High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Droplet-Based Microfluidic Design
2.2. Device Fabrication
2.3. Species Preparation
2.4. Species Identification
2.5. Droplet Generation
2.6. Traditional Batch Cultures
2.7. Growth Assay Inside Droplets
R2 = 0.9949
2.8. Growth Assay for Traditional Batch Cultures
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kellam, S.J.; Walker, J.M. Antibacterial activity from marine microalgae in laboratory culture. Br. Phycol. J. 1989, 24, 191–194. [Google Scholar] [CrossRef]
- Maadane, A.; Merghoub, N.; El Mernissi, N.; Ainane, T.; Amzazi, S. Antimicrobial activity of marine microalgae isolated from moroccan coastlines. J. Microbiol. Biotechnol. Food Sci. 2017, 6, 1257–1260. [Google Scholar] [CrossRef]
- Burkholder, P.R.; Burkholder, L.M.; Almodovar, L.R. Antibiotic activity of some marine algae of Puerto Rico. Bot. Mar. 1960, 2, 149–156. [Google Scholar] [CrossRef]
- Demirbas, A. Use of algae as biofuel sources. Energy Convers. Manag. 2010, 51, 2738–2749. [Google Scholar] [CrossRef]
- Saad, M.G.; Dosoky, N.S.; Zoromba, M.S.; Shafik, H.M. Algal Biofuels: Current Status and Key Challenges. Energies 2019, 12, 1920. [Google Scholar] [CrossRef]
- Raheem, A.; Prinsen, P.; Vuppaladadiyam, A.K.; Zhao, M.; Luque, R. A review on sustainable microalgae based biofuel and bioenergy production: Recent developments. J. Clean. Prod. 2018, 181, 42–59. [Google Scholar] [CrossRef]
- Priyadarshani, I.; Rath, B. Commercial and industrial applications of micro algae–A review. J. Algal Biomass Utln. 2012, 3, 89–100. [Google Scholar]
- Schnurr, P.J.; Allen, D.G. Factors affecting algae biofilm growth and lipid production: A review. Renew. Sustain. Energy Rev. 2015, 52, 418–429. [Google Scholar] [CrossRef]
- Wahidin, S.; Idris, A.; Shaleh, S.R.M. The influence of light intensity and photoperiod on the growth and lipid content of microalgae Nannochloropsis sp. Bioresour. Technol. 2013, 129, 7–11. [Google Scholar] [CrossRef]
- Lv, J.M.; Cheng, L.H.; Xu, X.H.; Zhang, L.; Chen, H.L. Enhanced lipid production of Chlorella vulgaris by adjustment of cultivation conditions. Bioresour. Technol. 2010, 101, 6797–6804. [Google Scholar] [CrossRef]
- Grima, E.M.; Belarbi, E.H.; Fernández, F.A.; Medina, A.R.; Chisti, Y. Recovery of microalgal biomass and metabolites: Process options and economics. Biotechnol. Adv. 2003, 20, 491–515. [Google Scholar] [CrossRef]
- Deenu, A.; Naruenartwongsakul, S.; Kim, S.M. Optimization and economic evaluation of ultrasound extraction of lutein from Chlorella vulgaris. Biotechnol. Bioprocess Eng. 2013, 18, 1151–1162. [Google Scholar] [CrossRef]
- Pratt, R.; Daniels, T.C.; Eiler, J.J.; Gunnison, J.B.; Kumler, W.D.; Oneto, J.F.; Strait, L.A.; Spoehr, H.A.; Hardin, G.J.; Milner, H.W.; et al. Chlorellin, an antibacterial substance from Chlorella. Science 1944, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, J.S.; Shin, C.S.; Park, S.C.; Kim, S.W. Effects of NO and SO2 on growth of highly-CO2,-tolerant microalgae. J. Microbiol. Biotechnol. 2000, 10, 338–343. [Google Scholar]
- Converti, A.; Casazza, A.A.; Ortiz, E.Y.; Perego, P.; Borghi, M. Del Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem. Eng. Process. 2009, 48, 1146–1151. [Google Scholar] [CrossRef]
- Mayo, A.W. Effects of temperature and pH on the kinetic growth of unialga Chlorella vulgaris cultures containing bacteria. Water Environ. Res. 1997, 69, 6472. [Google Scholar] [CrossRef]
- Szita, N.; Boccazzi, P.; Zhang, Z.; Boyle, P.; Sinskey, A.J.; Jensen, K.F. Development of a multiplexed microbioreactor system for high-throughput bioprocessing. Lab Chip 2005, 5, 819–826. [Google Scholar] [CrossRef]
- Sung, Y.J.; Kim, J.Y.H.; Choi, H.I.; Kwak, H.S.; Sim, S.J. Magnetophoretic sorting of microdroplets with different microalgal cell densities for rapid isolation of fast-growing strains. Sci. Rep. 2017, 7, 10390. [Google Scholar] [CrossRef]
- Deng, Y.L.; Kuo, M.Y.; Juang, Y.J. Development of flow through dielectrophoresis microfluidic chips for biofuel production: Sorting and detection of microalgae with different lipid contents. Biomicrofluidics 2014, 8, 064120. [Google Scholar] [CrossRef]
- Kwak, H.S.; Kim, J.Y.; Na, S.C.; Jeon, N.L.; Sim, S.J. Multiplex microfluidic system integrating sequential operations of microalgal lipid production. Analyst 2016, 141, 1218–1225. [Google Scholar] [CrossRef]
- Lee, D.H.; Bae, C.Y.; Han, J.I.; Park, J.K. In situ analysis of heterogeneity in the lipid content of single green microalgae in alginate hydrogel microcapsules. Anal. Chem. 2013, 85, 8749–8756. [Google Scholar] [CrossRef] [PubMed]
- Bodénès, P.; Wang, H.Y.; Lee, T.H.; Chen, H.Y.; Wang, C.Y. Biotechnology for Biofuels Microfluidic techniques for enhancing biofuel and biorefinery industry based on microalgae. Biotechnol. Biofuels 2019, 1–25. [Google Scholar] [CrossRef]
- Zheng, G.X.; Li, Y.J.; Qi, L.L.; Liu, X.M.; Wang, H.; Yu, S.P.; Wang, Y.H. Marine phytoplankton motility sensor integrated into a microfluidic chip for high-throughput pollutant toxicity assessment. Mar. Pollut. Bull. 2014, 84, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Rhituparna Payel, P. Lab on a Chip systems for environmental analysis. Master’s Thesis, University of Stavanger, Stavanger, Norway, 2014. [Google Scholar]
- Yang, Y.T.; Wang, C. Review of microfluidic photobioreactor technology for metabolic engineering and synthetic biology of cyanobacteria and microalgae. Micromachines 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Abgrall, P.; Gue, A.-M. lab-on-chip technologies: Making a microfluidic network and coupling it into a complete microsystem—A review. J. Micromech. Microeng. 2007, 17, R15–R49. [Google Scholar] [CrossRef]
- BG-11 Medium. Available online: https://utex.org/products/bg-11-medium (accessed on 6 March 2019).
- Fawley, M.W.; Fawley, K.P.; Buchheim, M.A. Molecular diversity among communities of freshwater microchlorophytes. Microb. Ecol. 2004, 48, 489–499. [Google Scholar] [CrossRef]
- Del Campo, E.M.; del Hoyo, A.; Royo, C.; Casano, L.M.; Álvarez, R.; Barreno, E. A single primer pair gives a specific ortholog amplicon in a wide range of Cyanobacteria and plastid-bearing organisms: Applicability in inventory of reference material from collections and phylogenetic analysis. Mol. Phylogenet. Evol. 2010, 57, 1323–1328. [Google Scholar] [CrossRef]
- Kim, H.S.; Waqued, S.C.; Nodurft, D.T.; Devarenne, T.P.; Yakovlev, V.V.; Han, A. Raman spectroscopy compatible PDMS droplet microfluidic culture and analysis platform towards on-chip lipidomics. Analyst 2017, 142, 1054–1060. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with Image J. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Issarapayup, K.; Powtongsook, S.; Pavasant, P. Flat panel airlift photobioreactors for cultivation of vegetative cells of microalga Haematococcus pluvialis. J. Biotechnol. 2009, 142, 227–232. [Google Scholar] [CrossRef]
- Absher, M. Hemocytometer counting. In Tissue Culture: Methods and Applications; Academic Press: New York, NY, USA, 1973; pp. 395–397. [Google Scholar]
- Wheeler, P.A.; Kokkinakis, S.A. Ammonium recycling limits nitrate use in the oceanic subarctic Pacific. Limnol. Oceanogr. 1990, 35, 1267–1278. [Google Scholar] [CrossRef]
- Shafik, H.M.; Saad, M.G.; El-Serehy, H.A. Impact of nitrogen regime on fatty acid profiles of Desmodesmus quadricaudatus and Chlorella sp. and ability to produce biofuel. Acta Bot. Hung. 2015, 57, 205–218. [Google Scholar] [CrossRef]
- Ogbonna, J.C.; Masui, H.; Tanaka, H. Sequential heterotrophic/autotrophic cultivation - An efficient method of producing Chlorella biomass for health food and animal feed. J. Appl. Phycol. 1997, 9, 359–366. [Google Scholar] [CrossRef]
- Yun, Y.S.; Park, J.M. Attenuation of monochromatic and polychromatic lights in Chlorella vulgaris suspensions. Appl. Microbiol. Biotechnol. 2001, 55, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Cepak, V.; Pribyl, P.; Vitova, M. The effect of light color on the nucleocytoplasmic and chloroplast cycle of the green chlorococcal alga Scenedesmus obliquus. Folia Microbiol. (Praha) 2006, 51, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Graham, P.J.; Sinton, D. Dual gradients of light intensity and nutrient concentration for full-factorial mapping of photosynthetic productivity. Lab Chip 2016, 16, 2785–2790. [Google Scholar] [CrossRef] [PubMed]
- Van Wagenen, J.; Miller, T.W.; Hobbs, S.; Hook, P.; Crowe, B.; Huesemann, M. Effects of light and temperature on fatty acid production in Nannochloropsis salina. Energies 2012, 5, 731–740. [Google Scholar] [CrossRef]
- Kautsky, H.; Hirsch, A. Neue versuche zur kohlensäureassimilation. Naturwissenschaften 1931, 19, 964. [Google Scholar] [CrossRef]
- Erickson, R.A.; Jimenez, R. Microfluidic cytometer for high-throughput measurement of photosynthetic characteristics and lipid accumulation in individual algal cells. Lab Chip 2013, 13, 2893–2901. [Google Scholar] [CrossRef]
- Daliry, S.; Hallajsani, A.; Mohammadi Roshandeh, J.; Nouri, H.; Golzary, A. Investigation of optimal condition for Chlorella vulgaris microalgae growth. Glob. J. Environ. Sci. Manag. 2017, 3, 217–230. [Google Scholar]
- Liu, Z.Y.; Wang, G.C.; Zhou, B.C. Effect of iron on growth and lipid accumulation in Chlorella vulgaris. Bioresour. Technol. 2008, 99, 4717–4722. [Google Scholar] [CrossRef] [PubMed]
- Scarsella, M.; Belotti, G.; De Filippis, P.; Bravi, M. Study on the optimal growing conditions of Chlorella vulgaris in bubble column photobioreactors. Chem. Eng. 2010, 20, 85–90. [Google Scholar]
- Jeanfils, J.; Canisius, M.F.; Burlion, N. Effect of high nitrate concentrations on growth and nitrate uptake by free-living and immobilized Chlorella vulgaris cells. J. Appl. Phycol. 1993, 5, 369–374. [Google Scholar] [CrossRef]
- Kim, H.S.; Weiss, T.L.; Thapa, H.R.; Devarenne, T.P.; Han, A. A microfluidic photobioreactor array demonstrating high-throughput screening for microalgal oil production. Lab Chip 2014, 14, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Dewan, A.; Kim, J.; McLean, R.H.; Vanapalli, S.A.; Karim, M.N. Growth kinetics of microalgae in microfluidic static droplet arrays. Biotechnol. Bioeng. 2012, 109, 2987–2996. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.J.; Kim, J.Y.; Bong, K.W.; Sim, S.J. Microdroplet photobioreactor for the photoautotrophic culture of microalgal cells. Analyst 2016, 141, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jeong, S.N.; Kim, B.; Kim, D.P.; Cho, Y.K. Rapid and automated quantification of microalgal lipids on a spinning disc. Anal. Chem. 2015, 87, 7865–7871. [Google Scholar] [CrossRef]
- Kuntanawat, P.; Ruenin, J.; Phatthanakun, R.; Kunhorm, P.; Surareungchai, W.; Sukprasong, S.; Chomnawang, N. An electrostatic microwell–based biochip for phytoplanktonic cell trapping. Biomicrofluidics 2014, 8, 034108. [Google Scholar] [CrossRef]
- Pan, J.; Stephenson, A.L.; Kazamia, E.; Huck, W.T.; Dennis, J.S.; Smith, A.G.; Abell, C. Quantitative tracking of the growth of individual algal cells in microdroplet compartments. Integr. Biol. 2011, 3, 1043–1051. [Google Scholar] [CrossRef]
- Perin, G.; Cimetta, E.; Monetti, F.; Morosinotto, T.; Bezzo, F. Novel micro-photobioreactor design and monitoring method for assessing microalgae response to light intensity. Algal Res. 2016, 19, 69–76. [Google Scholar] [CrossRef]


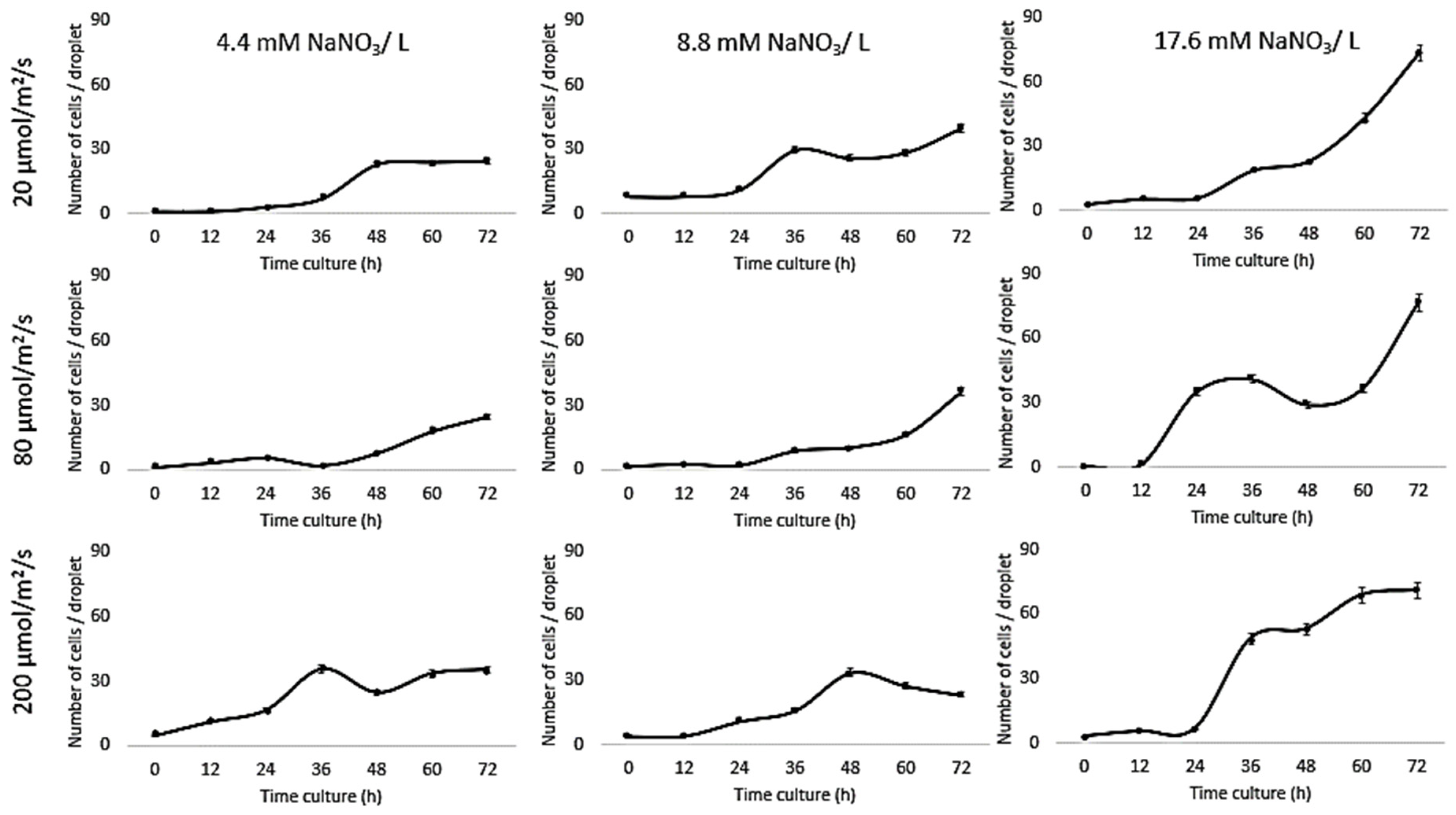
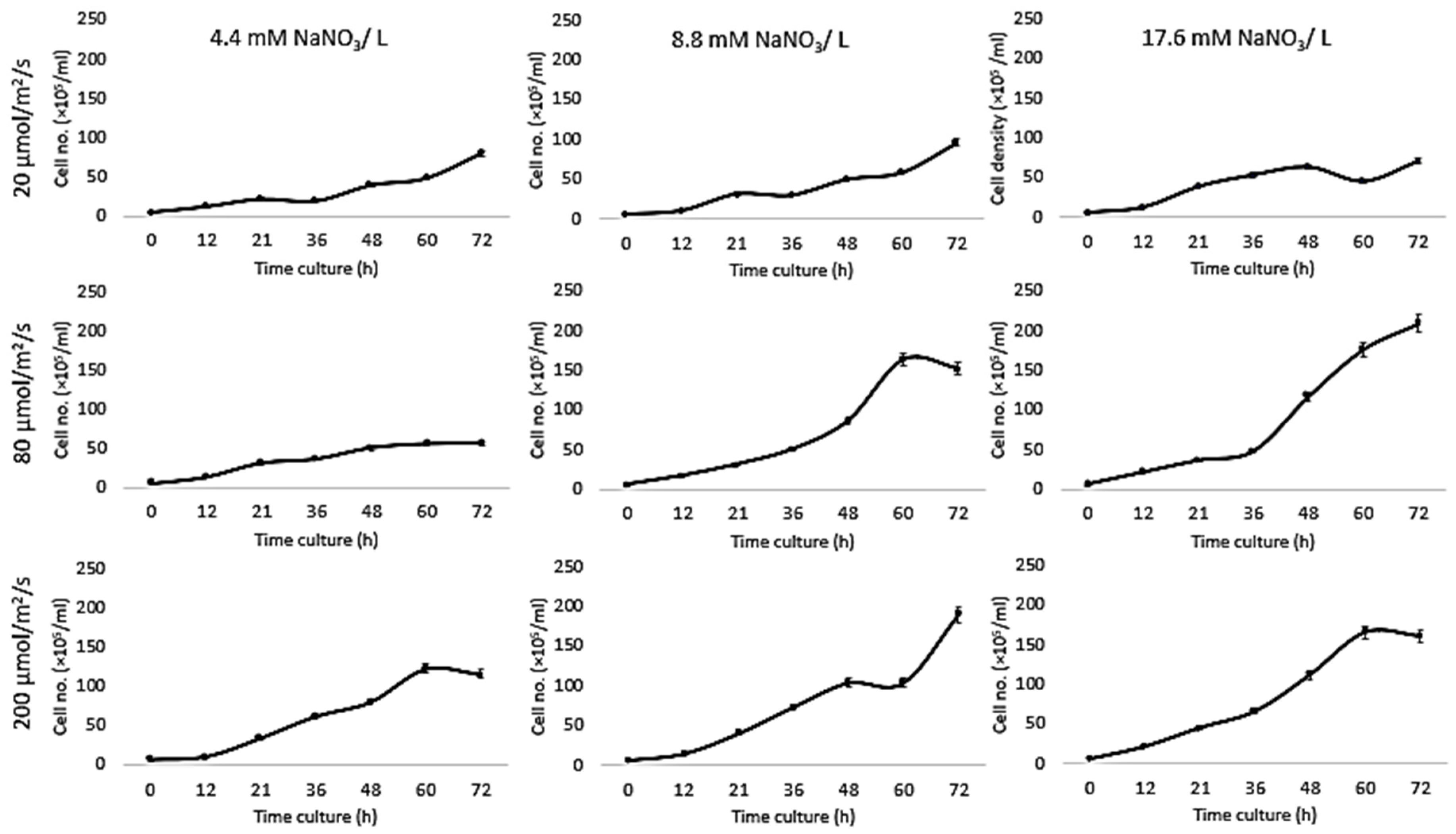

| Light Intensity (µmol/m2/s) | Nitrogen Condition (mM) | Max. Growth Rate (per h) | |
|---|---|---|---|
| Microfluidic Culture (mean/SD) | Traditional Batch Culture (×104) (mean/SD) | ||
| 20 | 4.4 | 0.10 ± 0.01 | 0.06 ± 0.002 |
| 8.8 | 0.08 ± 0 | 0.12 ± 0.02 | |
| 17.6 | 0.05 ± 0.01 | 0.12 ± 0.006 | |
| 80 | 4.4 | 0.10 ± 0.01 | 0.09 ± 0 |
| 8.8 | 0.07 ± 0.04 | 0.05 ± 0.015 | |
| 17.6 | 0.27 ± 0.05 | 0.11 ± 0.009 | |
| 200 | 4.4 | 0.06 ± 0.003 | 0.06 ± 0 |
| 8.8 | 0.06 ± 0.01 | 0.05 ± 0.007 | |
| 17.6 | 0.17 ± 0.01 | 0.09 ± 0.01 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saad, M.G.; Dosoky, N.S.; Khan, M.S.; Zoromba, M.S.; Mekki, L.; El-Bana, M.; Nobles, D.; Shafik, H.M. High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions. Biomolecules 2019, 9, 276. https://doi.org/10.3390/biom9070276
Saad MG, Dosoky NS, Khan MS, Zoromba MS, Mekki L, El-Bana M, Nobles D, Shafik HM. High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions. Biomolecules. 2019; 9(7):276. https://doi.org/10.3390/biom9070276
Chicago/Turabian StyleSaad, Marwa Gamal, Noura Sayed Dosoky, Muhammad Shuja Khan, Mohamed Shafick Zoromba, Laila Mekki, Magdy El-Bana, David Nobles, and Hesham Mohamed Shafik. 2019. "High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions" Biomolecules 9, no. 7: 276. https://doi.org/10.3390/biom9070276
APA StyleSaad, M. G., Dosoky, N. S., Khan, M. S., Zoromba, M. S., Mekki, L., El-Bana, M., Nobles, D., & Shafik, H. M. (2019). High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions. Biomolecules, 9(7), 276. https://doi.org/10.3390/biom9070276





