Abstract
At the end of its life cycle, the cellular slime mold Dictyostelium discoideum forms a fruiting body consisting of spores and a multicellular stalk. Originally, the chlorinated alkylphenone differentiation-inducing factors (DIFs) -1 and -3 were isolated as stalk cell inducers in D. discoideum. Later, DIFs and their derivatives were shown to possess several biologic activities including antitumor and anti-Trypanosoma properties. In this study, we examined the antibacterial activities of approximately 30 DIF derivatives by using several bacterial species. Several of the DIF derivatives strongly suppressed the growth of the Gram-positive bacteria Staphylococcus aureus, Bacillus subtilis, and Enterococcus faecalis and Enterococcus faecium, at minimum inhibitory concentrations (MICs) in the sub-micromolar to low-micromolar range. In contrast, none of the DIF derivatives evaluated had any noteworthy effect on the growth of the Gram-negative bacterium Escherichia coli (MIC, >100 µM). Most importantly, several of the DIF derivatives strongly inhibited the growth of methicillin-resistant S. aureus and vancomycin-resistant E. faecalis and E. faecium. Transmission electron microscopy revealed that treatment with DIF derivatives led to the formation of distinct multilayered structures consisting of cell wall or plasma membrane in S. aureus. The present results suggest that DIF derivatives are good lead compounds for developing novel antimicrobials.
1. Introduction
The cellular slime mold Dictyostelium discoideum has long been studied as an excellent model system in the fields of cell and developmental biology; at the end of its developmental process, this organism forms fruiting bodies, each consisting of spores and a multicellular stalk [1,2,3]. Differentiation-inducing factor 1 (DIF-1) (Figure 1A) is a chlorinated polyketide that induces stalk cell differentiation [4,5] and modulates cell chemotaxis during the development of D. discoideum [6]. DIF-3 (Figure 1A) is a metabolite of DIF-1 and has virtually no biologic activity in D. discoideum [5,6,7,8].
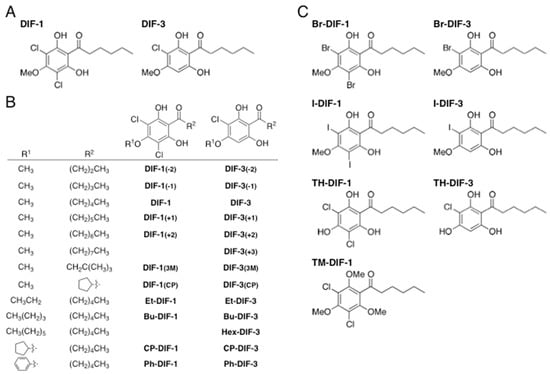
Figure 1.
(A) Chemical structures of Dictyostelium discoideum differentiation-inducing factor (DIF) 1 and DIF-3. (B, C) Chemical structures of DIF derivatives used in this study.
In addition to these developmental roles, DIF-1, DIF-3, and several of their derivatives (Figure 1B,C) exert multiple biologic activities [9]. In particular, several derivatives demonstrate strong anti-proliferative and anti-metastatic activities in tumor cells in vitro and in vivo [10,11,12,13,14,15,16,17,18,19,20,21,22]. Some DIF derivatives increase glucose consumption in non-transformed mammalian cells in vitro and in vivo [23,24,25], and others show immunomodulatory effects in T cells in vitro [26,27,28]. Furthermore, we found several DIF derivatives that display strong in vitro and in vivo effects against Trypanosoma cruzi, the protozoan parasite that causes Chagas disease (human American trypanosomiasis) [29]. Importantly, modifying the side chains of DIF derivatives might provide a means to isolate individual biologic activities [16,22,23,24,26,27,28], such that these biologics might become candidate lead compounds for developing novel anti-cancer, anti-diabetic, immunomodulatory, and anti-Trypanosoma drugs [9]. Moreover, the results of previous studies lead us to expect that some DIF derivatives may possess as yet unidentified biologic effects.
In this study, we investigated the antibacterial activities of DIF derivatives in vitro and show that several of them exert strong antibacterial effects against Gram-positive bacteria, including Staphylococcus aureus, Bacillus subtilis, Enterococcus faecalis and, Enterococcus faecium. In addition, several of the DIF derivatives evaluated strongly inhibited the growth of methicillin-resistant S. aureus and vancomycin-resistant enterococci (i.e., strains of E. faecalis and E. faecium). Our results support the investigation of such DIF derivatives as candidate lead compounds for developing novel antimicrobials.
2. Materials and Methods
2.1. Bacterial Species and Reagents
The Gram-positive bacteria methicillin-susceptible S. aureus (MSSA; strains 209P and ATCC29213), methicillin-resistant S. aureus (MRSA; MS29202 and ATCC43300), vancomycin-susceptible E. faecalis (VSE; ATCC29212), vancomycin-resistant E. faecalis (VRE: VanB; ATCC51299), vancomycin-resistant E. faecium (VRE: VanA; ATCC700221), and B. subtilis (ATCC6633) and the Gram-negative bacteria Escherichia coli (NIHJ) and Mycobacterium bovis (PG45; ATCC25523) were used in this study. DIF derivatives (Figure 1) were synthesized as previously described [16,22] and stored at −20 °C as 10 mM solutions in DMSO or ethanol (EtOH). AB0022A, Pf-1, and Pf-2 were synthesized as described [30,31] and stored at −20 °C as 10 mM solutions in DMSO. A combined solution of penicillin (5000 units/mL) and streptomycin (5 mg/mL) was purchased from Gibco (ThermoFisher Scientific, Waltham, MA, USA); vancomycin and oxacillin were obtained from Sigma-Aldrich (St. Louis, MO, USA); and cefoxitin and ampicillin were bought from Wako Pure Chemical Industries (Osaka, Japan). The hydrophobic index (ClogP) of each DIF derivative was calculated by using ChemDraw 16.0 software (PerkinElmer, Inc. Waltham, MA, USA).
2.2. Paper-Disc Diffusion Assay
We mixed 0.1 mL of S. aureus suspension (109 CFU/mL) in Mueller–Hinton broth (MHB) (0.2% (w/v) beef extract, 1.75% (w/v) casein hydrolysate, 0.15% (w/v) starch) into 10 mL of MHB agar (MHB containing 1.7% (w/v) agar; final bacterial density, 107 CFU/mL) and spread this suspension in a plastic culture dish (diameter, 100 mm). Paper discs (diameter, 8 mm; Advantec, Tokyo, Japan) impregnated with 2 µL of EtOH, DMSO, a 10 mM solution of the DIF or DIF derivative of interest in EtOH or DMSO (approx. 5.5–8 µg DIF/disc), or the purchased penicillin–streptomycin solution (10 units penicillin and 10 µg streptomycin/disc) were placed on the bacterial agar plate. In addition, paper discs (KB disc ‘EIKEN’; EIKEN Chemical Company, Tokyo, Japan) containing minomycin (30 µg/disc), imipenem (10 µg/disc), gentamicin (10 µg/disc), levofloxacin (5 µg/disc), or clindamycin (2 µg/disc) were placed on a MHB bacterial agar plate as controls. The plates were incubated for 20 h at 37 °C, after which the bacterial growth-inhibition zones around each disc was noted.
In addition, a diffusion assay without paper discs was performed for comparison. Briefly, 10 mL of S. aureus in MHB (107 CFU/mL) was spread on an MHB agar plate (diameter, 100 mm), after which 2 µL of each 10 mM DIF solution was dropped directly on the agar surface. The plates were incubated for 20 h at 37 °C, after which the bacterial growth-inhibition zones around the discs were noted.
2.3. Measurement of Minimum Inhibitory Concentration
Gram-positive and Gram-negative bacteria in MHB (5 × 105 CFU/mL; 0.1 mL/well) were incubated for 20 to 24 h at 37 °C in 96-well plates (Corning, NY, USA) in the presence of vehicle, various concentrations of serially diluted DIF derivatives, or known antibiotics; MIC was defined as the lowest concentration of the additives that inhibited visible bacterial growth.
M. bovis was grown in Hayflick modified PPLO (pleuropneumonia-like organisms) broth (0.5% (w/v) glucose, 5% (w/v) beef heart infusion, 2.5% (w/v) yeast extract, 1% (w/v) peptone, 0.5% (w/v) NaCl, 15% (v/v) heat-inactivated horse serum, 50 µg/mL of ampicillin) and used at concentrations of 104 to 107 CFU/mL for the minimum inhibitory concentration (MIC) assay as described; tiamulin (Santa Cruz Biotechnology, Dallas, TX, USA) and enrofloxacin (Sigma-Aldrich) were used as positive-control antibiotics.
2.4. Transmission Electron Microscopy
Specimens for transmission electron microscopy were prepared according to standard procedures [32]. Briefly, S. aureus (strain ATCC29213) in MHB (1–5 × 108 CFU/mL; 2 mL per well) was incubated for 1.5 h at 37 °C in 6-well plates (Corning) in the presence of 0.2% DMSO (vehicle) or 4 µM DIF derivative. After incubation, the bacteria were transferred to centrifuge tubes, collected by centrifugation (4000 × g, 1 min), fixed overnight at 4 °C in 0.5 mL of 2.5% (v/v) glutaraldehyde in 100 mM phosphate buffer (pH 7.4) (LSI Medience Corporation, Tokyo, Japan), and postfixed in 1% (w/v) OsO4 in 100 mM phosphate buffer. Fixed specimens were dehydrated through a graded series of ethanol and embedded in Epon812 (Oken-Shoji, Tokyo, Japan). Ultrathin sections were cut by using an ultramicrotome (model UC6; Leica, Wetzlar, Germany) and stained with uranyl acetate and lead citrate. These sections were examined under a transmission electron microscope (model HT7700; Hitachi High-Tech, Tokyo, Japan).
3. Results
3.1. Antimicrobial Effects of DIFs and Their Derivatives on S. aureus in Agar Plates
We first examined the effects of DIF-1, DIF-3 (Figure 1A), and their derivatives (Figure 1B,C) on the growth of methicillin-susceptible S. aureus (MSSA: strain 209P) (Figure 2A) and methicillin-resistant S. aureus (MRSA: strain MS29202) (Figure 2B) by using a paper-disc diffusion assay. In MSSA (Figure 2A), a large ring free of bacterial growth formed around the disc containing penicillin and streptomycin, as expected. Similar growth-free rings developed around the discs containing DIF-1 and several DIF-1 derivatives but not DIF-3. In MRSA-containing plates (Figure 2B), penicillin and streptomycin failed to inhibit bacterial growth. However, DIF-1 and several DIF-1 derivatives, but not DIF-3, again produced zones free of bacterial growth. Although the MRSA strain we used was susceptible to minomycin, it was resistant to imipenem, levofloxacin, and clindamycin and was marginally resistant to gentamicin (Figure 2C). These results indicate that various DIFs and their derivatives possess antimicrobial activity against S. aureus. In addition, the mechanism underlying the antimicrobial action of DIFs likely differs (at least in part) from those of the known antibiotics that we used.
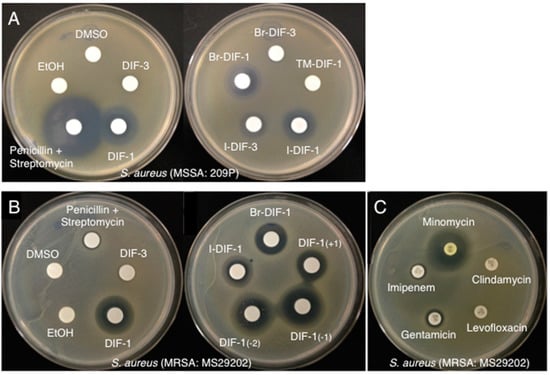
Figure 2.
(A) Effects of DIF derivatives on the growth of methicillin-susceptible Staphylococcus aureus (MSSA) on agar. Paper discs were placed on Mueller–Hinton broth (MHB) agar plates that contained MSSA (209P); paper discs were impregnated with 2 µL of either ethanol (EtOH; vehicle), DMSO (vehicle), a 10 mM solution of the indicated DIF or DIF derivative, or a mixture of penicillin (5000 units/mL) and streptomycin (5 mg/mL). After incubation for 20 h at 37 °C, bacterial growth-inhibition zones around the discs were noted. (B, C) Effects of DIF derivatives on the growth of methicillin-resistant S. aureus (MRSA) on agar. Paper discs were placed on MHB agar plates that contained MRSA (MS29202); these discs were impregnated with 2 µL of either EtOH (vehicle), DMSO (vehicle), a 10 mM solution of the indicated DIF or DIF derivative, or a mixture of penicillin and streptomycin (B). In addition, paper discs containing the indicated antibiotics were placed on MRSA agar plates (C). After incubation for 20 h at 37 °C, bacterial growth-inhibition zones around the discs were noted.
3.2. MIC Values of DIF Derivatives in S. aureus, B. subtilis, E. coli, and M. bovis
We next examined the effects of DIF derivatives on the growth of the Gram-positive bacteria MSSA (strain 209P), MRSA (MS29202), and B. subtilis (ATCC6633) and on the Gram-negative bacterium E. coli (NIHJ), and MIC values of DIF derivatives were determined (Table 1). Several of the DIF derivatives showed strong antibacterial activity against the Gram-positive bacteria, yielding MIC values of less than 1 µM. However, none of the DIF derivatives that we tested inhibited the growth of E. coli and M. bovis (MIC, >100 µM) (Table 1); M. bovis was sensitive to the antibiotics tiamulin (MIC, 0.5 µM) and enrofloxacin (0.7 µM).

Table 1.
ClogP and minimum inhibitory concentration (MIC) (µM) values of DIFs and DIF derivatives against MSSA, MRSA, Bacillus subtilis, Escherichia coli, and Mycobacterium bovis.
Note that the MIC values of DIF-3 against MSSA and MRSA were comparable to those of DIF-1 (Table 1), even though DIF-3 lacked antibacterial activity against these strains in the paper-disc diffusion assay (Figure 2). These results suggest that DIF-3 and its derivatives exert antibacterial activity in solution but become ineffective on solid media (e.g., paper disc, agar). To examine this hypothesis, we compared the effects of DIF-1, DIF-3, and three DIF-3 derivatives on the growth of MSSA and MRSA by using DIF-impregnated paper discs or by spotting the DIF solutions directly on the surface of the bacteria-containing agar (Figure 3). Again, DIF-1—but not DIF-3 or its derivatives—showed a strong antibacterial activity against MSSA and MRSA in the paper-disc diffusion assay (Figure 3A,C). However, when the solutions were dropped directly on the agar, all of the DIF derivatives we tested showed antibacterial activity against the bacteria (Figure 3B,C). These results suggest that DIF-3 and its derivatives are likely to diffuse poorly through some matrixes, including agar and paper.
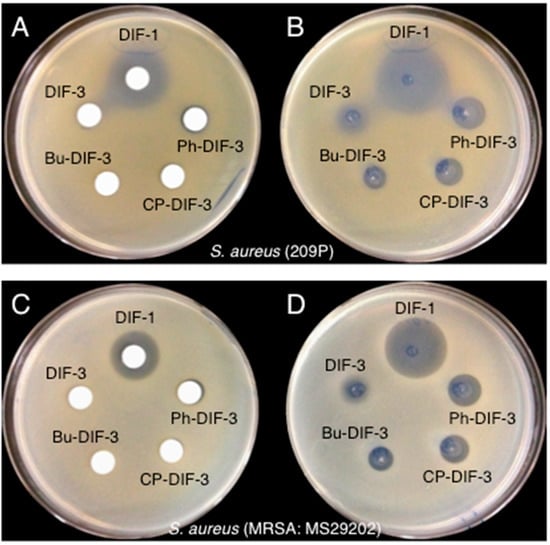
Figure 3.
Effects of DIF derivatives on the growth of MSSA and MRSA on agar. Paper discs were placed on MHB agar plates that contained (A) MSSA (strain 209P) or (C) MRSA (MS29202); each paper disc was impregnated with 2 µL of a 10 mM solution of the indicated DIF derivative. In parallel, 2 µL of a 10 mM solution of the indicated DIF derivative was placed directly on MHB agar plates that contained (B) MSSA (209P) or (D) MRSA (MS29202). After incubation for 20 h at 37 °C, bacterial growth-inhibition zones around the discs were noted.
3.3. MIC Values of DIF Derivatives in S. aureus, E. faecalis, and E. faecium
We then further assessed the potential antibacterial activities of representative DIF derivatives (Figure 1) on the growth of MSSA (strain ATCC29213), MRSA (ATCC43300), VSE (ATCC29212), and VREs (ATCC51299 and ATCC700221); the latter two strains carry the vancomycin-resistant genes vanB and vanA, respectively (Table 2). Again, several DIF derivatives showed strong antibacterial activity against MSSA and MRSA (S. aureus strains different from those in Table 1), with MIC values of less than 2 µM (Table 2). In addition, various DIF derivatives demonstrated strong antibacterial effects against VSE and two VRE strains, with associated MIC values of less than 2 µM (Table 2); note that the MRSA strain was highly resistant to cefoxitin and oxacillin, and the two VREs were highly or intermediately resistant to vancomycin and ampicillin (Table 2).

Table 2.
MIC (µM) values of DIFs and derivatives against MSSA, MRSA, VSE, and VRE.
These results indicate that DIF derivatives may possess strong antibacterial activities against a broad range of Gram-positive bacteria, including known drug-resistant bacteria such as MRSAs and VREs.
3.4. Effects of DIF Derivatives on the Ultracellular Structure of S. aureus
To help elucidate the mechanism underlying the antibacterial activity of DIF derivatives, we examined the effects of Ph-DIF-1, Ph-DIF-3, and Bu-DIF-3 on the ultracellular structure of MSSA (strain ATCC29213). Transmission electron microscopy revealed distinct multilayered structures consisting of cell wall and/or plasma membrane in the DIF-treated MSSA cells (Figure 4). This finding suggests that the tested DIF derivatives suppress the growth of S. aureus by hindering cell wall formation.
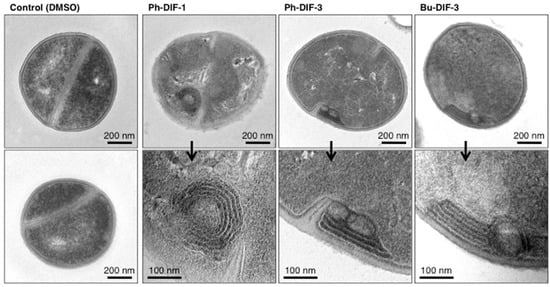
Figure 4.
Effects of DIF derivatives on the ultracellular structure of S. aureus (MSSA). MSSA (ATCC29213) was incubated for 1.5 h at 37 °C in MHB in the presence of 0.2% DMSO (vehicle) or 4 µM of Ph-DIF-1, Ph-DIF-3, or Bu-DIF-3. The cells were collected, treated as described in the Materials and Methods section, and observed under a transmission electron microscope. Two representative photos of the DMSO-treated control MSSA and two representative photos of each DIF-treated MSSA are shown. The DIF-treated MSSA samples each show distinct multilayered structures composed of cell wall and/or plasma membrane.
3.5. Comparison of the Antibacterial Effects of Chlorinated Dibenzofurans and DIFs on S. aureus
The antibacterial substance AB0022A, a chlorinated dibenzofuran (Figure 5A), was isolated from D. purpureum [30] and is similar in structure to DIF-1. Like DIF-1 and several of its derivatives, AB0022A inhibits the growth of Gram-positive bacteria; its MIC values against S. aureus (strain IID671), MRSA (IID1677), E. faecalis (ATCC29212), and B. subtilis (ATCC6633) are 1.56 µg/mL (3.4 µM), 3.13 µg/mL (6.8 µM), 50 µg/mL (109 µM), and 0.78 µg/mL (1.7 µM), respectively [30], and thus are at least somewhat comparable to those of DIF derivatives (Table 1 and Table 2). In addition, two other chlorinated dibenzofurans, Pf-1 and Pf-2 (Figure 5A), have been isolated from the cellular slime mold Polysphondylium filamentosum [31], but their antibacterial activities have not previously been examined.
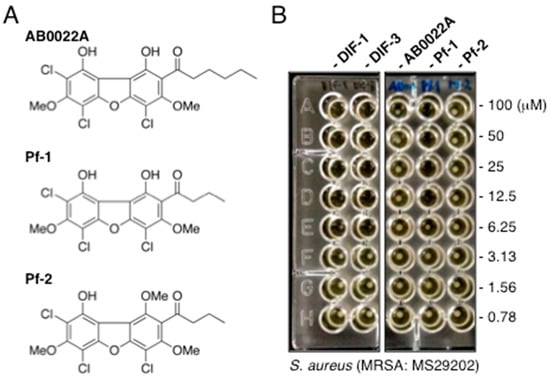
Figure 5.
(A) Chemical structures of the chlorinated dibenzofurans AB0022A, Pf-1, and Pf-2. (B) Effects of chlorinated dibenzofurans and DIFs on the growth of S. aureus (MRSA). MRSA (MS29202) was incubated for 20 h at 37 °C in the presence of the indicated concentrations of DIF-1, DIF-3, AB0022A, Pf-1, or Pf-2, after which colony formation was noted for determining the MICs of the compounds.
When we assessed the MIC values of these dichlorinated dibenzofurans in MRSA (strain MS29202), neither AB0022A nor Pf-2 had any noteworthy inhibitory effect (MIC, >100 µM) on the growth of this strain (Figure 5B). In contrast, Pf-1 suppressed the growth of this MRSA at a MIC value of 12.5 µM, which was comparable to those of DIF-1 and DIF-3 (25 and 12.5 µM, respectively). The results suggest that DIF derivatives such as CP-DIF-3 and Ph-DIF-3 (Table 1 and Table 2) are likely better lead antibacterial drugs than the chlorinated dibenzofurans we evaluated.
4. Discussion
Most of the many antibiotics (antimicrobials) identified since the discovery of penicillin in the fungus Penicillium notatum have been isolated from fungi and Actinobacteria [33,34,35,36]. However, due to intensive use, overuse, or the use of subtherapeutic concentrations of antibiotics, the number of drug-resistant bacteria, such as MRSAs and VREs, is growing. A search for new antibiotic molecules and bio-resources that produce novel antimicrobials is required [9,35,36,37].
The cellular slime mold D. discoideum has long been used as a model organism in the study of cell and developmental biology. However, slime molds have recently been identified as excellent sources of potential lead compounds for drug discovery and the development of novel medicines [9,38]. In this regard, we have focused on elucidating the biologic and pharmacologic activities (such as antitumor and antiparasitic effects) of the D. discoideum-derived DIFs and their derivative compounds in various types of eukaryotic cells [9,16,29].
In the present study, we showed that several D. discoideum DIF derivatives possess antibacterial activity against Gram-positive bacteria, including S. aureus, E. faecalis, E. faecium, and B. subtilis. In contrast, these DIF derivatives did not inhibit the growth of Mycoplasma or the Gram-negative bacterium E. coli (Table 1 and Table 2). Most importantly, several of the DIF derivatives we evaluated strongly suppressed the growth of MRSAs and VREs (Table 1 and Table 2). Note that the MRSA strain MS29202 was resistant not only to penicillin and streptomycin (Figure 2B) but also to clindamycin, levofloxacin, and imipenem (Figure 2C), whereas the MRSA strain (ATCC43300) was resistant to both cefoxitin and oxacillin (Table 2). In addition, the VRE isolates E. faecalis (which harbors the vancomycin-resistance gene vanB) and E. faecium (containing vanA) were resistant to both vancomycin and ampicillin (Table 2). Although they are primarily opportunistic pathogens, E. faecalis and E. faecium cause the great majority of enterococcal infections, and isolates that carry drug-resistance genes such as vanA or vanB can cause serious infections [39,40]. Overall, our results suggest that the mechanism underlying the antibacterial actions of DIF derivatives likely differ (at least in part) from those of the known antibiotics we assessed. Consequently, DIF derivatives are promising lead compounds for novel antimicrobials against a broad range of Gram-positive bacteria, including known drug-resistant strains.
Although the antibacterial activities of the DIF derivatives varied among the Gram-positive strains of bacteria examined in this study (Table 1 and Table 2), the structure–activity relationship analysis revealed that compounds with longer alkyl chains at the acyl group (e.g., DIF-3(+2) and DIF-3(+3)) and compounds with larger alkyl groups in place of the methyl group (e.g., Bu-DIF-3, Hex-DIF-3, and Ph-DIF-3) had greater antibacterial activities against Gram-positive strains than did those with shorter alkyl chains or smaller alkyl groups, and that compounds with one chlorine substituted in the benzene ring had greater antibacterial activities against Gram-positive strains than did those with two chlorines substituted in the benzene ring. In contrast, there was no clear overall relationship between hydrophobicity (assessed as ClogP value) and antibacterial activity (Table 1).
Regarding the mechanism through which DIF derivatives exert their antibacterial effects, DIF derivatives have mitochondrial uncoupling activity in mammalian cells [17,18]. Given that mitochondria and bacteria are similar in many aspects, these previous findings suggest that DIF derivatives may affect bacterial proton transport (or intracellular pH) and thereby suppress bacterial growth. However, the strength of the mitochondrial uncoupling properties of DIF derivatives [17,18] does not necessarily correlate with the strength of their antibacterial activities (Table 1 and Table 2). Furthermore, in the present study, we showed that in S. aureus, Ph-DIF-1, Ph-DIF-3, and Bu-DIF-3 induce the formation of distinct multilayered structures composed of cell wall or membrane (Figure 4). Although the relationship between the antibacterial effects of DIFs and the DIF-induced formation of these structures is unclear currently, this observation may be a clue for elucidating the mechanism underlying the antibacterial activity of DIF derivatives.
5. Conclusions
In this study, we showed that several derivatives of DIF-1 and DIF-3, chlorinated polyketides found in D. discoideum possess strong antimicrobial activities against Gram-positive bacteria including MRSAs and VREs. Our results suggest that the DIF derivatives are good lead compounds for developing novel antimicrobials.
6. Patents
The following authors hold a patent related to this article:
Kubohara, Y.; Kikuchi, H.; Oshima, Y. Antibacterial drugs. Japanese Patent No. 6478378, 15 February 2019.
Author Contributions
Conceptualization, Y.K.; formal analysis, Y.K., Y.S., H.I., K.T., Y.O. and H.K.; funding acquisition, Y.K. and Y.O.; investigation, Y.K., Y.S., H.I., K.T., Y.O. and H.K.; project administration, Y.K.; resources, Y.O. and H.K.; Supervision, Y.K.; writing—original draft preparation, Y.K. and H.K.; writing—review and editing, Y.K., Y.S., K.T., Y.O., and H.K.
Funding
This work was supported in part by JSPS KAKENHI Grants (nos. 24590110 and 15K07964 to YK and YO) and by grants from the Institute for Fermentation, Osaka (IFO) (to YK), the Hamaguchi Foundation for Advancement of Biochemistry (to YK), the Shimabara Science Promotion Foundation (to YK), and the Joint Research Program of Juntendo University, Faculty of Health and Sports Science (to YK).
Acknowledgments
We thank the members of the Laboratory of Morphology and Image Analysis (Research Support Center, Juntendo University Graduate School of Medicine) for their technical assistance with microscopy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Raper, K.B. Dictyostelium discoideum, A new species of slime mold from decaying forest leaves. J. Agr. Res. 1935, 55, 289–316. [Google Scholar]
- Newell, P.C. The development of the cellular slime mould Dictyostelium discoideum: A model system for the study of cellular differentiation. Essays Biochem. 1971, 7, 87–126. [Google Scholar] [PubMed]
- Darmon, M.; Brachet, P.; Pereira da Silva, L.H. Chemotactic signals induce cell differentiation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 1975, 72, 3163–3166. [Google Scholar] [CrossRef]
- Morris, H.R.; Taylor, G.W.; Masento, M.S.; Jermyn, K.A.; Kay, R.R. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature 1987, 328, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Kay, R.R.; Berks, M.; Traynor, D. Morphogen hunting in Dictyostelium discoideum. Development 1989, 107 Suppl. 81–90. [Google Scholar]
- Kuwayama, H.; Kubohara, Y. Differentiation-inducing factor-1 and -2 function also as modulators for Dictyostelium chemotaxis. PLoS ONE 2009, 4, e6658. [Google Scholar] [CrossRef] [PubMed]
- Morris, H.R.; Masento, M.S.; Taylor, G.W.; Jermyn, K.A.; Kay, R.R. Structure elucidation of two differentiation inducing factors (DIF-2 and DIF-3) from the cellular slime mould Dictyostelium discoideum. Biochem. J. 1988, 249, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Kay, R.R.; Flatman, P.; Thompson, C.R.L. DIF signalling and cell fate. Semin. Cell Dev. Biol. 1999, 10, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Kubohara, Y.; Kikuchi, H. Dictyostelium: An important source of structural and functional diversity in drug discovery. Cells 2019, 8, 6. [Google Scholar] [CrossRef]
- Asahi, K.; Sakurai, A.; Takahashi, N.; Kubohara, Y.; Okamoto, K.; Tanaka, Y. DIF-1, morphogen of Dictyostelium discoideum, induces the erythroid differentiation in murine and human leukemia cells. Biochem. Biophys. Res. Commun. 1995, 208, 1036–1039. [Google Scholar] [CrossRef]
- Kubohara, Y.; Saito, Y.; Tatemoto, K. Differentiation-inducing factor of D. discoideum raises intracellular calcium concentration and suppresses cell growth in rat pancreatic AR42J cells. FEBS Lett. 1995, 359, 119–122. [Google Scholar] [CrossRef]
- Kubohara, Y. DIF-1, putative morphogen of D. discoideum, suppresses cell growth and promotes retinoic acid-induced cell differentiation in HL-60. Biochem. Biophys. Res. Commun. 1997, 236, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Kubohara, Y. Effects of differentiation-inducing factors (DIFs) of Dictyostelium discoideum on the human leukemia K562 cells: DIF-3 is the most potent anti-leukemic agent. Eur. J. Pharmacol. 1999, 381, 57–62. [Google Scholar] [CrossRef]
- Kanai, M.; Konda, Y.; Nakajima, T.; Izumi, Y.; Nanakin, A.; Kanda, N.; Kubohara, Y.; Chiba, T. Differentiation-inducing factor-1 (DIF-1) inhibits STAT3 activity involved in gastric cancer cell proliferation via MEK-ERK dependent pathway. Oncogene 2003, 22, 548–554. [Google Scholar] [CrossRef]
- Takahashi-Yanaga, F.; Taba, Y.; Miwa, Y.; Kubohara, Y.; Watanabe, Y.; Hirata, M.; Morimoto, S.; Sasaguri, T. Dictyostelium differentiation-inducing factor-3 activates glycogen synthase kinase-3β and degrades cyclin D1 in mammalian cells. J. Biol. Chem. 2003, 278, 9663–9670. [Google Scholar] [CrossRef]
- Gokan, N.; Kikuchi, H.; Nakamura, K.; Oshima, Y.; Hosaka, K.; Kubohara, Y. Structural requirements of Dictyostelium differentiation-inducing factors for their stalk-cell-inducing activity in Dictyostelium cells and anti-proliferative activity in K562 human leukemic cells. Biochem. Pharmacol. 2005, 70, 676–685. [Google Scholar] [CrossRef]
- Kubohara, Y.; Kikuchi, H.; Matsuo, Y.; Oshima, Y.; Homma, Y. Mitochondria are the target organelle of differentiation-inducing factor-3, an anti-tumor agent isolated from Dictyostelium discoideum. PLoS ONE 2013, 8, e72118. [Google Scholar] [CrossRef]
- Kubohara, Y.; Komachi, M.; Homma, Y.; Kikuchi, H.; Oshima, Y. Derivatives of Dictyostelium differentiation-inducing factors inhibit lysophosphatidic acid–stimulated migration of murine osteosarcoma LM8 cells. Biochem. Biophys. Res. Commun. 2015, 463, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Kubokura, N.; Takahashi-Yanaga, F.; Arioka, M.; Yoshihara, T.; Igawa, K.; Tomooka, K.; Morimoto, S.; Nakatsu, Y.; Tsuzuki, T.; Nakabeppu, Y.; et al. Differentiation-inducing factor-3 inhibits intestinal tumor growth in vitro and in vivo. J. Pharmacol. Sci. 2015, 127, 446–455. [Google Scholar] [CrossRef]
- Oladimeji, P.; Kubohara, Y.; Kikuchi, H.; Oshima, Y.; Rusch, C.; Skerl, R.; Diakonova, M. A derivative of differentiation-inducing factor-3 inhibits PAK1 activity and breast cancer cell proliferation. Int. J. Cancer Clinic. Res. 2015, 2, 1–6. [Google Scholar] [CrossRef]
- Arioka, M.; Takahashi-Yanaga, F.; Kubo, M.; Igawa, K.; Tomooka, K.; Sasaguri, T. Anti-tumor effects of differentiation-inducing factor-1 in malignant melanoma: GSK-3-mediated inhibition of cell proliferation and GSK-3-independent suppression of cell migration and invasion. Biochem Pharmacol. 2017, 138, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Kikuchi, H.; Nguyen, V.H.; Oshima, Y.; Ishigaki, H.; Nakajima-Shimada, J.; Kubohara, Y. Biological activities of novel derivatives of differentiation-inducing factor-3 from Dictyostelium discoideum. Biol. Pharm. Bull. 2017, 40, 1941–1947. [Google Scholar] [CrossRef]
- Omata, W.; Shibata, H.; Nagasawa, M.; Kojima, I.; Kikuchi, H.; Oshima, Y.; Hosaka, K.; Kubohara, Y. Dictyostelium differentiation-inducing factor-1 induces glucose transporter 1 translocation and promotes glucose uptake in mammalian cells. FEBS J. 2007, 274, 3392–3404. [Google Scholar] [CrossRef] [PubMed]
- Kubohara, Y.; Kikuchi, H.; Oshima, Y. Exploitation of the derivatives of Dictyostelium differentiation-inducing factor-1, which promote glucose consumption in mammalian cells. Life Sci. 2008, 83, 608–612. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, R.; Nakamura, A.; Takahashi, K.; Kikuchi, H.; Oshima, Y.; Kubohara, Y. Oral administration of Dictyostelium differentiation-inducing factor 1 lowers blood glucose levels in streptozotocin-induced diabetic rats. Life Sci. 2016, 155, 56–62. [Google Scholar] [CrossRef]
- Takahashi, K.; Murakami, M.; Hosaka, K.; Kikuchi, H.; Oshima, Y.; Kubohara, Y. Regulation of IL-2 production in Jurkat cells by Dictyostelium-derived factors. Life Sci. 2009, 85, 438–443. [Google Scholar] [CrossRef]
- Takahashi, K.; Murakami, M.; Kikuchi, H.; Oshima, Y.; Kubohara, Y. Derivatives of Dictyostelium differentiation-inducing factors promote mitogen-activated IL-2 production via AP-1 in Jurkat cells. Life Sci. 2011, 88, 480–485. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Kikuchi, H.; Kubohara, Y.; Takahashi, K.; Katou, Y.; Oshima, Y. Development of novel DIF-1 derivatives that selectively suppress innate immune responses. Bioorg. Med. Chem. 2015, 23, 4311–4315. [Google Scholar] [CrossRef] [PubMed]
- Nakajima-Shimada, J.; Hatabu, T.; Hosoi, Y.; Onizuka, Y.; Kikuchi, H.; Oshima, Y.; Kubohara, Y. Derivatives of Dictyostelium discoideum differentiation-inducing factor-3 suppress the activities of Trypanosoma cruzi in vitro and in vivo. Biochem. Pharmacol. 2013, 85, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Sawada, T.; Aono, M.; Asakawa, S.; Ito, A.; Awano, K. Structure determination and total synthesis of a novel antibacterial substance, AB0022A, produced by a cellular slime mold. J. Antibiot. 2000, 53, 959–966. [Google Scholar] [CrossRef]
- Kikuchi, H.; Kubohara, Y.; Nguyen, V.H.; Katou, Y.; Oshima, Y. Novel chlorinated dibenzofurans isolated from the cellular slime mold, Polysphondylium filamentosum, and their biological activities. Bioorg. Med. Chem. 2013, 21, 4628–4633. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, S.; Yamaguchi, J.; Suzuki, C.; Sasaki, M.; Kazuno, S.; Uchiyama, Y. Small GTPase Rab1B is associated with ATG9A vesicles and regulates autophagosome formation. FASEB J. 2017, 31, 3757–3773. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Fernandes, P. Antibacterial discovery and development–the failure of success? Nat. Biotechnol. 2006, 24, 1497–1503. [Google Scholar] [CrossRef]
- Mohr, K.I. History of antibiotics research. Curr. Top. Microbiol. Immunol. 2016, 398, 237–272. [Google Scholar]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Barnett, R.; Stallforth, P. Natural products from social amoebae. Chem. Eur. J. 2018, 24, 4202–4214. [Google Scholar] [CrossRef]
- Shlaes, D.M.; Etter, L.; Gutmann, L. Synergistic killing of vancomycin-resistant enterococci of classes A, B, and C by combinations of vancomycin, penicillin, and gentamicin. Antimirob. Agents Chemother. 1991, 35, 776–779. [Google Scholar] [CrossRef]
- Gold, H.S. Vancomycin-resistant Enterococci: Mechanisms and clinical observations. Clin. Infect. Dis. 2001, 33, 210–219. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).