Model of Early Stage Intermediate in Respect to Its Final Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of a Nonredundant Protein Set
2.2. Identification of Structural Codes
2.3. Identification of Structures Compliant with a 3D Gaussian Distribution
2.4. Calculation of Contingency Tables
- Designation of structural codes for entire domains;
- Generation of sequence–structure pairs by moving a 4-position window along the chain; four amino acid symbols and four structural code symbols were read for each window position.
- In the case of domains not compliant with the fuzzy oil drop model, the window selection procedure additionally used the assessment of compliance of subsequent chain positions.
- The corresponding counter in the contingency table was increased for each sequence–structure pair.
2.5. Correlation Coefficient
3. Results
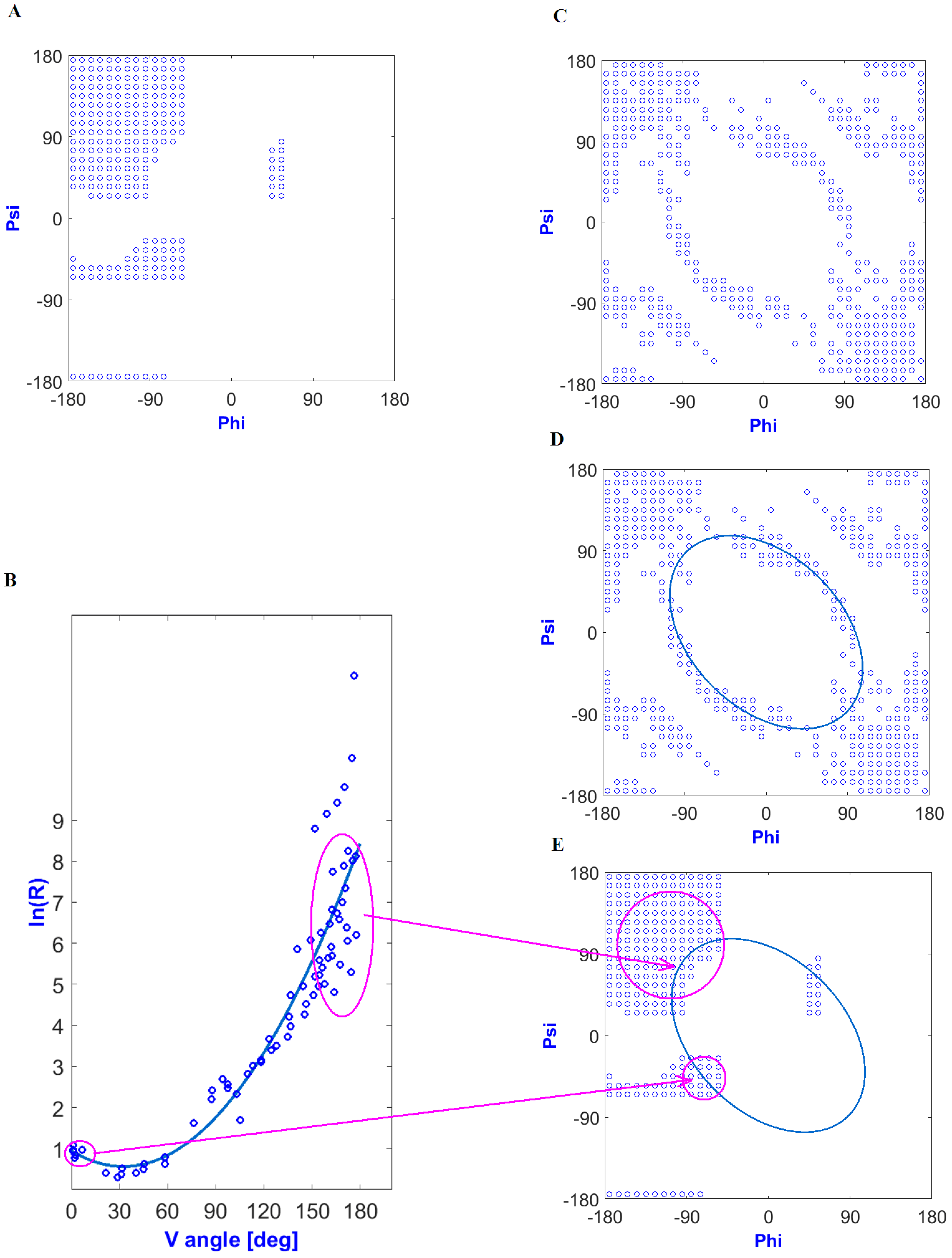
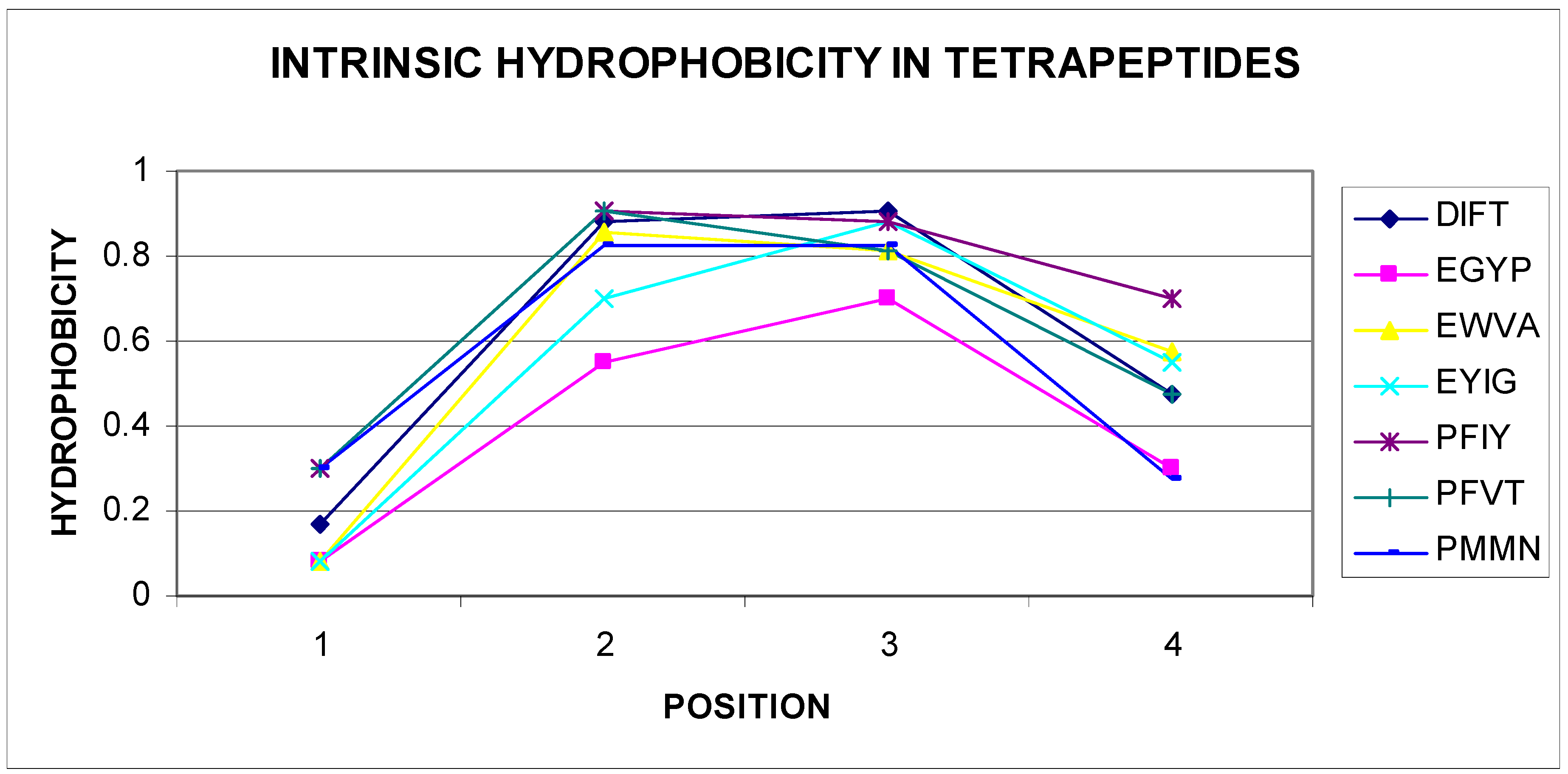
3.1. The Dependence of the Probability Distribution of a Given Tetrapeptide Sequence in a Compatible or Incompatible Form with a Micellar Hydrophobicity Distribution in the Final Structural Form of the Proteins
3.2. The Dependence of the Probability Distribution of a Given Set of Tetrapeptide Structural Codes in a Form Compatible or Incompatible with a Micellar Hydrophobicity Distribution in the Final Structural Form of the Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Creighton, T.E. Protein folding. Biochem. J. 1990, 270, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Creighton, T.E. How important is the molten globule for correct protein folding? Trends. Biochem. Sci. 1997, 22, 6–10. [Google Scholar] [CrossRef]
- Darby, N.J.; Creighton, T.E. Folding proteins. Nature 1990, 344, 715–716. [Google Scholar] [CrossRef] [PubMed]
- Creighton, T.E.; Darby, N.J.; Kemmink, J. The roles of partly folded intermediates in protein folding. Faseb. J. 1996, 10, 110–118. [Google Scholar] [CrossRef]
- Creighton, T.E. Characterizing intermediates in protein folding. Curr. Biol. 1991, 1, 8–10. [Google Scholar] [CrossRef]
- Ewbank, J.J.; Creighton, T.E. Protein folding by stages. Curr. Biol. 1992, 2, 347–349. [Google Scholar] [CrossRef]
- Creighton, T.E. Intermediates in the refolding of reduced ribonuclease A. J. Mol. Biol. 1979, 129, 411–431. [Google Scholar] [CrossRef]
- Creighton, T.E.; Cothia, C. Selecting buried residues. Nature 1989, 339, 14–15. [Google Scholar] [CrossRef]
- Bystroff, C.; Shao, Y. Modeling protein holding pathways. In Practical Bioinformatics; Bujnicki, J.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 95–122. [Google Scholar]
- Yang, J.; Zhang, Y. Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinform. 2015, 52, 5.8.1–5.8.15. [Google Scholar] [CrossRef]
- Zhou, H.; Skolnick, J. Improving threading algorithms for remote homology modeling by combining fragment and template comparisons. Proteins 2010, 78, 2041–2048. [Google Scholar] [CrossRef]
- Kolinski, A.; Skolnick, J. Monte Carlo simulations of protein folding. I. Lattice model and interaction scheme. Proteins 1994, 18, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.T.; Moult, J. Genetic algorithms for protein structure prediction. Curr. Opin. Struct. Biol. 1996, 6, 227–231. [Google Scholar] [CrossRef]
- Saunders, J.A.; Gibson, K.D.; Scheraga, H.A. Ab initio folding of multiple-chain proteins. Biocomputing 2002, 7, 601–612. [Google Scholar] [CrossRef]
- Czaplewski, C.; Karczynska, A.; Sieradzan, A.K.; Liwo, A. UNRES server for physics-based coarse-grained simulations and prediction of protein structure, dynamics and thermodynamics. Nucleic. Acids. Res. 2018, 46, W304–W309. [Google Scholar] [CrossRef] [PubMed]
- Liwo, A.; He, Y.; Scheraga, H.A. Coarse-grained force field: General folding theory. Phys. Chem. Chem. Phys. 2011, 13, 16890–16901. [Google Scholar] [CrossRef] [PubMed]
- Liwo, A.; Khalili, M.; Scheraga, H.A. Ab initio simulations of protein-folding pathways by molecular dynamics with the united-residue model of polypeptide chains. Proc. Natl. Acad. Sci. USA 2005, 102, 2362–2367. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic. Acids. Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef]
- Ovchinnikov, S.; Park, H.; Kim, D.E.; DiMaio, F.; Baker, D. Protein structure prediction using Rosetta in CASP12. Proteins 2018, 86, 113–121. [Google Scholar] [CrossRef]
- Chivian, D.; Kim, D.E.; Malmström, L.; Bradley, P.; Robertson, T.; Murphy, P.; Strauss, C.E.; Bonneau, R.; Rohl, C.A.; Baker, D. Automated prediction of CASP-5 structures using the Robetta server. Proteins 2003, 53, 524–533. [Google Scholar] [CrossRef]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Schwede, T.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)—round x. Proteins 2013, 82, 1–6. [Google Scholar] [CrossRef]
- Protein Structure Prediction Center. Available online: http://predictioncenter.org/ (accessed on 1 October 2019).
- Roterman, I. The geometrical analysis of peptide backbone structure and its local deformations. Biochimie 1995, 77, 204–216. [Google Scholar] [CrossRef]
- Roterman, I. Modelling the optimal simulation path in the peptide chain folding--studies based on geometry of alanine heptapeptide. J. Biol. 1995, 177, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Alejster, P.; Jurkowski, W.; Roterman, I. Structural information involved in the interpretation of the stepwise protein folding process. In Protein Folding in Silico; Roterman-Konieczna, I., Ed.; Woodhead Publishing (currently Elsevier): Oxford, UK; Cambridge, UK; Philadelphia, PA, USA; New Delhi, India, 2012; pp. 39–54. [Google Scholar] [CrossRef]
- Roteman, I.; Banach, M.; Konieczny, L. Application of the Fuzzy Oil Drop Model Describes Amyloid as a Ribbonlike Micelle. Entropy 2017, 19, 167. [Google Scholar] [CrossRef]
- Kalinowska, B.; Banach, M.; Konieczny, L.; Roterman, I. Application of Divergence Entropy to Characterize the Structure of the Hydrophobic Core in DNA Interacting Proteins. Entropy 2015, 17, 1477–1507. [Google Scholar] [CrossRef]
- Banach, M.; Konieczny, L.; Roterman, I. Proteins structured as spherical micelles. In From Globular Proteins to Amyloids; Roterman-Konieczna, I., Ed.; Elsevier: Amsterdam, The Netherlands; Oxford, UK; Cambridge, MA, USA, 2019; pp. 55–68. ISBN 978-O-O8-102981-7. [Google Scholar]
- Banach, M.; Konieczny, L.; Roterman, I. Composite structures. In From Globular Proteins to Amyloids; Roterman-Konieczna, I., Ed.; Elsevier: Amsterdam, The Netherlands; Oxford, UK; Cambridge, MA, USA, 2019; pp. 117–134. ISBN 978-O-O8-102981-7. [Google Scholar]
- Banach, M.; Konieczny, L.; Roterman, I. Use of the “fuzzy oil drop” model to identify the complexation area in protein homodimers. In Protein Folding in Silico; Roterman-Konieczna, I., Ed.; Woodhead Publishing (currently Elsevier): Oxford, UK; Cambridge, UK; Philadelphia, PA, USA; New Delhi, India, 2012; pp. 95–122. [Google Scholar] [CrossRef]
- Banach, M.; Konieczny, L.; Roterman, I. Ligand-binding site recognition. In Protein Folding in Silico; Roterman-Konieczna, I., Ed.; Woodhead Publishing (currently Elsevier): Oxford, UK; Cambridge, UK; Philadelphia, PA, USA; New Delhi, India, 2012; pp. 79–94. [Google Scholar] [CrossRef]
- Dułak, D.; Gadzała, M.; Banach, M.; Ptak, M.; Wiśniowski, Z.; Konieczny, L.; Roterman, I. Filamentous aggregates of tau proteins fulfil standard amyloid criteria provided by the fuzzy oil drop (FOD) model. Int. J. Mol. Sci. 2018, 19, 2910. [Google Scholar] [CrossRef]
- Kalinowska, B.; Krzykalski, A.; Roterman, I. Contingency Table Browser—Prediction of early stage protein structure. Bioinformation 2015, 11, 486–488. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic. Acids. Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Available online: ftp://resources.rcsb.org/sequence/clusters (accessed on 1 October 2019).
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome. Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Dawson, N.L.; Lewis, T.E.; Das, S.; Lees, J.G.; Lee, D.; Ashford, P.; Orengo, C.A.; Sillitoe, I. CATH: An expanded resource to predict protein function through structure and sequence. Nucleic. Acids. Res. 2017, 45, D289–D295. [Google Scholar] [CrossRef]
- Available online: http://www.cathdb.info/download (accessed on 20 August 2019).
- Kullback, S.; Leibler, R.A. On Information and Sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
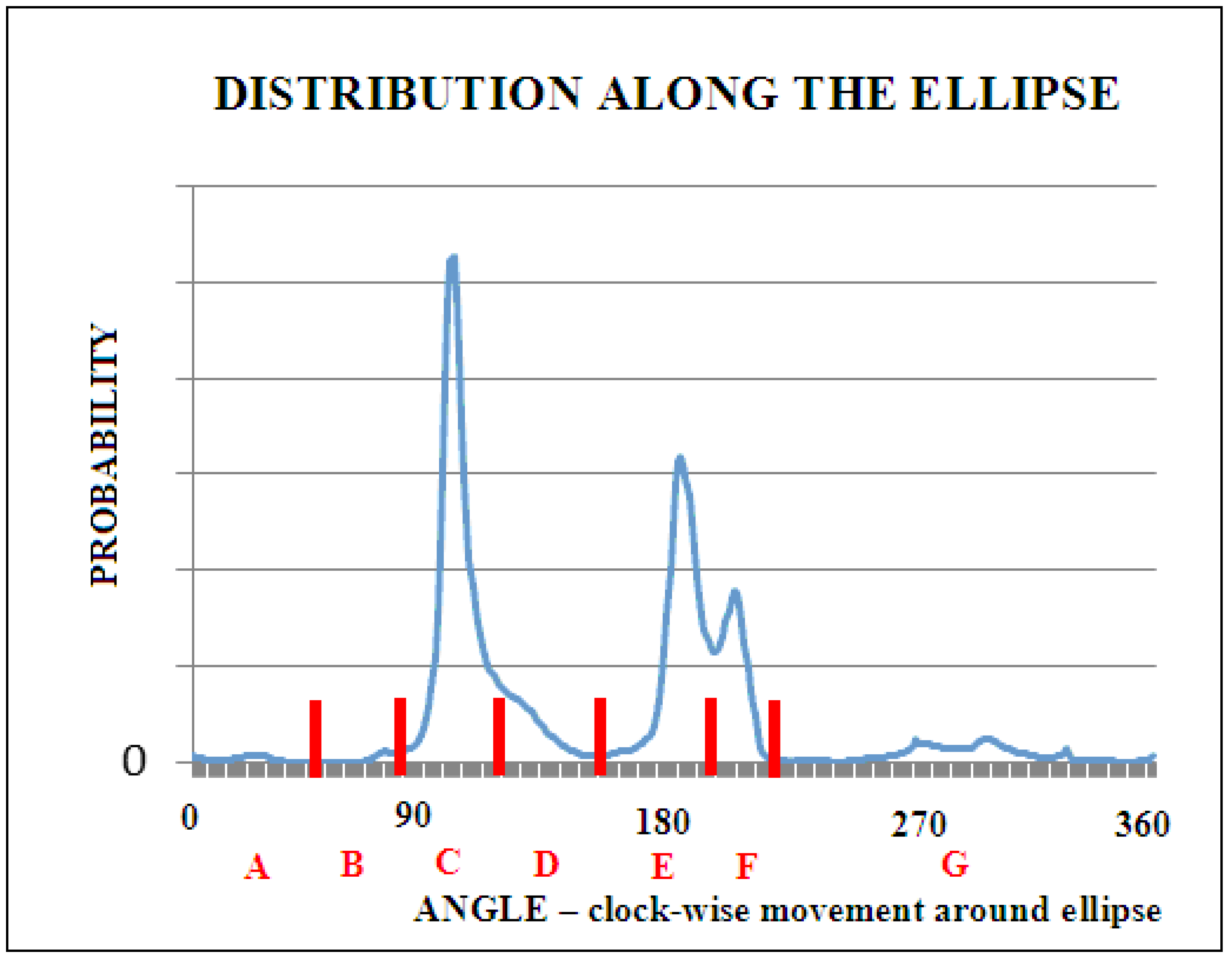

| Code | From | To |
|---|---|---|
| A | 0 | 50 |
| B | 51 | 85 |
| C | 86 | 110 |
| D | 111 | 150 |
| E | 151 | 193 |
| F | 194 | 225 |
| G | 226 | 359 |
| A | C | D | E | F | G | H | I | K | L | N | P | Q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AENN AGHE AGYP AIIP AKNT ATGY AYGL AYPV | CVAS | DADE DAVP DFIV DFSK DIFL DIFT DIKF DKAG DKIC DLGS DLNP DPLD DPNG DPVP DVSG DYVF | EFYT EGYP EKFN EKKS EKNI ELYL ENVD EPKP ETPL EWVA EYEF EYIG | FGAD FGEP FIKN FQVV FRPG FVEV FVRL FVRN FGAD FGEP FIKN FQVV FRPG FVEV FVRL FVRN | GADE GAPE GFDI GHLK GNEV GNIN GNPV GSPI GSRL GTPA GTPN GTYI GWRL GYAV GYEI GYEV GYQL | HAEN HAKG HIVE HLDV HVAF | IAPV IAVD IEPI IEVQ IFFK IFNG IFTE IGSN IITY IKVN INIG INLH IPLK IPVA ISVW IVFD IVHR IVQF | KDFT KETF KLPA KLSL KLTK KLYS KLYY KVGI KYKL KYYA | LATP LDLQ LDSK LFDD LFLS LGEN LGLQ LHTN LHVH LIHG LNLR LPHV LPVY LRYD LSGH LSTP LSVG LTLY LVSY LWVE | NADS NEGA NGTR NKVL NPKV NVVC NVVG | PAVG PDDP PFIY PFLF PFVT PLKF PMMN PPPE PQGF PVLG | QIST QLEI QSLH |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabian, P.; Stapor, K.; Roterman, I. Model of Early Stage Intermediate in Respect to Its Final Structure. Biomolecules 2019, 9, 866. https://doi.org/10.3390/biom9120866
Fabian P, Stapor K, Roterman I. Model of Early Stage Intermediate in Respect to Its Final Structure. Biomolecules. 2019; 9(12):866. https://doi.org/10.3390/biom9120866
Chicago/Turabian StyleFabian, Piotr, Katarzyna Stapor, and Irena Roterman. 2019. "Model of Early Stage Intermediate in Respect to Its Final Structure" Biomolecules 9, no. 12: 866. https://doi.org/10.3390/biom9120866
APA StyleFabian, P., Stapor, K., & Roterman, I. (2019). Model of Early Stage Intermediate in Respect to Its Final Structure. Biomolecules, 9(12), 866. https://doi.org/10.3390/biom9120866






