Evolutionary Analyses of Sequence and Structure Space Unravel the Structural Facets of SOD1
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Space Analysis
2.1.1. Data Pre-Processing
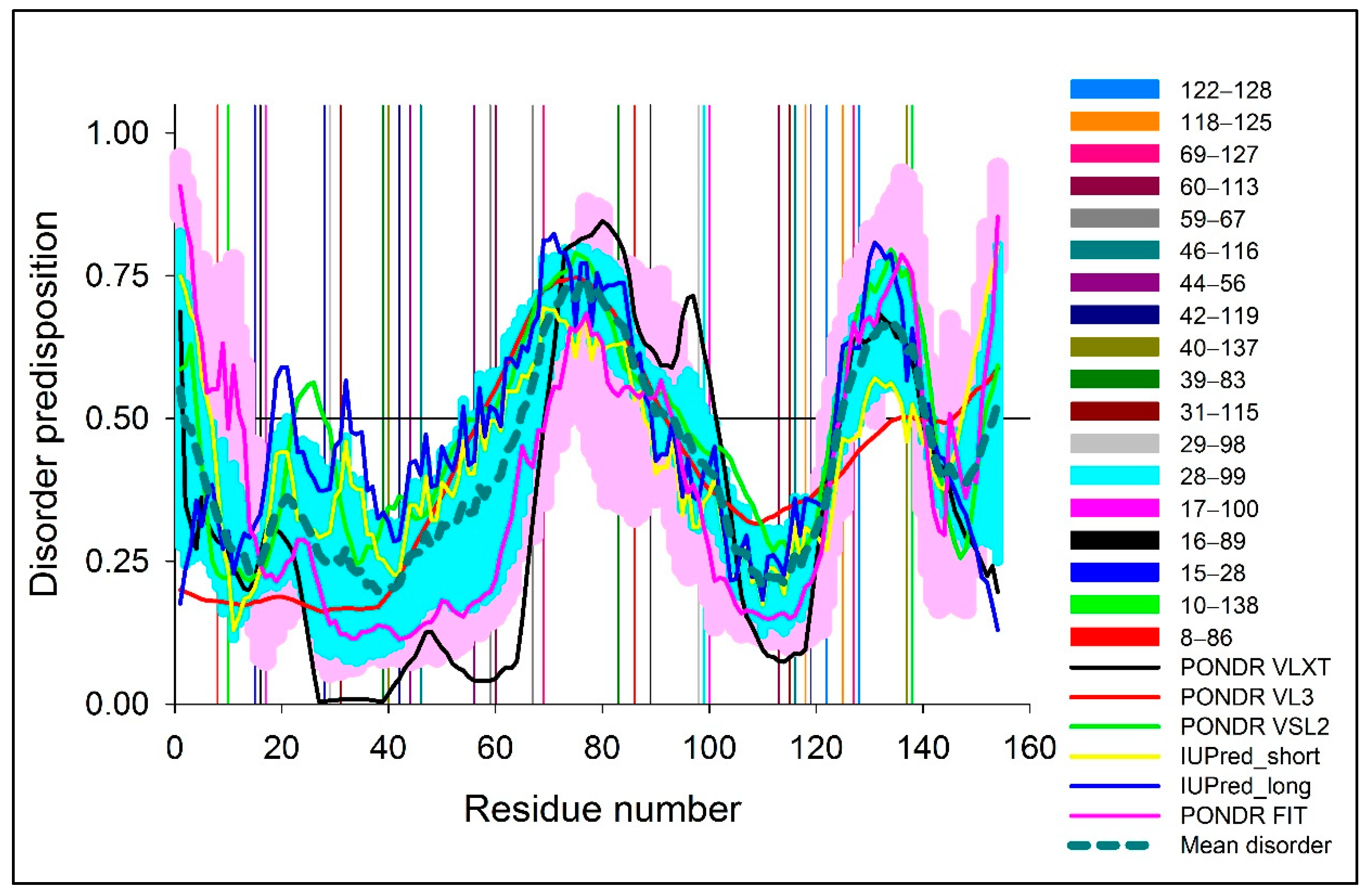
2.1.2. Computational Analysis of the Intrinsic Disorder Predisposition of Human SOD1
2.1.3. Coupling Study
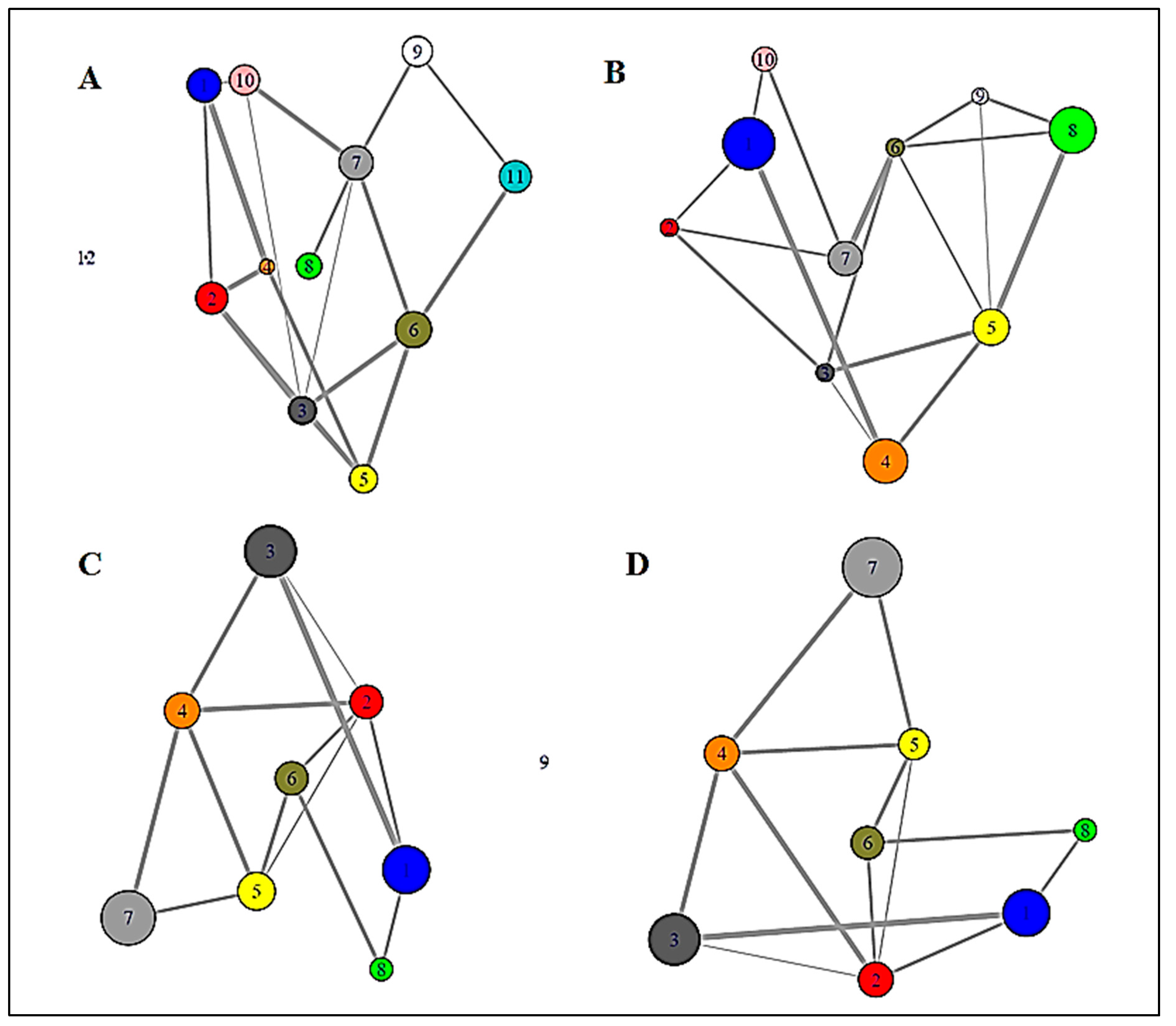
2.1.4. Graph Theoretical Modelling
2.2. Structure Network Analysis
2.2.1. Model Building
2.2.2. Structure Network Analysis
3. Results
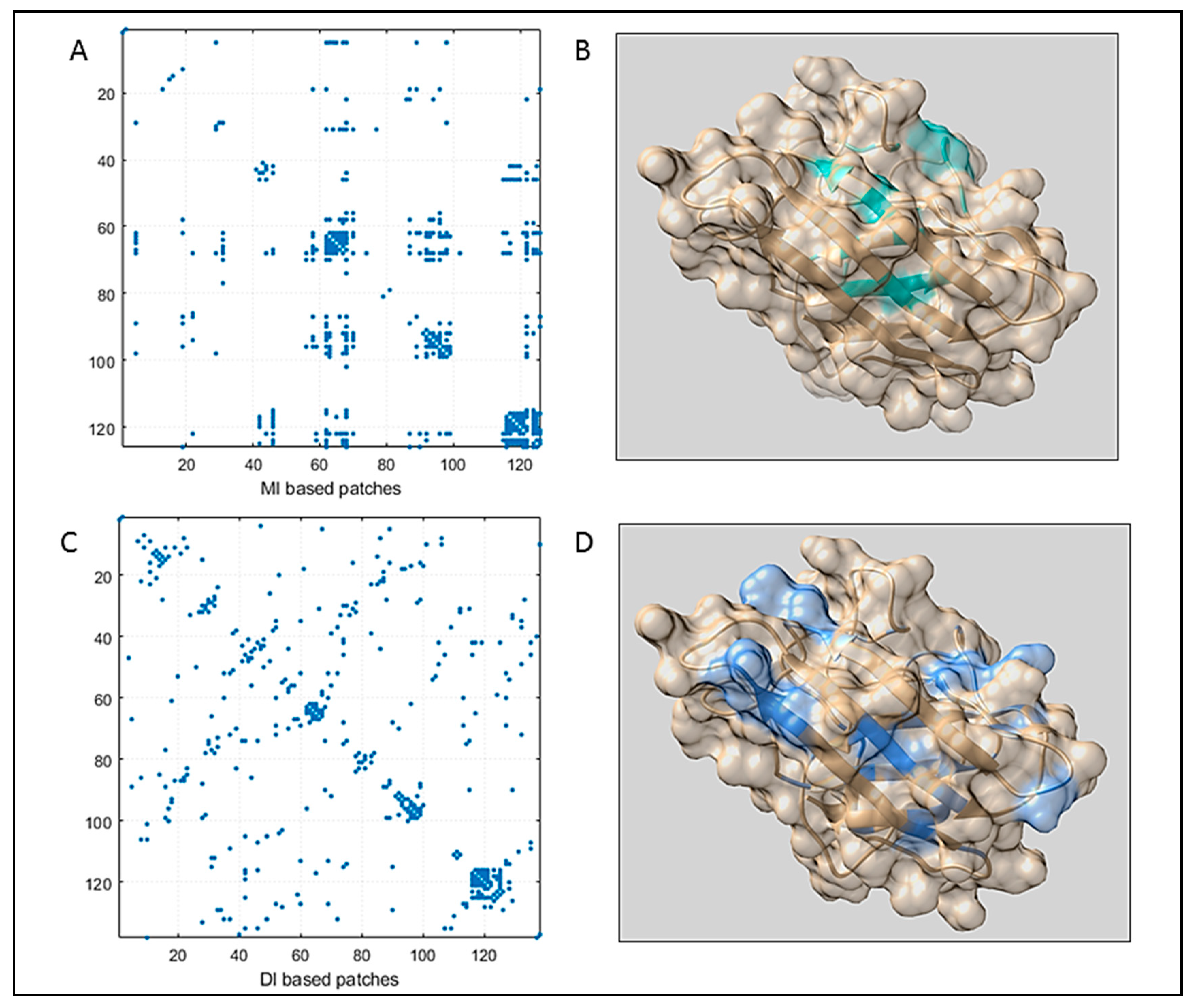
3.1. Sequence Space Analysis
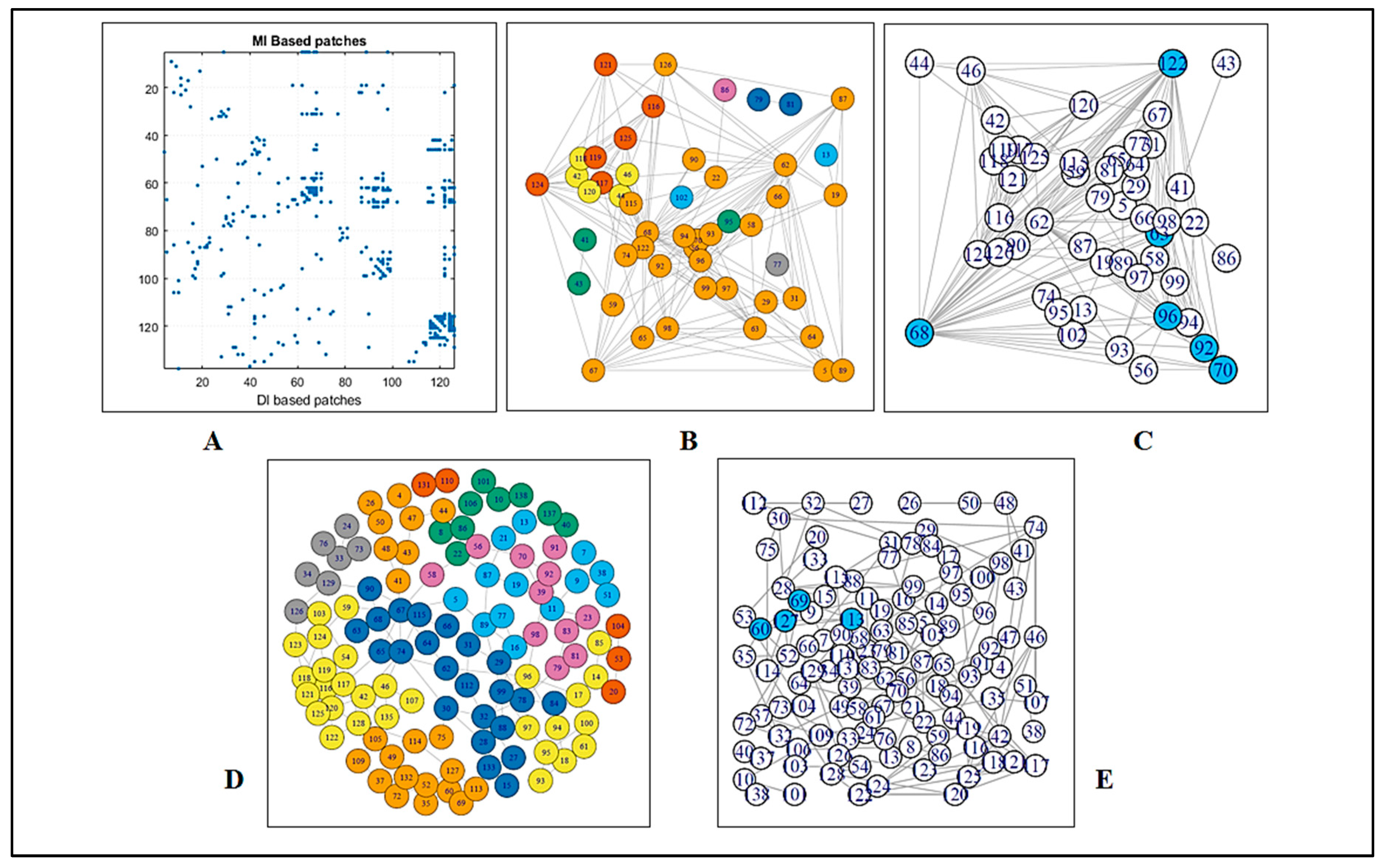
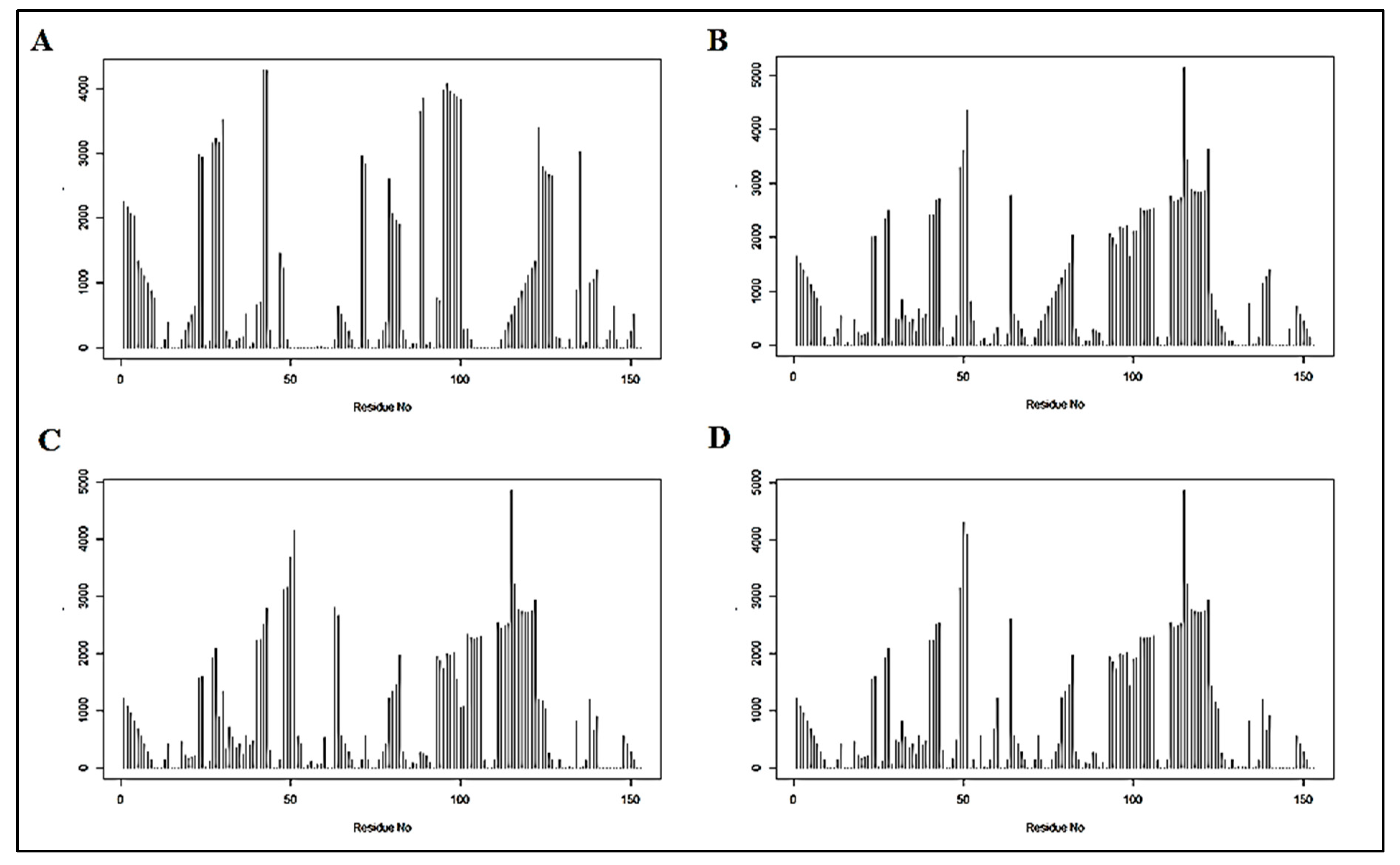
3.1.1. Coupling Analysis
3.1.2. Graph Theoretical Modelling
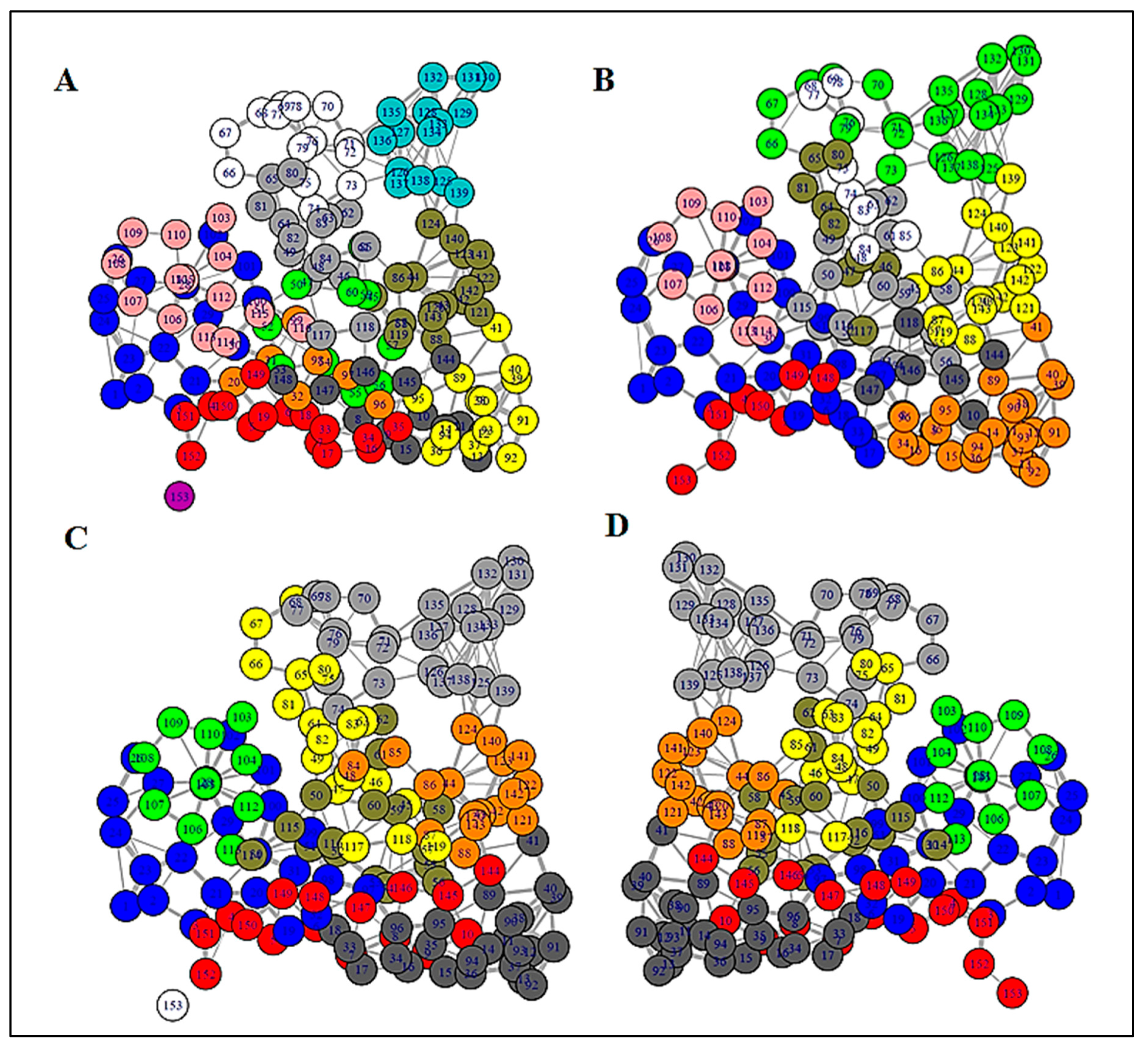
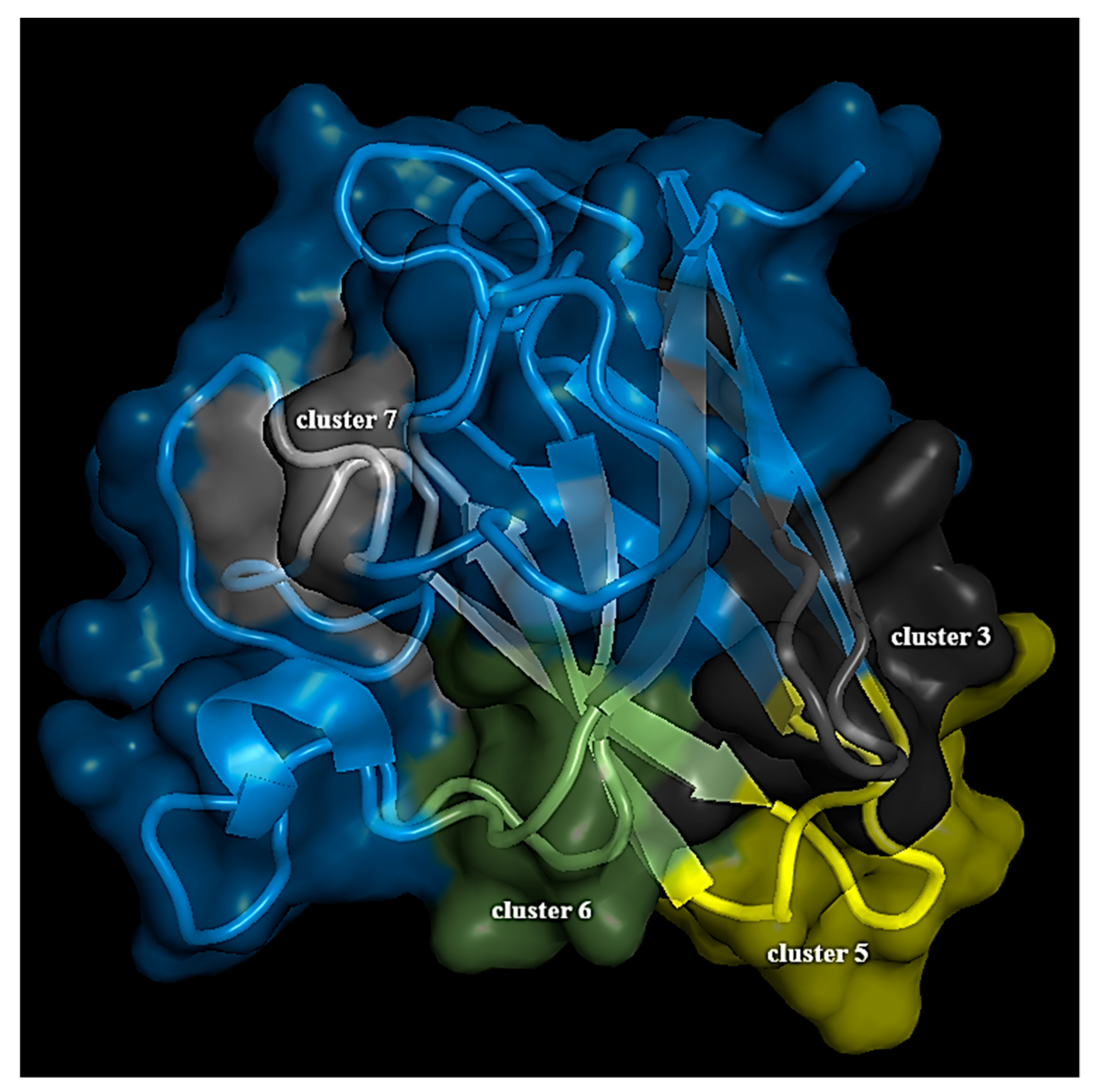
3.2. Structure Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
References
- Chan, H.S.; Dill, K.A. The protein folding problem. Phys. Today 1993, 46, 24–32. [Google Scholar] [CrossRef]
- Benzie, I.F.F. Evolution of antioxidant defence mechanisms. Eur. J. Nutr. 2000, 39, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Mccord, J.M. The evolution of free radicals and oxidative stress. Am. J. Med. 2000, 108, 652–659. [Google Scholar] [CrossRef]
- Fridovich, I. Biological effects of the superoxide radical. Arch. Biochem. Biophys. 1986, 247, 1–11. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxygen Stress and Superoxide Dismutases. Plant Physiol. 1993, 101, 7–12. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Isozaki, Y. The story of O2. Science 2008, 322, 540–542. [Google Scholar] [CrossRef]
- Sessions, A.L.; Doughty, D.M.; Welander, P.V.; Summons, R.E.; Newman, D.K. The Continuing Puzzle of the Great Oxidation Event. Curr. Boil. 2009, 19, R567–R574. [Google Scholar] [CrossRef]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free. Radic. Boil. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Borgstahl, G.E.; Parge, H.E.; Hickey, M.J.; Beyer, W.F., Jr.; Hallewell, R.A.; Tainer, J.A. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell 1992, 71, 107–118. [Google Scholar] [CrossRef]
- Bowling, A.C.; Schulz, J.B.; Brown, R.H.; Beal, M.F. Superoxide Dismutase Activity, Oxidative Damage, and Mitochondrial Energy Metabolism in Familial and Sporadic Amyotrophic Lateral Sclerosis. J. Neurochem. 1993, 61, 2322–2325. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Boca, M.; Girotto, S.; Martinelli, M.; Valentine, J.S.; Vieru, M. SOD1 and Amyotrophic Lateral Sclerosis: Mutations and Oligomerization. PLoS ONE 2008, 3, e1677. [Google Scholar] [CrossRef] [PubMed]
- Valentine, J.S.; Doucette, P.A.; Potter, S.Z. Copper-Zinc Superoxide Dismutase and Amyotrophic Lateral Sclerosis. Annu. Rev. Biochem. 2005, 74, 563–593. [Google Scholar] [CrossRef] [PubMed]
- Shaw, B.; Valentine, J. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem. Sci. 2007, 32, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Banci, L.; Bertini, I.; Martinelli, M.; Furukawa, Y.; O’Halloran, T.V. The Unusually Stable Quaternary Structure of Human Cu,Zn-Superoxide Dismutase 1 Is Controlled by Both Metal Occupancy and Disulfide Status. J. Boil. Chem. 2004, 279, 47998–48003. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Cantini, F.; Kozyreva, T.; Massagni, C.; Palumaa, P.; Rubino, J.T.; Zovo, K. Human superoxide dismutase 1 (hSOD1) maturation through interaction with human copper chaperone for SOD1 (hCCS). Proc. Natl. Acad. Sci. USA 2012, 109, 13555–13560. [Google Scholar] [CrossRef]
- Chowdhury, S.; Banerjee, A.; Chattopadhyay, K. Metal ion co-factors sculpt the heterogeneity of conformational landscape in Superoxide Dismutase. Eur. Biophys. J. Biophys. Lett. 2017, 46, S344. [Google Scholar]
- Sangwan, S.; Zhao, A.; Adams, K.L.; Jayson, C.K.; Sawaya, M.R.; Guenther, E.L.; Pan, A.C.; Ngo, J.; Moore, D.M.; Soriaga, A.B.; et al. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc. Natl. Acad. Sci. USA 2017, 114, 8770–8775. [Google Scholar] [CrossRef]
- Zhu, C.; Beck, M.V.; Griffith, J.D.; Deshmukh, M.; Dokholyan, N.V. Large SOD1 aggregates, unlike trimeric SOD1, do not impact cell viability in a model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2018, 115, 4661–4665. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein disorder prediction: Implications for structural proteomics. Structure 2003, 11, 1453–1459. [Google Scholar] [CrossRef]
- Alexander, M.D.; Traynor, B.J.; Miller, N.; Corr, B.; Frost, E.; McQuaid, S.; Brett, F.M.; Green, A.; Hardiman, O. “True” sporadic ALS associated with a novel SOD-1 mutation, Annals of Neurology: Official Journal of the American Neurological Association and the Child. Neurol. Soc. 2002, 52, 680–683. [Google Scholar]
- Srinivasan, E.; Rajasekaran, R. Computational Investigation on Electrostatic Loop Mutants Instigating Destabilization and Aggregation on Human SOD1 Protein Causing Amyotrophic Lateral Sclerosis. Protein J. 2019, 38, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, H.; Logan, D.T.; Mu, X.; Danielsson, J.; Oliveberg, M. The Cost of Long Catalytic Loops in Folding and Stability of the ALS-Associated Protein SOD. J. Am. Chem. Soc. 2018, 140, 16570–16579. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Sen, S.; Banerjee, A.; Uversky, V.N.; Maulik, U.; Chattopadhyay, K. Network mapping of the conformational heterogeneity of SOD1 by deploying statistical cluster analysis of FTIR spectra. Cell. Mol. Life Sci. 2019, 76, 4145–4154. [Google Scholar] [CrossRef] [PubMed]
- Culik, R.M.; Sekhar, A.; Nagesh, J.; Deol, H.; Rumfeldt, J.A.O.; Meiering, E.M.; Kay, L.E. Effects of maturation on the conformational free-energy landscape of SOD. Proc. Natl. Acad. Sci. USA 2018, 115, E2546–E2555. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins: Struct. Funct. Bioinform. 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmók, V.; Tompa, P.; Simon, I. The Pairwise Energy Content Estimated from Amino Acid Composition Discriminates between Folded and Intrinsically Unstructured Proteins. J. Mol. Boil. 2005, 347, 827–839. [Google Scholar] [CrossRef]
- Walsh, I.; Giollo, M.; Di Domenico, T.; Ferrari, C.; Zimmermann, O.; Tosatto, S.C. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics 2014, 31, 201–208. [Google Scholar] [CrossRef]
- Fan, X.; Kurgan, L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J. Biomol Struct Dyn. 2014, 32, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Kurgan, L. On the complementarity of the consensus-based disorder prediction. Pac. Symp. Biocomput. 2012, 176–187. [Google Scholar]
- Dunn, S.; Gloor, G.; Wahl, L. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Biochemistry 2007, 24, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.; Hamer, R.; Reinert, G.; Deane, C.M. Mutual information and variants for protein domain-domain contact prediction. BMC Res. Notes 2012, 5, 472. [Google Scholar] [CrossRef]
- Morcos, F.; Pagnani, A.; Lunt, B.; Bertolino, A.; Marks, D.S.; Sander, C.; Zecchina, R.; Onuchic, J.N.; Hwa, T.; Weigt, M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA 2011, 108, E1293–E1301. [Google Scholar] [CrossRef]
- Auclair, J.R.; Brodkin, H.R.; D’Aquino, J.A.; Petsko, G.A.; Ringe, D.; Agar, J.N. Structural Consequences of Cysteinylation of Cu/Zn-Superoxide Dismutase. Biochemistry 2013, 52, 6145–6150. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Skjærven, L.; Yao, X.-Q.; Scarabelli, G.; Grant, B.J. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinform. 2014, 15, 399. [Google Scholar] [CrossRef]
- Bahar, I.; Rader, A.J. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Boil. 2005, 15, 586–592. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Boccaletti, S.; Latora, V.; Moreno, Y.; Chavez, M.; Hwang, D.-U. Complex networks: Structure and dynamics. Phys. Rep. 2006, 424, 175–308. [Google Scholar] [CrossRef]
- Hirano, M.; Fujii, J.; Nagai, Y.; Sonobe, M.; Okamoto, K.; Araki, H.; Taniguchi, N.; Ueno, S. A New Variant Cu/Zn Superoxide Dismutase (Val7→Glu) Deduced from Lymphocyte mRNA Sequences from Japanese Patients with Familial Amyotrophic Lateral Sclerosis. Biochem. Biophys. Res. Commun. 1994, 204, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Ogasawara, M.; Matsubara, Y.; Narisawa, K.; Nakamura, S.; Itoyama, Y.; Abe, K. Familial amyotrophic lateral sclerosis (ALS) in Japan associated with H46R mutation in Cu/Zn superoxide dismutase gene: A possible new subtype of familial ALS. J. Neurol. Sci. 1994, 126, 77–83. [Google Scholar] [CrossRef]
- Elam, J.S.; Taylor, A.B.; Strange, R.; Antonyuk, S.A.; Doucette, P.A.; Rodriguez, J.; Hasnain, S.S.; Hayward, L.J.; Valentine, J.S.; Yeates, T.O.; et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Mol. Boil. 2003, 10, 461–467. [Google Scholar] [CrossRef]
- Banci, L.; Cantini, F.; Kozyreva, T.; Rubino, J.T. Mechanistic Aspects of hSOD1 Maturation from the Solution Structure of Cu I -Loaded hCCS Domain 1 and Analysis of Disulfide-Free hSOD1 Mutants. ChemBioChem 2013, 14, 1839–1844. [Google Scholar] [CrossRef]
- Furukawa, Y.; Kaneko, K.; Yamanaka, K.; O’Halloran, T.V.; Nukina, N. Complete Loss of Post-translational Modifications Triggers Fibrillar Aggregation of SOD1 in the Familial Form of Amyotrophic Lateral Sclerosis. J. Boil. Chem. 2008, 283, 24167–24176. [Google Scholar] [CrossRef]
- Hörnberg, A.; Logan, D.T.; Marklund, S.L.; Oliveberg, M. The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase. J. Mol. Boil. 2007, 365, 333–342. [Google Scholar] [CrossRef]
- Del Grande, A.; Luigetti, M.; Conte, A.; Mancuso, I.; Lattante, S.; Marangi, G.; Stipa, G.; Zollino, M.; Sabatelli, M. A novel L67P SOD1 mutation in an Italian ALS patient. Amyotroph. Lateral Scler. 2011, 12, 150–152. [Google Scholar] [CrossRef]
- Andersen, P.M.; Sims, K.B.; Xin, W.W.; Kiely, R.; O’Neill, G.; Ravits, J.; Pioro, E.; Harati, Y.; Brower, R.D.; Levine, J.S.; et al. Sixteen novel mutations in the Cu/Zn superoxide dismutase gene in amyotrophic lateral sclerosis: A decade of discoveries, defects and disputes. Amyotroph. Lateral Scler. 2003, 4, 62–73. [Google Scholar] [CrossRef]
- DiDonato, M.; Craig, L.; Huff, M.E.; Thayer, M.M.; Cardoso, R.M.; Kassmann, C.J.; Lo, T.P.; Bruns, C.K.; Powers, E.T.; Kelly, J.W.; et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Boil. 2003, 332, 601–615. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Boil. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
| Mutant Name | Mutations |
|---|---|
| apo-SOD1 | H63F |
| Cu-SOD1 | H120F |
| Zn-SOD1 | H71F |
| L4_SOD1 | N65S, L67P, G72S, D76Y, H80A |
| L7_SOD1 | D124G, D125H, L126S, S134N, N139K, L144F |
| L4S_SOD1 | G72S, D76Y, H80A, L84F, A89T, D90A, G93C, A95G |
| Residue Number (Mean PDS) | Coupling Pairs (Mean PDS) | MI Score |
|---|---|---|
| 29 (0.25 ± 0.16) | 98 (0.43 ± 0.11) | 0.64260 |
| 42 (0.21 ± 0.10) | 117 (0.24 ± 0.09) | 0.703328 |
| 44 (0.26 ± 0.13) | 122 (0.36 ± 0.05) | 0.593079 |
| 46 (0.27 ± 0.11) | 115 (0.23 ± 0.09) | 0.577762 |
| 58 (0.38 ± 0.18) | 68 (0.60 ± 0.12) | 0.615481 |
| 62 (0.44 ± 0.19) | 116 (0.26 ± 0.10) | 0.583716 |
| 67 (0.56 ± 0.14) | 124 (0.48 ± 0.06) | 0.608395 |
| 70 (0.68 ± 0.10) | 96 (0.47 ± 0.11) | 0.603931 |
| 99 (0.42 ± 0.10) | 122 (0.36 ± 0.05) | 0.603343 |
| 118 (0.27 ± 0.09) | 125 (0.52 ± 0.07) | 0.6989 |
| Residue Number (Mean PDS) | Coupling Pairs (Mean PDS) | DI Score |
|---|---|---|
| 8 (0.33 ± 0.12) | 86 (0.59 ± 0.05) | 0.108738 |
| 10 (0.28 ± 0.09) | 138 (0.63 ± 0.09) | 0.122847 |
| 15 (0.24 ± 0.05) | 28 (0.25 ± 0.16) | 0.125331 |
| 16 (0.25 ± 0.05) | 89 (0.54 ± 0.04) | 0.109716 |
| 17 (0.27 ± 0.07) | 100 (0.41 ± 0.09) | 0.0900074 |
| 28 (0.25 ± 0.16) | 99 (0.42 ± 0.10) | 0.131024 |
| 29 (0.25 ± 0.16) | 98 (0.43 ± 0.11) | 0.095733 |
| 31 (0.25 ± 0.16) | 115 (0.23 ± 0.09) | 0.0851411 |
| 39 (0.21 ± 0.11) | 83 (0.67 ± 0.09) | 0.0948818 |
| 40 (0.21 ± 0.11) | 137 (0.62 ± 0.12) | 0.249519 |
| 42 (0.21 ± 0.10) | 119 (0.28 ± 0.07) | 0.105276 |
| 44 (0.26 ± 0.13) | 56 (0.34 ± 0.17) | 0.0880899 |
| 46 (0.27 ± 0.11) | 116 (0.26 ± 0.10) | 0.0962272 |
| 59 (0.38 ± 0.19) | 67 (0.56 ± 0.14) | 0.107917 |
| 60 (0.39 ± 0.19) | 113 (0.22 ± 0.08) | 0.156108 |
| 69 (0.65 ± 0.13) | 127 (0.58 ± 0.09) | 0.126032 |
| 118 (0.27 ± 0.09) | 125 (0.52 ± 0.07) | 0.100409 |
| 122 (0.36 ± 0.05) | 128 (0.58 ± 0.09) | 0.156898 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chowdhury, S.; Sanyal, D.; Sen, S.; Uversky, V.N.; Maulik, U.; Chattopadhyay, K. Evolutionary Analyses of Sequence and Structure Space Unravel the Structural Facets of SOD1. Biomolecules 2019, 9, 826. https://doi.org/10.3390/biom9120826
Chowdhury S, Sanyal D, Sen S, Uversky VN, Maulik U, Chattopadhyay K. Evolutionary Analyses of Sequence and Structure Space Unravel the Structural Facets of SOD1. Biomolecules. 2019; 9(12):826. https://doi.org/10.3390/biom9120826
Chicago/Turabian StyleChowdhury, Sourav, Dwipanjan Sanyal, Sagnik Sen, Vladimir N. Uversky, Ujjwal Maulik, and Krishnananda Chattopadhyay. 2019. "Evolutionary Analyses of Sequence and Structure Space Unravel the Structural Facets of SOD1" Biomolecules 9, no. 12: 826. https://doi.org/10.3390/biom9120826
APA StyleChowdhury, S., Sanyal, D., Sen, S., Uversky, V. N., Maulik, U., & Chattopadhyay, K. (2019). Evolutionary Analyses of Sequence and Structure Space Unravel the Structural Facets of SOD1. Biomolecules, 9(12), 826. https://doi.org/10.3390/biom9120826






