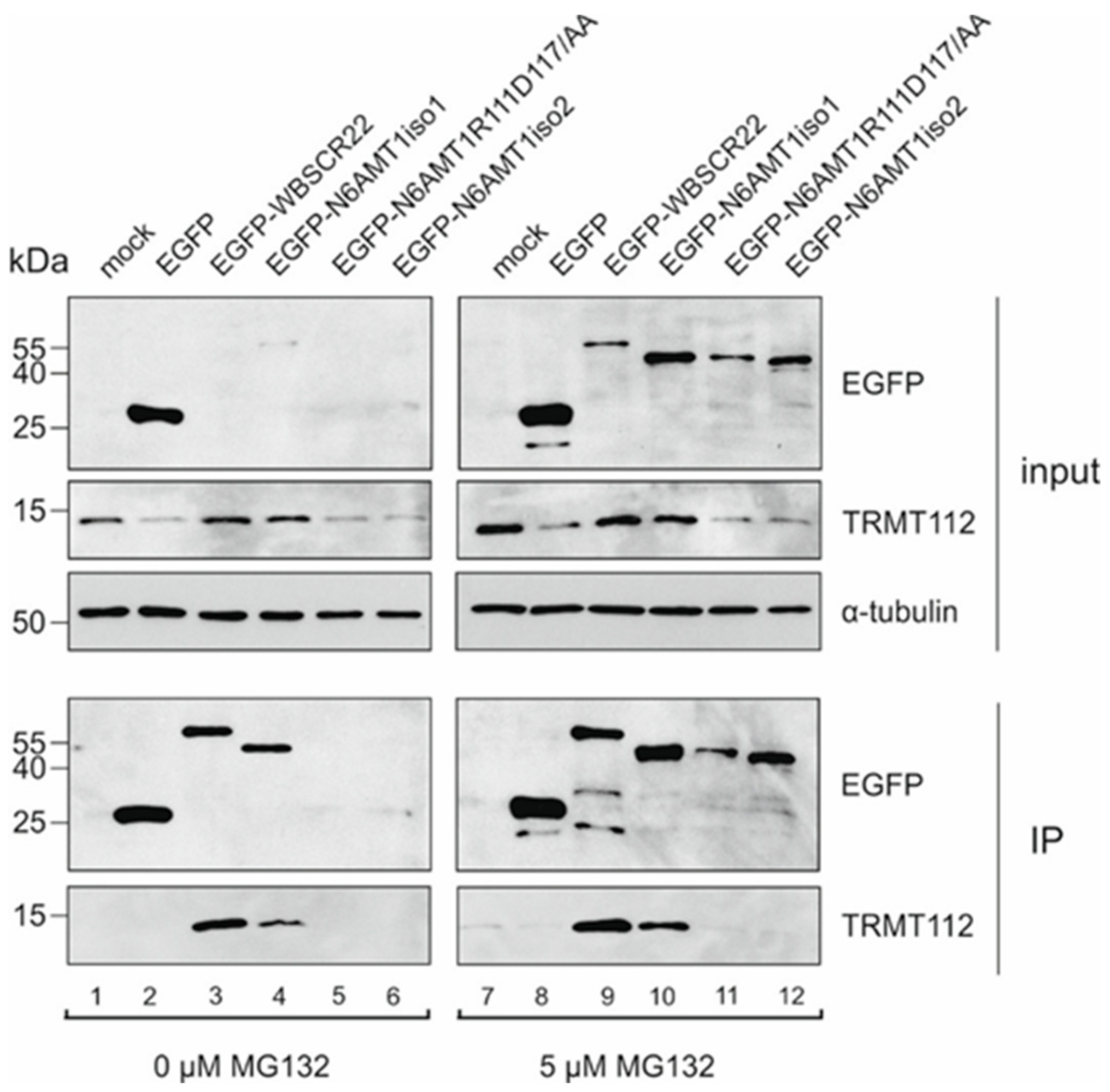
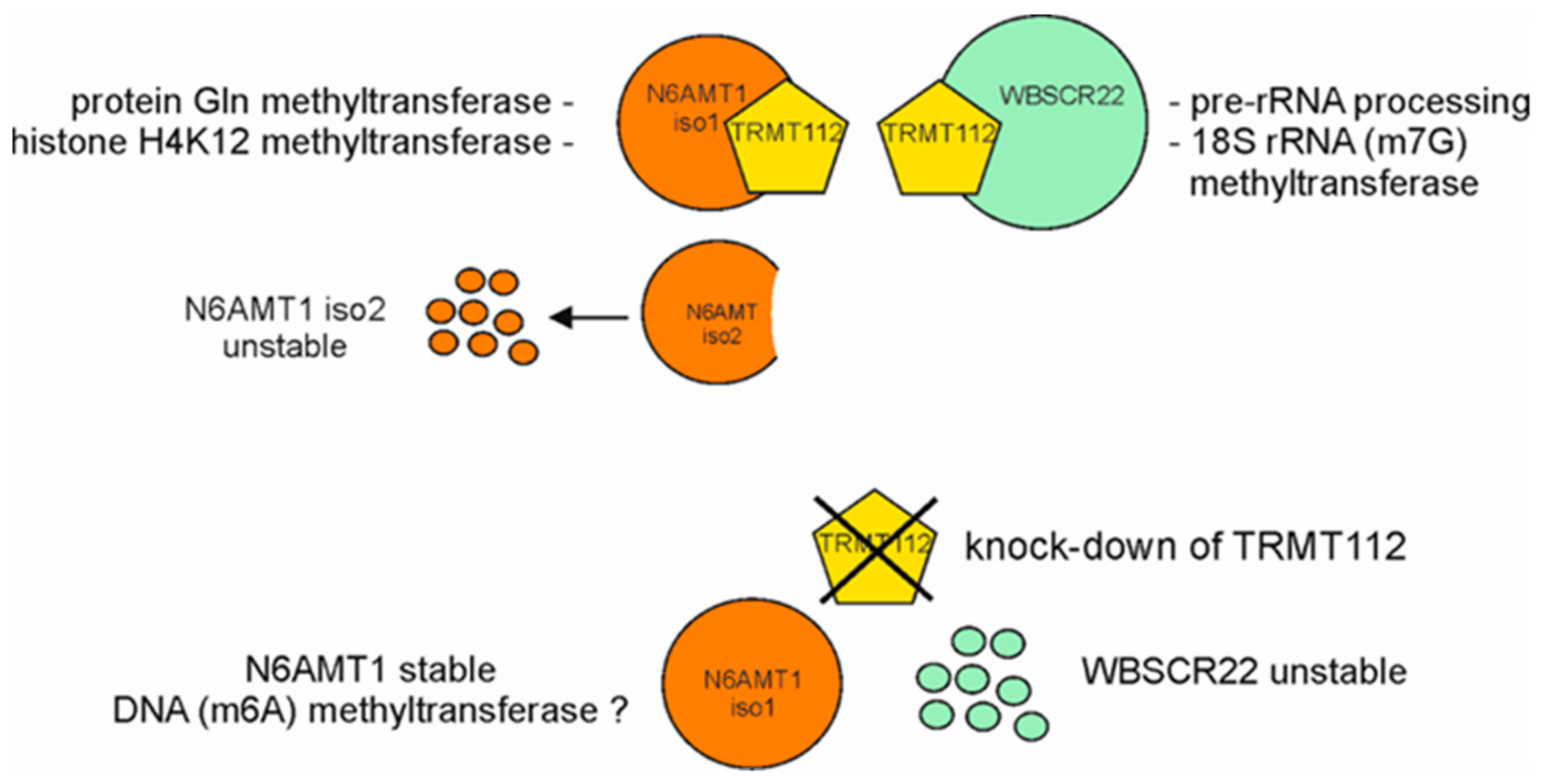
3.1. The Stability of N6AMT1 Isoforms Is Regulated by Proteasomes
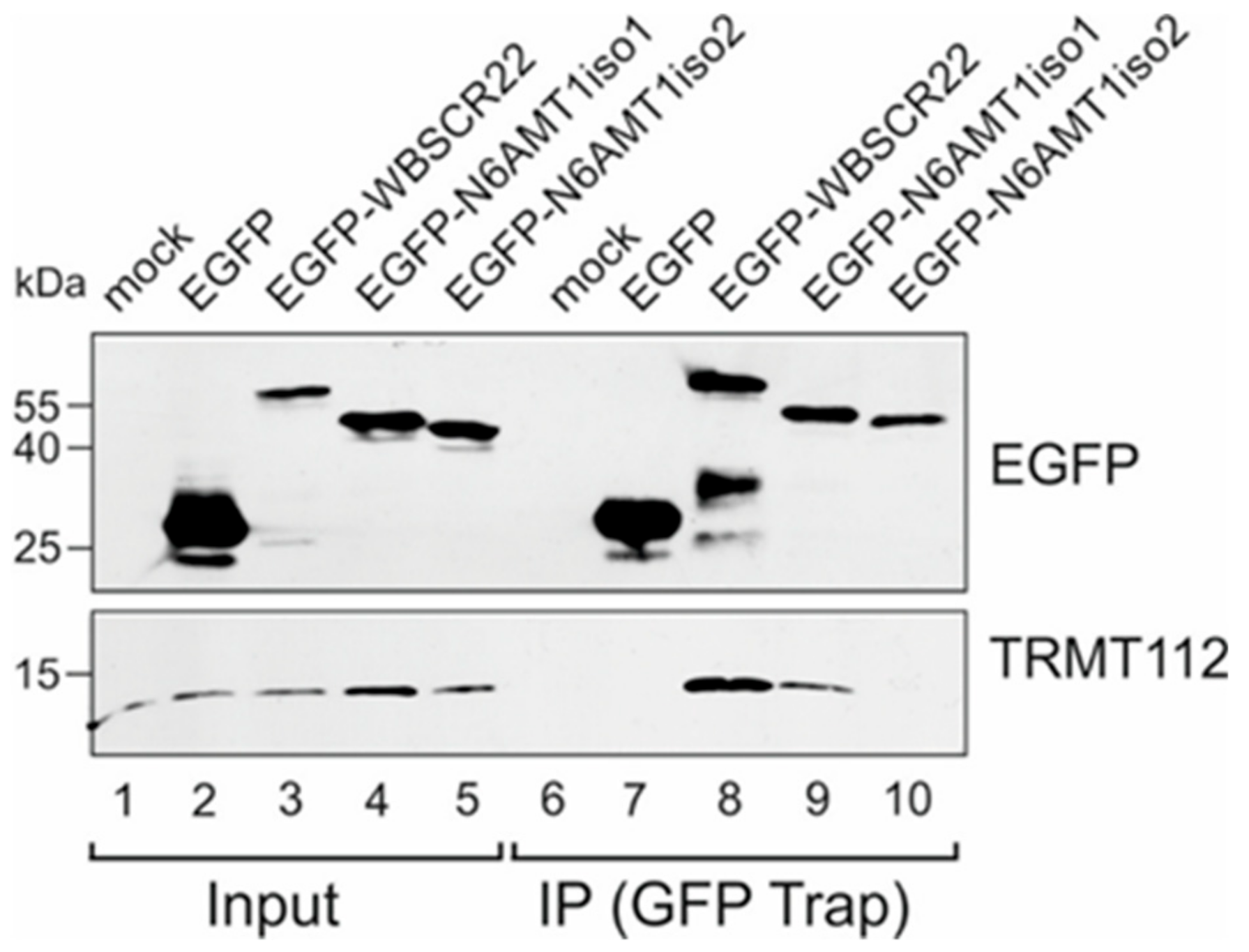
3.2. N6AMT1 iso2 Does Not Interact with TRMT112
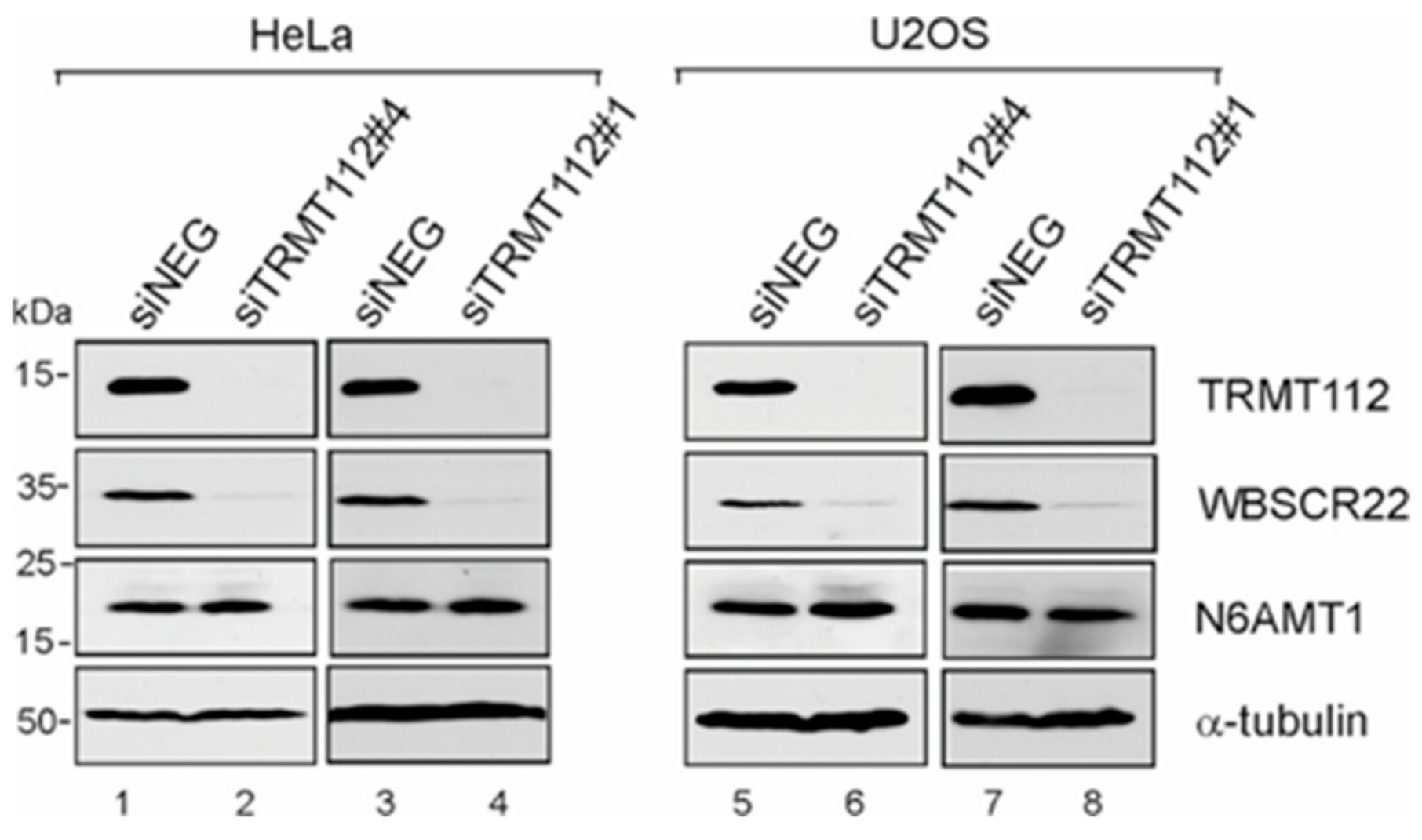
3.3. WBSCR22 and N6AMT1 Proteins Are Differently Regulated by Down-Regulation of TRMT112
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Petrossian, T.; Clarke, S. Uncovering the human methyltransferasome. Mol. Cell. Proteomics 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; McMillian, F. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Jones, P.; Issa, J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Roberti, A.; Valdes, A.; Torrecillas, R.; Fraga, M.; Fernandez, A. Epigenetics in cancer therapy and nanomedicine. Clin. Epigenetics 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Gujar, H.; Weisenberger, D.; Liang, G. The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome. Genes 2019, 10, 172. [Google Scholar] [CrossRef]
- Bourgeois, G.; Létoquart, J.; van Tran, N.; Graille, M. Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus. Biomolecules 2017, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Van Tran, N.; Muller, L.; Ross, R.; Lestini, R.; Létoquart, J.; Ulryck, N.; Limbach, P.; de Crécy-Lagard, V.; Cianférani, S.; Graille, M. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018, 46, 8483–8499. [Google Scholar] [CrossRef]
- Zorbas, C.; Nicolas, E.; Wacheul, L.; Huvelle, E.; Heurgué-Hamard, V.; Lafontaine, D. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 2015, 26, 2080–2095. [Google Scholar] [CrossRef]
- Õunap, K.; Leetsi, L.; Matsoo, M.; Kurg, R. The Stability of Ribosome Biogenesis Factor WBSCR22 Is Regulated by Interaction with TRMT112 via Ubiquitin-Proteasome Pathway. PLoS ONE 2015, 10, e0133841. [Google Scholar] [CrossRef]
- Figaro, S.; Scrima, N.; Buckingham, R.; Heurgué-Hamard, V. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008, 582, 2352–2356. [Google Scholar] [CrossRef] [Green Version]
- Õunap, K.; Käsper, L.; Kurg, A.; Kurg, R. The human WBSCR22 protein is involved in the biogenesis of the 40S ribosomal subunits in mammalian cells. PLoS ONE 2013, 8, e75686. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Kretschmer, J.; Bohnsack, M. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 2015, 21, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Jangani, M.; Poolman, T.; Matthews, L.; Yang, N.; Farrow, S.; Berry, A.; Hanley, N.; Williamson, A.; Whetton, A.; Donn, R.; et al. The methyltransferase WBSCR22/Merm1 enhances glucocorticoid receptor function and is regulated in lung inflammation and cancer. J. Biol. Chem. 2014, 289, 8931–8946. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Arai, H.; Fujita, A. The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer Res. 2011, 71, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Brophy, J.; Chan, C.; Atmore, K.; Begley, U.; Paules, R.; Dedon, P.; Begley, T.; Samson, L. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010, 30, 2449–2459. [Google Scholar] [CrossRef]
- Songe-Møller, L.; van den Born, E.; Leihne, V.; Vågbø, C.; Kristoffersen, T.; Krokan, H.; Kirpekar, F.; Falnes, P.; Klungland, A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol 2010, 30, 1814–1827. [Google Scholar] [CrossRef]
- Van den Born, E.; Vågbø, C.; Songe-Møller, L.; Leihne, V.; Lien, G.; Leszczynska, G.; Malkiewicz, A.; Krokan, H.; Kirpekar, F.; Klungland, A.; et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat. Commun. 2011, 2. [Google Scholar] [CrossRef]
- Purushothaman, S.; Bujnicki, J.; Grosjean, H.; Lapeyre, B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005, 25, 4359–4370. [Google Scholar] [CrossRef]
- Bourgeois, G.; Marcoux, J.; Saliou, J.; Cianférani, S.; Graille, M. Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 2017, 45, 1971–1982. [Google Scholar] [CrossRef]
- Metzger, E.; Wang, S.; Urban, S.; Willmann, D.; Schmidt, A.; Offermann, A.; Allen, A.; Sum, M.; Obier, N.; Cottard, F.; et al. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat Struct Mol Biol. 2019, 26, 361–371. [Google Scholar] [CrossRef]
- Xiao, C.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.; Luo, F.; et al. N6-Methyladenine DNA Modification in the Human Genome. Mol. Cell. 2018, 71, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Kusevic, D.; Kudithipudi, S.; Jeltsch, A. Substrate Specificity of the HEMK2 Protein Glutamine Methyltransferase and Identification of Novel Substrates. J. Biol. Chem. 2016, 291, 6124–6133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liger, D.; Mora, L.; Lazar, N.; Figaro, S.; Henri, J.; Scrima, N.; Buckingham, R.; van Tilbeurgh, H.; Heurgué-Hamard, V.; Graille, M. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011, 39, 6249–6259. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Aleshin, M.; Jo, W.; Dills, R.; Kalman, D.; Vulpe, C.; Smith, M.; Zhang, L. Involvement of N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) in arsenic biomethylation and its role in arsenic-induced toxicity. Environ. Health Perspect. 2011, 119, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Ratel, D.; Ravanat, J.; Charles, M.; Platet, N.; Breuillaud, L.; Lunardi, J.; Berger, F.; Wion, D. Undetectable levels of N6-methyl adenine in mouse DNA: Cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006, 580, 3179–3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Létoquart, J.; Huvelle, E.; Wacheul, L.; Bourgeois, G.; Zorbas, C.; Graille, M.; Heurgué-Hamard, V.; Lafontaine, D. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. USA 2014, 111, E5518–E5526. [Google Scholar] [CrossRef]
- Létoquart, J.; van Tran, N.; Caroline, V.; Aleksandrov, A.; Lazar, N.; van Tilbeurgh, H.; Liger, D.; Graille, M. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 2015, 43, 10989–11002. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Nie, S.; Li, B.; Yang, Z.; Xu, Z.; Fei, J.; Lin, C.; Zeng, R.; Xu, G. Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality. Mol. Cell. Biol. 2010, 30, 4245–4253. [Google Scholar] [CrossRef]
- Li, X.; Zhao, Q.; Wei, W.; Lin, Q.; Magnan, C.; Emami, M.; Wearick-Silva, L.; Viola, T.; Marshall, P.; Yin, J.; et al. The DNA modification N6-methyl-2’-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction. Nat. Neurosci. 2019, 22, 534–544. [Google Scholar] [CrossRef]
- Wu, T.; Wang, T.; Seetin, M.; Lai, Y.; Zhu, S.; Lin, K.; Liu, Y.; Byrum, S.; Mackintosh, S.; Zhong, M.; et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016, 532, 329–333. [Google Scholar] [CrossRef]
- Yao, B.; Cheng, Y.; Wang, Z.; Li, Y.; Chen, L.; Huang, L.; Zhang, W.; Chen, D.; Wu, H.; Tang, B.; et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; Johnson, A. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol. Biol. Cell 2012, 23, 4313–4322. [Google Scholar] [CrossRef] [PubMed]
- Van Tran, N.; Ernst, F.; Hawley, B.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.; Bohnsak, M.; Jaffrey, S.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tress, M.; Abascal, F.; Valencia, A. Alternative Splicing May Not Be the Key to Proteome Complexity. Trends Biochem. Sci. 2017, 42, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.; Morettin, A.; Côté, J. Role of PRMTs in cancer: Could minor isoforms be leaving a mark? World J. Biol. Chem. 2014, 5, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Goulet, I.; Gauvin, G.; Boisvenue, S.; Côté, J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 2007, 282, 33009–33021. [Google Scholar] [CrossRef]
- Patounas, O.; Papacharalampous, I.; Eckerich, C.; Markopoulos, G.; Kolettas, E.; Fackelmayer, F. A novel splicing isoform of protein arginine methyltransferase 1 (PRMT1) that lacks the dimerization arm and correlates with cellular malignancy. J. Cell. Biochem. 2018, 119, 2110–2123. [Google Scholar] [CrossRef]
- Ostler, K.; Davis, E.; Payne, S.; Gosalia, B.; Expósito-Céspedes, J.; Le Beau, M.; Godley, L. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene 2007, 26, 5553–5563. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, S.; Van Emburgh, B.; Shan, J.; Su, Z.; Fields, C.; Vieweg, J.; Hamazaki, T.; Schwartz, P.; Terada, N.; Robertson, K. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol. Cancer Res. 2009, 10, 1622–1634. [Google Scholar] [CrossRef]
- Duymich, C.; Charlet, J.; Yang, X.; Jones, P.; Liang, G. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leetsi, L.; Õunap, K.; Abroi, A.; Kurg, R. The Common Partner of Several Methyltransferases TRMT112 Regulates the Expression of N6AMT1 Isoforms in Mammalian Cells. Biomolecules 2019, 9, 422. https://doi.org/10.3390/biom9090422
Leetsi L, Õunap K, Abroi A, Kurg R. The Common Partner of Several Methyltransferases TRMT112 Regulates the Expression of N6AMT1 Isoforms in Mammalian Cells. Biomolecules. 2019; 9(9):422. https://doi.org/10.3390/biom9090422
Chicago/Turabian StyleLeetsi, Lilian, Kadri Õunap, Aare Abroi, and Reet Kurg. 2019. "The Common Partner of Several Methyltransferases TRMT112 Regulates the Expression of N6AMT1 Isoforms in Mammalian Cells" Biomolecules 9, no. 9: 422. https://doi.org/10.3390/biom9090422





