E6 Oncoproteins from High-Risk Human Papillomavirus Induce Mitochondrial Metabolism in a Head and Neck Squamous Cell Carcinoma Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Stable Transfections
2.4. Chemicals
2.5. Cell Viability
2.6. Cell Respirometry.
2.7. Complex I (CI) Linked Respiration and Complex IV (CIV) Activity
2.8. Western Blot Analysis
2.9. ROS Quantification
2.10. Protein Carbonyls
2.11. GSH and GSSG Quantification
2.12. Antioxidant Enzyme Activity Assays
2.13. Statistical Analysis
3. Results
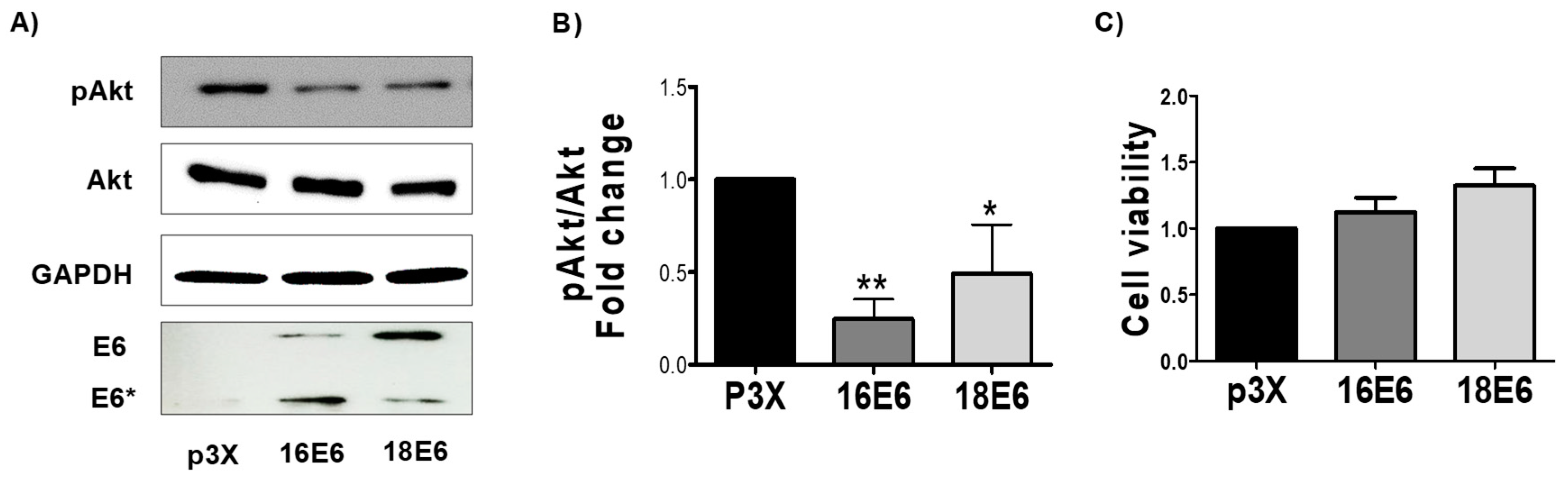
3.1. E6 Oncoproteins Reduce pAkt
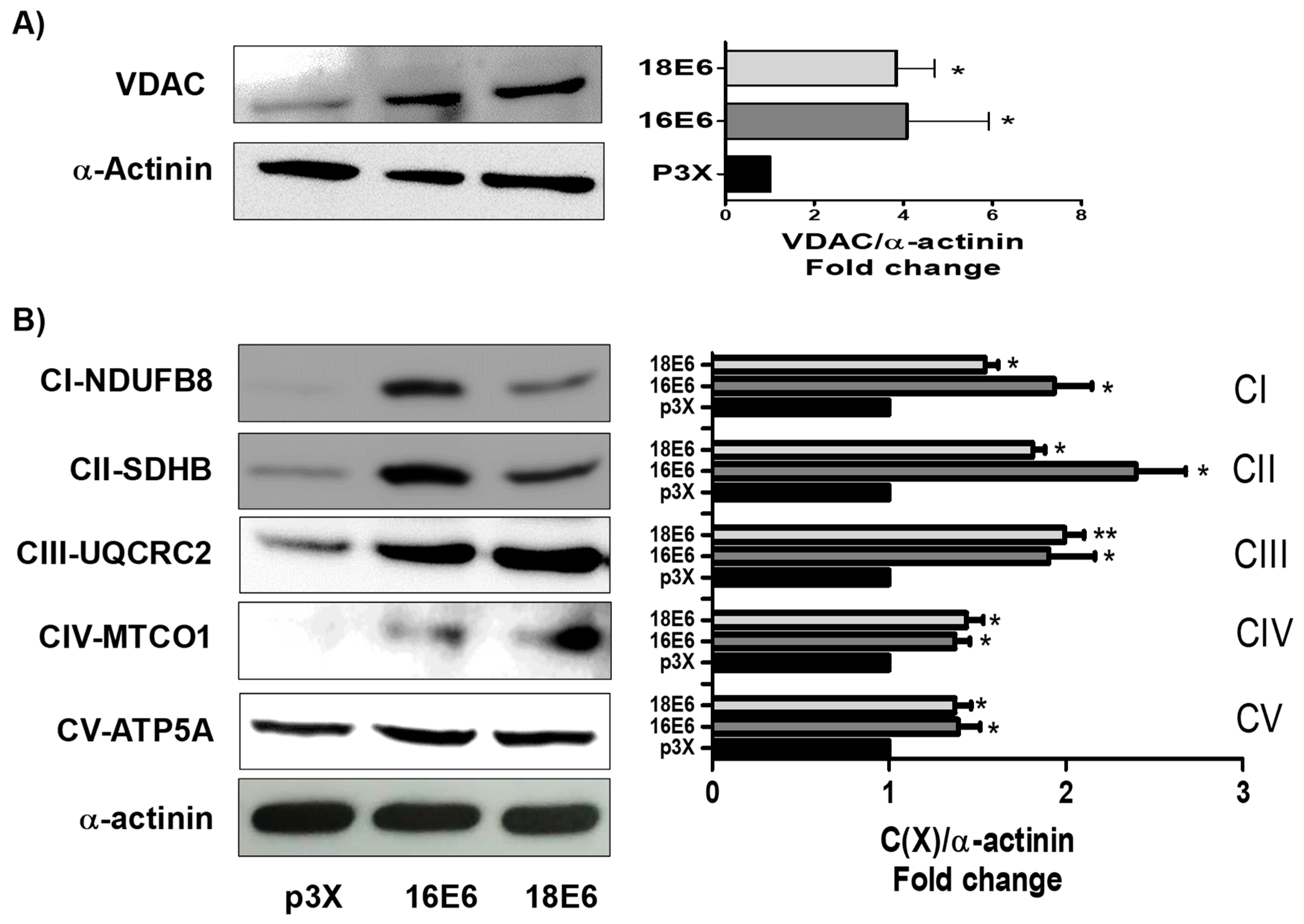
3.2. E6 Oncoproteins Increase Mitochondrial Proteins
3.3. E6 Oncoproteins Promote Mitochondrial Electron Transport System Activity
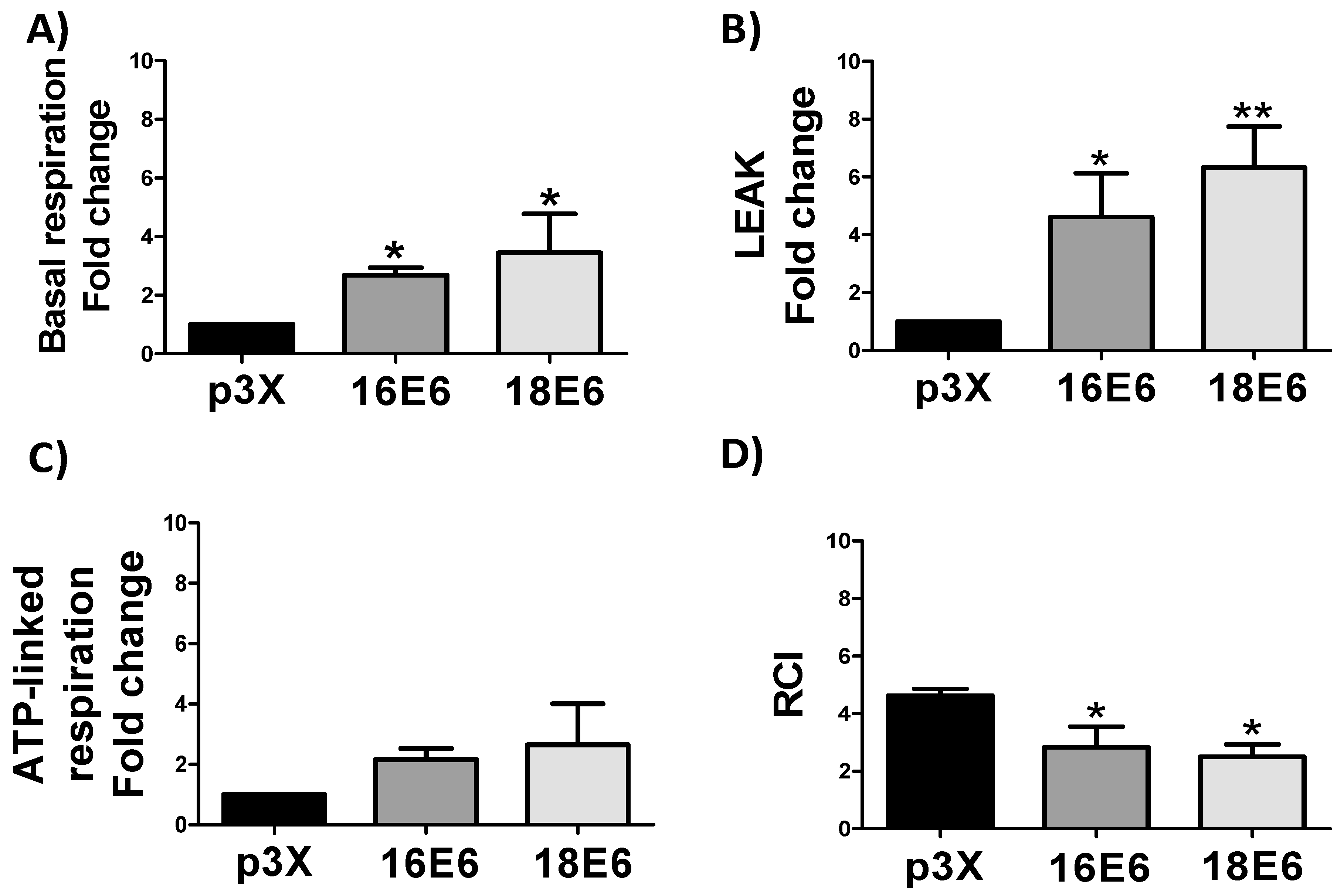
3.4. E6 Proteins Increase Cellular Respiration Rates, Inducing Mitochondrial Decoupling
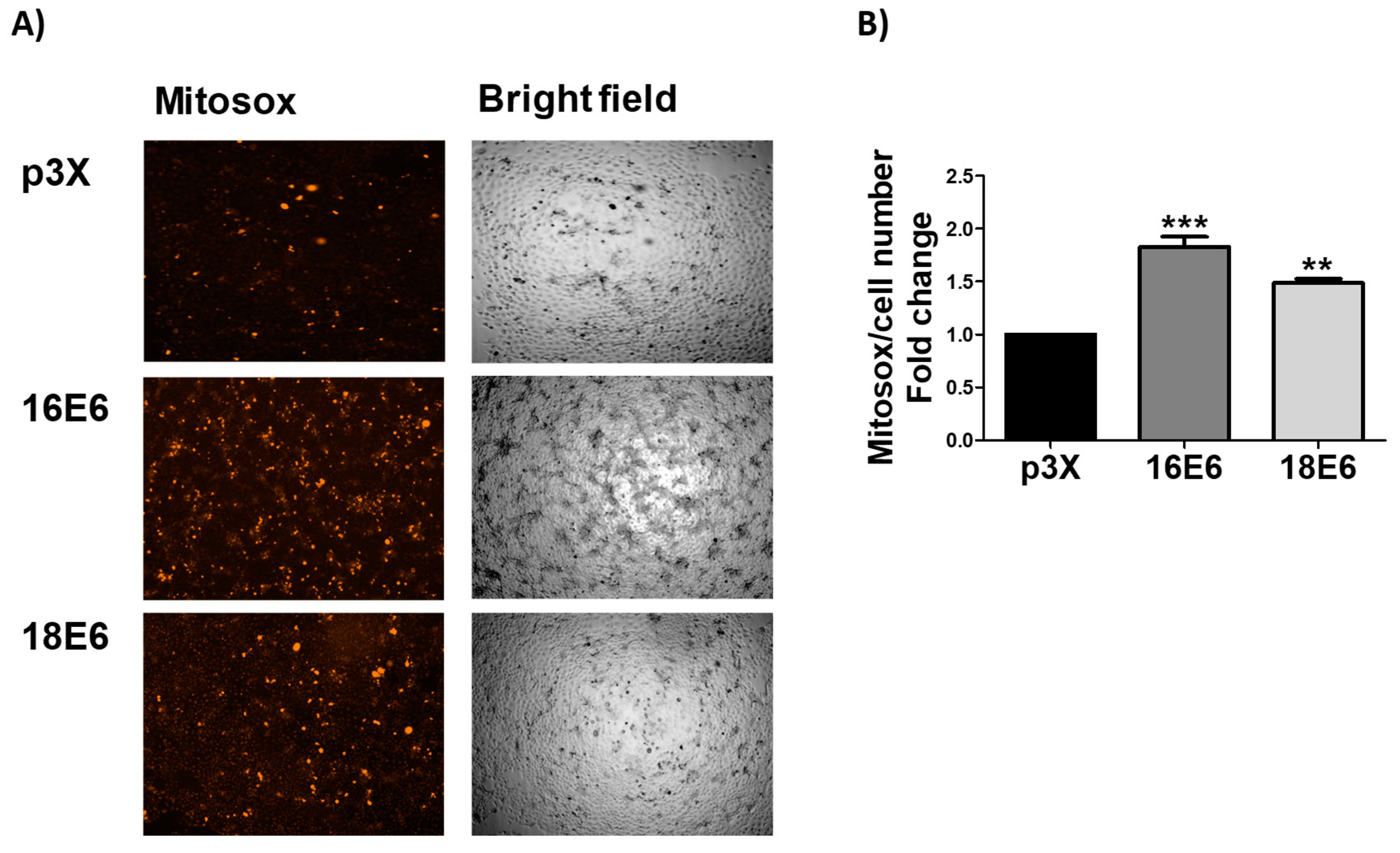
3.5. E6 Promotes Mitochondrial ROS Production
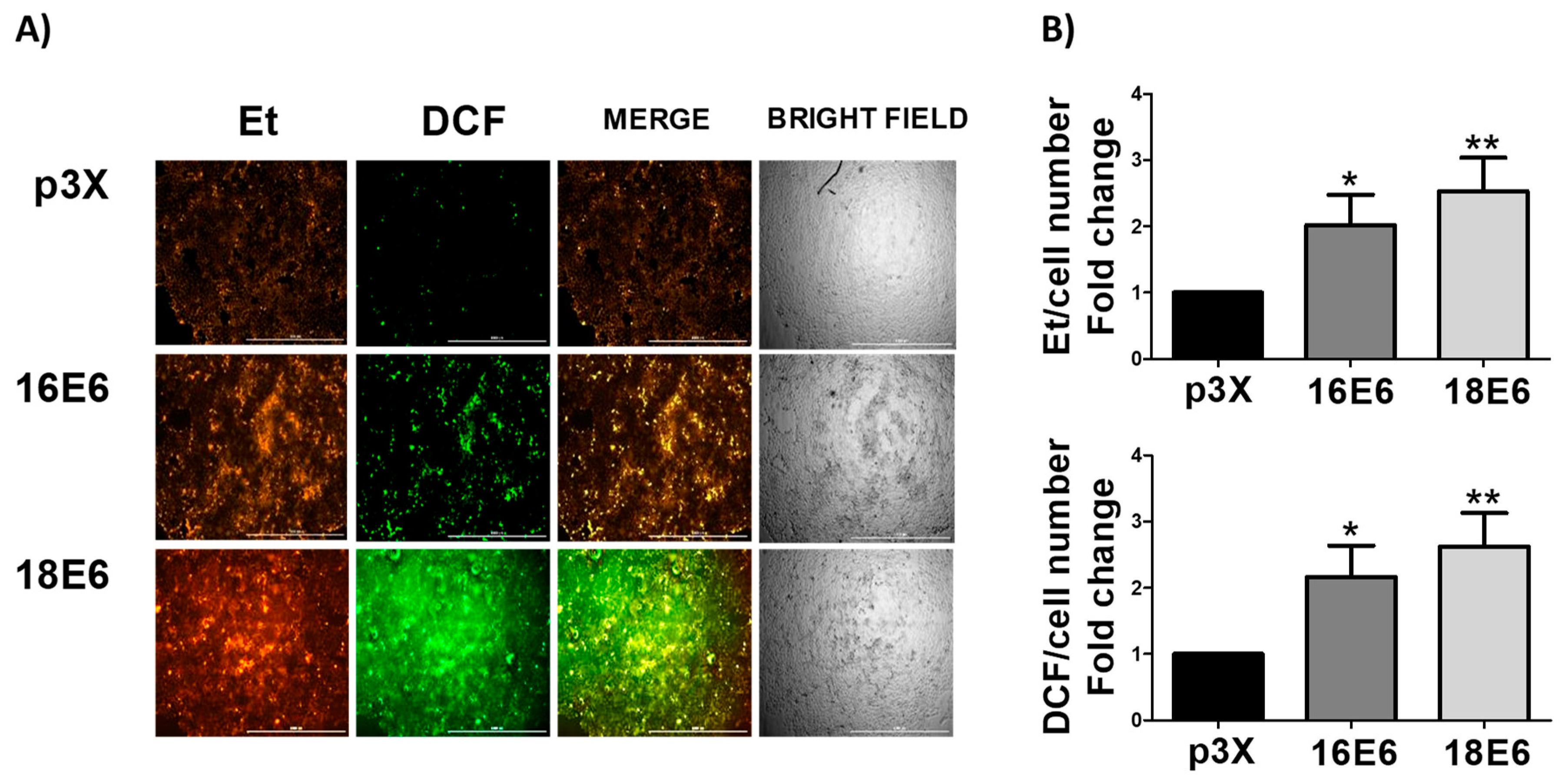
3.6. E6 Promotes the Increase in ROS Levels in Whole Cells
3.7. E6 oncoproteins from HPV16 and HPV18 Induce Oxidative Stress through the Increase in Protein Carbonyls and GSSG and the Decrease in GSH and GSH/GSSG Ratio
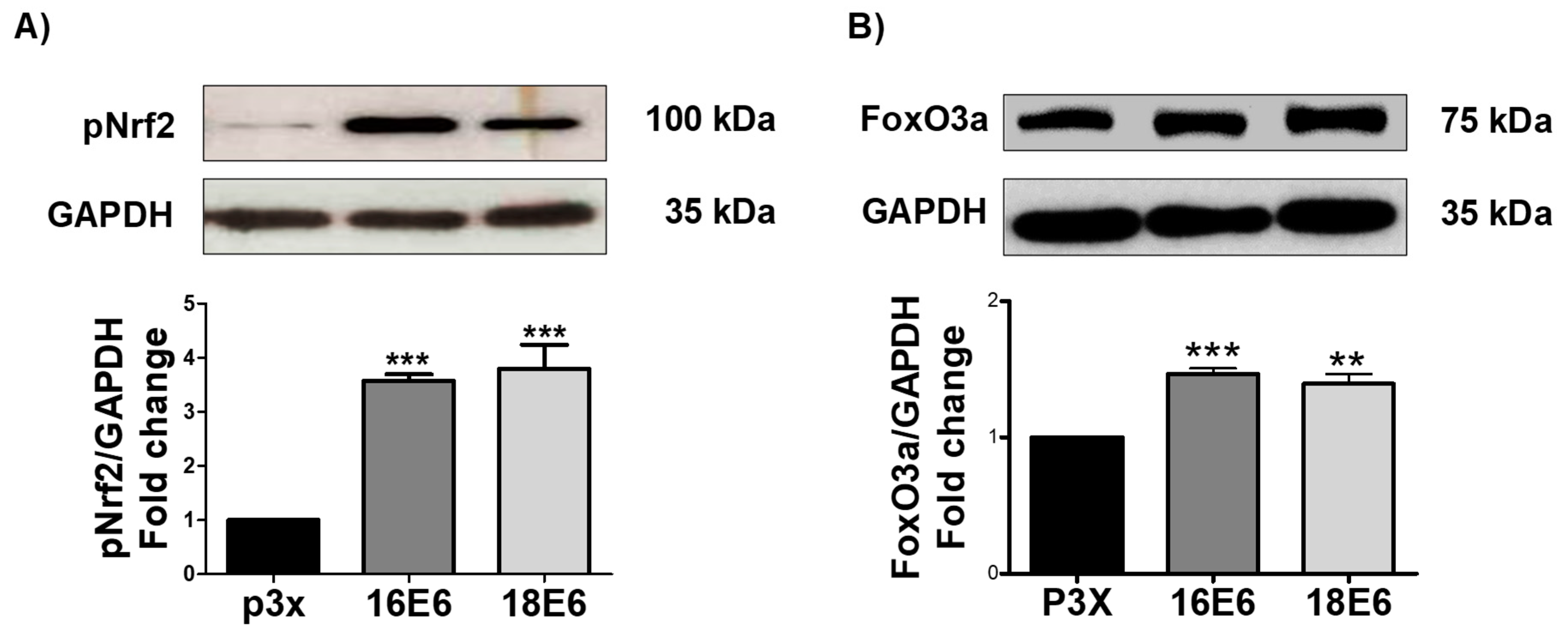
3.8. E6 from HPV16 and HPV18 Increase pNrf2 and FoxO3a Levels
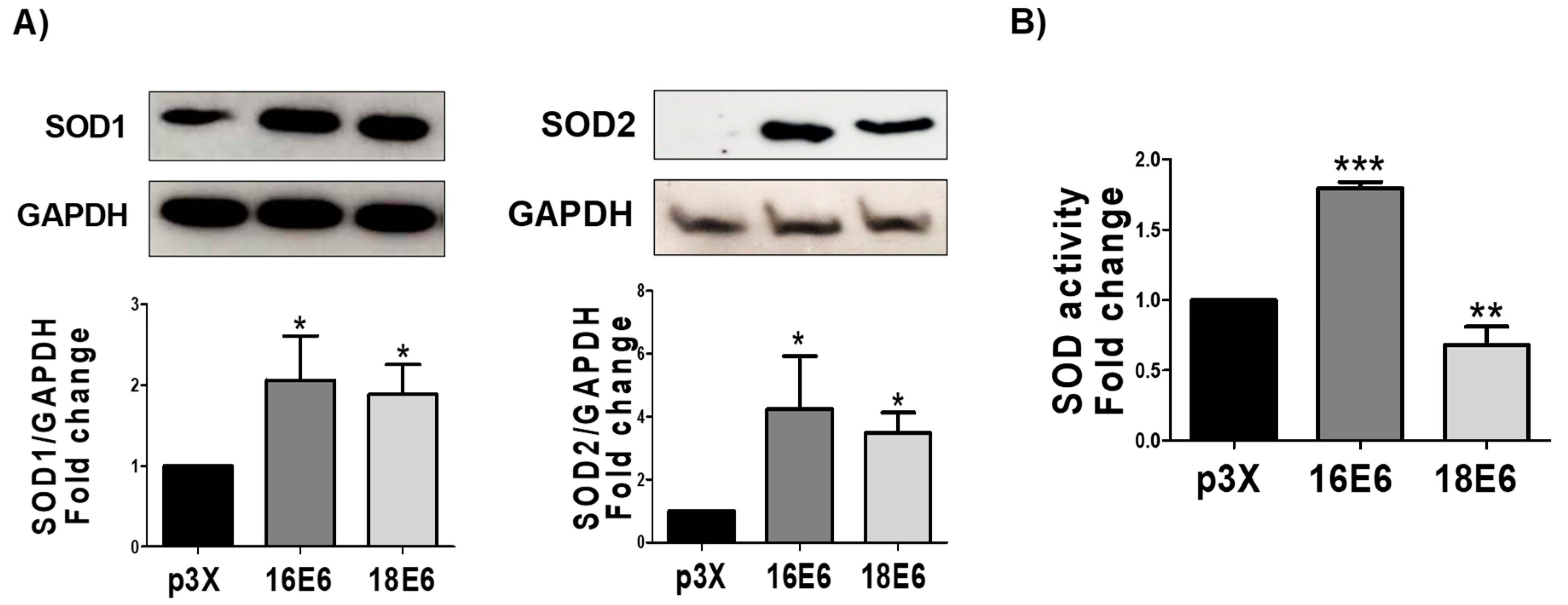
3.9. E6 Oncoproteins Increase SOD1 and SOD2 Levels and Activity
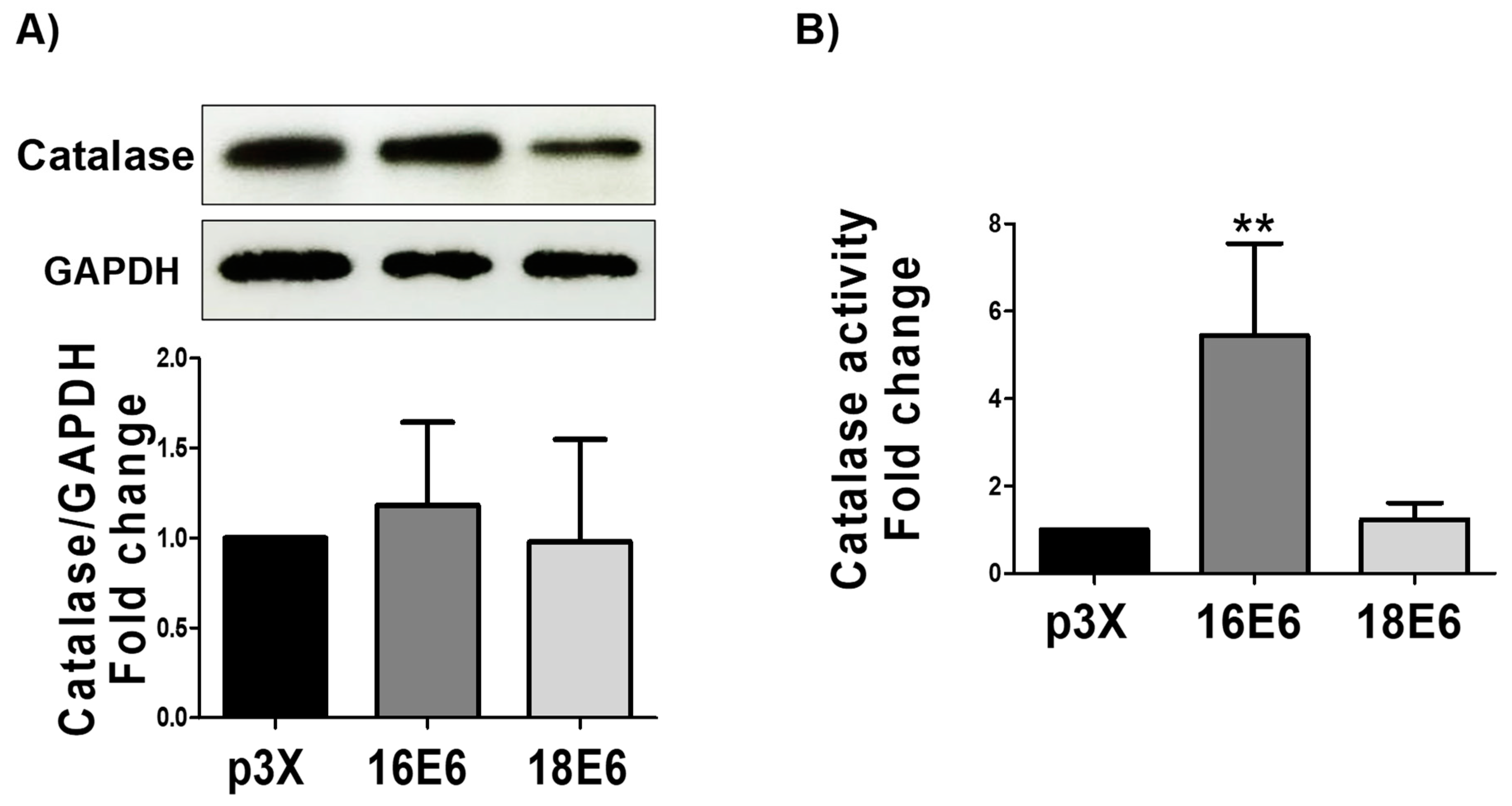
3.10. 16E6 Increases Catalase Activity without Changing Catalase Levels
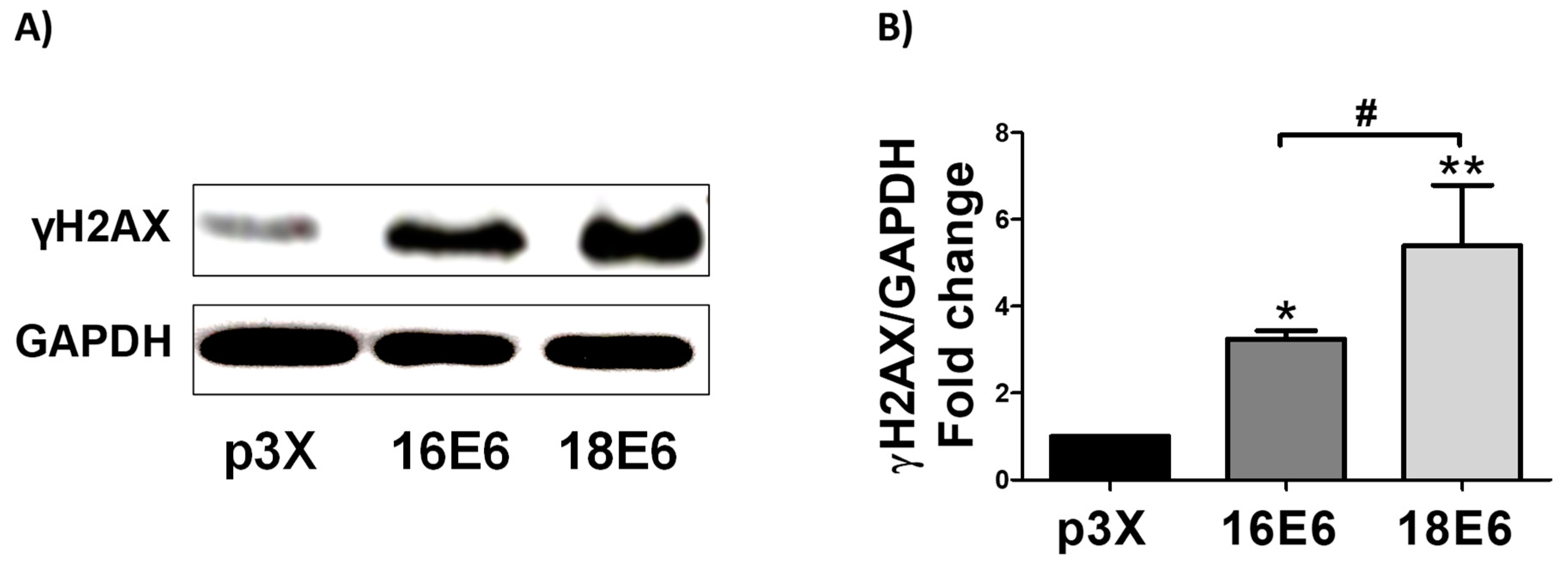
3.11. 16E6 and 18E6 Oncoproteins Induce DNA Damage
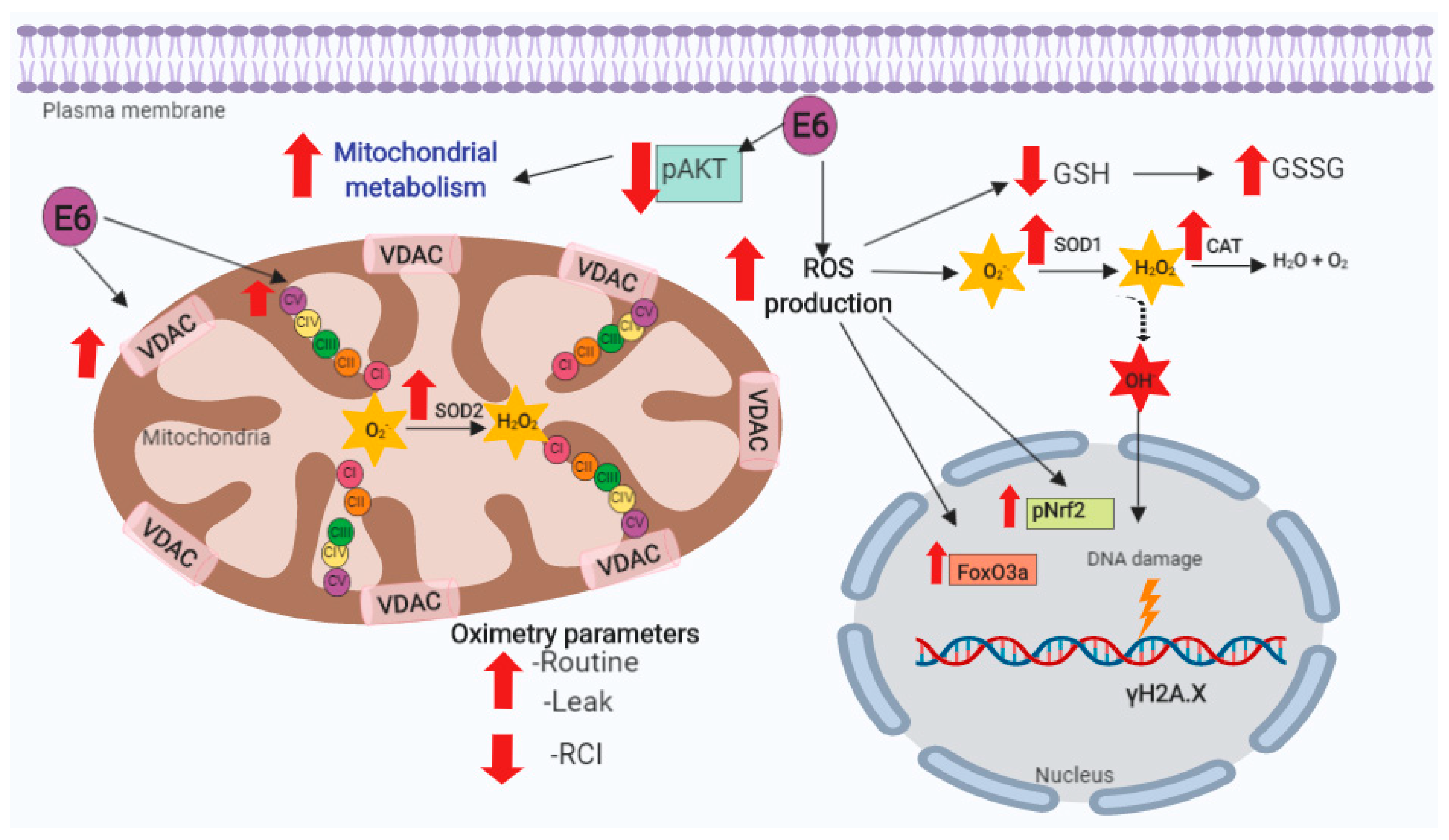
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef] [PubMed]
- Pytynia, K.B.; Dahlstrom, K.R.; Sturgis, E.M. Epidemiology of HPV-associated oropharyngeal cancer. Oral Oncol. 2014, 50, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Dok, R.; Nuyts, S. HPV Positive Head and Neck Cancers: Molecular Pathogenesis and Evolving Treatment Strategies. Cancers (Basel) 2016, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Jou, A.; Hess, J. Epidemiology and Molecular Biology of Head and Neck Cancer. Oncol Res. Treat. 2017, 40, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.; Young, R.J.; Rischin, D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin. Cancer Biol. 2018, 52, 228–240. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.-X.; Zhang, J.; Wang, J.; et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef]
- Mims, J.; Bansal, N.; Bharadwaj, M.S.; Chen, X.; Molina, A.J.; Tsang, A.W.; Furdui, C.M. Energy metabolism in a matched model of radiation resistance for head and neck squamous cell cancer. Radiat. Res. 2015, 183, 291–304. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Najy, A.J.; Huang, W.; Sethi, S.; Snyder, M.; Sakr, W.; Dyson, G.; Hüttemann, M.; Lee, I.; Ali-Fehmi, R.; et al. HPV-associated differential regulation of tumor metabolism in oropharyngeal head and neck cancer. Oncotarget 2017, 8, 51530–51541. [Google Scholar] [CrossRef]
- Cruz-Gregorio, A.; Martínez-Ramírez, I.; Pedraza-Chaverri, J.; Lizano, M. Reprogramming of Energy Metabolism in Response to Radiotherapy in Head and Neck Squamous Cell Carcinoma. Cancers (Basel) 2019, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.-X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.-S.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Badaracco, G.; Venuti, A.; Sedati, A.; Marcante, M.L. HPV16 and HPV18 in genital tumors: Significantly different levels of viral integration and correlation to tumor invasiveness. J. Med. Virol. 2002, 67, 574–582. [Google Scholar] [CrossRef]
- Romanczuk, H.; Howley, P.M. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. USA 1992, 89, 3159–3163. [Google Scholar] [CrossRef]
- Mantovani, F.; Banks, L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 2001, 20, 7874–7887. [Google Scholar] [CrossRef]
- Boyer, S.N.; Wazer, D.E.; Band, V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996, 56, 4620–4624. [Google Scholar]
- Thomas, M.; Pim, D.; Banks, L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999, 18, 7690–7700. [Google Scholar] [CrossRef]
- Cruz-Gregorio, A.; Manzo-Merino, J.; Gonzaléz-García, M.C.; Pedraza-Chaverri, J.; Medina-Campos, O.N.; Valverde, M.; Rojas, E.; Rodríguez-Sastre, M.A.; García-Cuellar, C.M.; Lizano, M. Human Papillomavirus Types 16 and 18 Early-expressed Proteins Differentially Modulate the Cellular Redox State and DNA Damage. Int. J. Biol. Sci. 2018, 14, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Gregorio, A.; Manzo-Merino, J.; Lizano, M. Cellular redox, cancer and human papillomavirus. Virus Res. 2018, 246, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Morgillo, F.; Tuccillo, C.; Ciardiello, F.; Loguercio, C. Chronic inflammation and oxidative stress in human carcinogenesis. Int. J. Cancer 2007, 121, 2381–2386. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, C.L.; Perevoshchikova, I.V.; Hey-Mogensen, M.; Orr, A.L.; Brand, M.D. Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biol. 2013, 1, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.-O.; Sánchez-Ramos, C.; Prieto-Arroyo, I.; Urbánek, P.; Steinbrenner, H.; Monsalve, M. Redox regulation of FoxO transcription factors. Redox Biol. 2015, 6, 51–72. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Aparicio-Trejo, O.E.; Reyes-Fermín, L.M.; Briones-Herrera, A.; Tapia, E.; León-Contreras, J.C.; Hernández-Pando, R.; Sánchez-Lozada, L.G.; Pedraza-Chaverri, J. Protective effects of N-acetyl-cysteine in mitochondria bioenergetics, oxidative stress, dynamics and S-glutathionylation alterations in acute kidney damage induced by folic acid. Free Radic. Biol. Med. 2019, 130, 379–396. [Google Scholar] [CrossRef]
- Reyes-Fermín, L.M.; Avila-Rojas, S.H.; Aparicio-Trejo, O.E.; Tapia, E.; Rivero, I.; Pedraza-Chaverri, J. The Protective Effect of Alpha-Mangostin against Cisplatin-Induced Cell Death in LLC-PK1 Cells is Associated to Mitochondrial Function Preservation. Antioxidants (Basel) 2019, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Pedraza-Chaverrí, J.; Reyes-Fermín, L.M.; Nolasco-Amaya, E.G.; Orozco-Ibarra, M.; Medina-Campos, O.N.; González-Cuahutencos, O.; Rivero-Cruz, I.; Mata, R. ROS scavenging capacity and neuroprotective effect of alpha-mangostin against 3-nitropropionic acid in cerebellar granule neurons. Exp. Toxicol. Pathol. 2009, 61, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Williams, J.A.; Stadtman, E.R.; Shacter, E. Carbonyl assays for determination of oxidatively modified proteins. Meth. Enzymol. 1994, 233, 346–357. [Google Scholar] [PubMed]
- Rahman, I.; Kode, A.; Biswas, S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006, 1, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase in vitro. Meth. Enzymol. 1984, 105, 121–126. [Google Scholar] [PubMed]
- Elstrom, R.L.; Bauer, D.E.; Buzzai, M.; Karnauskas, R.; Harris, M.H.; Plas, D.R.; Zhuang, H.; Cinalli, R.M.; Alavi, A.; Rudin, C.M.; et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004, 64, 3892–3899. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.B.; Hay, N. Is Akt the “Warburg kinase”?—Akt-energy metabolism interactions and oncogenesis. Semin. Cancer Biol. 2009, 19, 25–31. [Google Scholar] [CrossRef]
- De Rosa, V.; Iommelli, F.; Monti, M.; Fonti, R.; Votta, G.; Stoppelli, M.P.; Del Vecchio, S. Reversal of Warburg Effect and Reactivation of Oxidative Phosphorylation by Differential Inhibition of EGFR Signaling Pathways in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 5110–5120. [Google Scholar] [CrossRef]
- Zhao, M.; Zhou, Y.; Liu, S.; Li, L.; Chen, Y.; Cheng, J.; Lu, Y.; Liu, J. Control release of mitochondria-targeted antioxidant by injectable self-assembling peptide hydrogel ameliorated persistent mitochondrial dysfunction and inflammation after acute kidney injury. Drug Deliv 2018, 25, 546–554. [Google Scholar] [CrossRef]
- Wang, C.; Li, L.; Liu, S.; Liao, G.; Li, L.; Chen, Y.; Cheng, J.; Lu, Y.; Liu, J. GLP-1 receptor agonist ameliorates obesity-induced chronic kidney injury via restoring renal metabolism homeostasis. PLoS ONE 2018, 13, e0193473. [Google Scholar] [CrossRef]
- Svensson, K.; Schnyder, S.; Cardel, B.; Handschin, C. Loss of Renal Tubular PGC-1α Exacerbates Diet-Induced Renal Steatosis and Age-Related Urinary Sodium Excretion in Mice. PLoS ONE 2016, 11, e0158716. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yuan, Q.; Xu, T.; Yao, L.; Feng, J.; Ma, J.; Wang, L.; Lu, C.; Wang, D. Pioglitazone Improves Mitochondrial Function in the Remnant Kidney and Protects against Renal Fibrosis in 5/6 Nephrectomized Rats. Front. Pharmacol. 2017, 8, 545. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Stallons, L.J.; Collier, J.B.; Chavin, K.D.; Schnellmann, R.G. Suppression of mitochondrial biogenesis through toll-like receptor 4-dependent mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signaling in endotoxin-induced acute kidney injury. J. Pharmacol. Exp. Ther. 2015, 352, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Aparicio-Trejo, O.E.; Tapia, E.; Sánchez-Lozada, L.G.; Pedraza-Chaverri, J. Mitochondrial bioenergetics, redox state, dynamics and turnover alterations in renal mass reduction models of chronic kidney diseases and their possible implications in the progression of this illness. Pharmacol. Res. 2018, 135, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gnaiger, E. Mitochondrial Pathways and Respiratory Control. An. Introduction to OXPHOS Analysis; OROBOROS INSTRUMENTS Corp.: Innsbruck, Austria, 2014; ISBN 9783950239966. [Google Scholar]
- Makrecka-Kuka, M.; Krumschnabel, G.; Gnaiger, E. High-Resolution Respirometry for Simultaneous Measurement of Oxygen and Hydrogen Peroxide Fluxes in Permeabilized Cells, Tissue Homogenate and Isolated Mitochondria. Biomolecules 2015, 5, 1319–1338. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.-Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Antunes, F.; Canali, R.; Rettori, D.; Cadenas, E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 2003, 278, 5557–5563. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef]
- Osellame, L.D.; Blacker, T.S.; Duchen, M.R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 2012, 26, 711–723. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Holmström, K.M.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.C.; Woo, J.; Moutasim, K.; Mellone, M.; Frampton, S.J.; Mead, A.; Ahmed, W.; Wood, O.; Robinson, H.; Ward, M.; et al. HPV, tumour metabolism and novel target identification in head and neck squamous cell carcinoma. Br. J. Cancer 2019, 120, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Sewell, A.; Brown, B.; Biktasova, A.; Mills, G.B.; Lu, Y.; Tyson, D.R.; Issaeva, N.; Yarbrough, W.G. Reverse-phase protein array profiling of oropharyngeal cancer and significance of PIK3CA mutations in HPV-associated head and neck cancer. Clin. Cancer Res. 2014, 20, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Lee, J.H.; Wilke, W.W.; Quon, H.; Smith, G.; Maity, A.; Buatti, J.M.; Spitz, D.R. Radiation response in two HPV-infected head-and-neck cancer cell lines in comparison to a non-HPV-infected cell line and relationship to signaling through AKT. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Evans, W.; Filippova, M.; Filippov, V.; Bashkirova, S.; Zhang, G.; Reeves, M.E.; Duerksen-Hughes, P. Overexpression of HPV16 E6* Alters β-Integrin and Mitochondrial Dysfunction Pathways in Cervical Cancer Cells. Cancer Genom. Proteom. 2016, 13, 259–273. [Google Scholar]
- Paget-Bailly, P.; Meznad, K.; Bruyère, D.; Perrard, J.; Herfs, M.; Jung, A.C.; Mougin, C.; Prétet, J.-L.; Baguet, A. Comparative RNA sequencing reveals that HPV16 E6 abrogates the effect of E6*I on ROS metabolism. Sci. Rep. 2019, 9, 5938. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial Reactive Oxygen Species (ROS) and ROS-Induced ROS Release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Baffy, G. Mitochondrial uncoupling in cancer cells: Liabilities and opportunities. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 655–664. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Kyoung Kim, H.; Kyoung Kim, Y.; Song, I.-H.; Baek, S.-H.; Lee, S.-R.; Hye Kim, J.; Kim, J.-R. Down-regulation of a forkhead transcription factor, FOXO3a, accelerates cellular senescence in human dermal fibroblasts. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Tseng, A.H.H.; Shieh, S.-S.; Wang, D.L. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic. Biol. Med. 2013, 63, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Brunet, A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005, 24, 7410–7425. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.; Chun, E.; Howell, J.C.; Sengupta, T.; Chen, D.; Kim, H. MicroRNA-30b-mediated regulation of catalase expression in human ARPE-19 cells. PLoS ONE 2012, 7, e42542. [Google Scholar] [CrossRef] [PubMed]
- Chiantore, M.V.; Mangino, G.; Iuliano, M.; Zangrillo, M.S.; De Lillis, I.; Vaccari, G.; Accardi, R.; Tommasino, M.; Columba Cabezas, S.; Federico, M.; et al. Human papillomavirus E6 and E7 oncoproteins affect the expression of cancer-related microRNAs: Additional evidence in HPV-induced tumorigenesis. J. Cancer Res. Clin. Oncol. 2016, 142, 1751–1763. [Google Scholar] [CrossRef] [PubMed]
- Boon, S.S.; Banks, L. High-risk human papillomavirus E6 oncoproteins interact with 14-3-3ζ in a PDZ binding motif-dependent manner. J. Virol. 2013, 87, 1586–1595. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Simpson, D.A.; Zhou, Y.; Mitra, A.; Mitchell, D.L.; Cordeiro-Stone, M.; Kaufmann, W.K. Human papilloma virus type16 E6 deregulates CHK1 and sensitizes human fibroblasts to environmental carcinogens independently of its effect on p53. Cell Cycle 2009, 8, 1775–1787. [Google Scholar] [CrossRef]
- Bhakta-Guha, D.; Efferth, T. Hormesis: Decoding Two Sides of the Same Coin. Pharmaceuticals (Basel) 2015, 8, 865–883. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Tan, C.L.; Gunaratne, J.; Quek, L.S.; Nei, W.; Thierry, F.; Bellanger, S. Localization of HPV-18 E2 at Mitochondrial Membranes Induces ROS Release and Modulates Host Cell Metabolism. PLoS ONE 2013, 8, e75625. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Gregorio, A.; Aranda-Rivera, A.K.; Aparicio-Trejo, O.E.; Coronado-Martínez, I.; Pedraza-Chaverri, J.; Lizano, M. E6 Oncoproteins from High-Risk Human Papillomavirus Induce Mitochondrial Metabolism in a Head and Neck Squamous Cell Carcinoma Model. Biomolecules 2019, 9, 351. https://doi.org/10.3390/biom9080351
Cruz-Gregorio A, Aranda-Rivera AK, Aparicio-Trejo OE, Coronado-Martínez I, Pedraza-Chaverri J, Lizano M. E6 Oncoproteins from High-Risk Human Papillomavirus Induce Mitochondrial Metabolism in a Head and Neck Squamous Cell Carcinoma Model. Biomolecules. 2019; 9(8):351. https://doi.org/10.3390/biom9080351
Chicago/Turabian StyleCruz-Gregorio, Alfredo, Ana Karina Aranda-Rivera, Omar Emiliano Aparicio-Trejo, Iris Coronado-Martínez, José Pedraza-Chaverri, and Marcela Lizano. 2019. "E6 Oncoproteins from High-Risk Human Papillomavirus Induce Mitochondrial Metabolism in a Head and Neck Squamous Cell Carcinoma Model" Biomolecules 9, no. 8: 351. https://doi.org/10.3390/biom9080351
APA StyleCruz-Gregorio, A., Aranda-Rivera, A. K., Aparicio-Trejo, O. E., Coronado-Martínez, I., Pedraza-Chaverri, J., & Lizano, M. (2019). E6 Oncoproteins from High-Risk Human Papillomavirus Induce Mitochondrial Metabolism in a Head and Neck Squamous Cell Carcinoma Model. Biomolecules, 9(8), 351. https://doi.org/10.3390/biom9080351







