Abstract
The COP9 (constitutive photomorphogenesis 9) signalosome (CSN) is an evolutionarily conserved protein complex which regulates various growth and developmental processes. However, the role of CSN during environmental stress is largely unknown. Using Arabidopsis as model organism, we used CSN hypomorphic mutants to study the role of the CSN in plant responses to environmental stress and found that heat stress specifically enhanced the growth of csn5a-1 but not the growth of other hypomorphic photomorphogenesis mutants tested. Following heat stress, csn5a-1 exhibits an increase in cell size, ploidy, photosynthetic activity, and number of lateral roots and an upregulation of genes connected to the auxin response. Immunoblot analysis revealed an increase in deneddylation of CUL1 but not CUL3 following heat stress in csn5a-1, implicating improved CUL1 activity as a basis for the improved growth of csn5a-1 following heat stress. Studies using DR5::N7-VENUS and DII-VENUS reporter constructs confirm that the heat-induced growth is due to an increase in auxin signaling. Our results indicate that CSN5A has a specific role in deneddylation of CUL1 and that CSN5A is required for the recovery of AUX/IAA repressor levels following recurrent heat stress to regulate auxin homeostasis in Arabidopsis.
1. Introduction
The COP9 (constitutive photomorphogenesis 9) signalosome (CSN) is an evolutionarily conserved multiprotein complex comprised of eight subunits (CSN1 to CSN8) [1]. CSN was discovered in Arabidopsis as a repressor of light-regulated development [2,3]. Complete loss of any subunit results in pleiotropic constitutively photomorphogenic/de-etolated/fusca (cop/det/fus) phenotype with characteristic stunted growth, open cotyledons in dark-grown seedling, short hypocotyl, and anthocyanin pigment accumulation [3,4].
Other than photomorphogenesis, the CSN regulates a number of hormonal signaling pathways through its action as a deneddylase regulating ubiquitin-mediated protein stability [5]. CSN regulates responses to auxin, jasmonate, and gibberellic acid, as well as flower development, through its regulation of Cullin-RING ubiquitin E3 ligases (CRLs) such as the SKP1, Cullin and F-box-containing protein (SCF) complexes SCFTIR1, SCFCOI1, SCFSLI1, SCFCFK1, and SCFUFO [6,7,8,9,10]. The CSN also regulates other CRLs such as those containing CUL3 (cullin 3) and CUL4 [11,12].
CSN plays a critical role in protecting plants from biotic stress by regulating N gene-mediating resistance to tobacco mosaic virus [13] and jasmonic acid-dependent plant defense response [14]. CSN is also involved in double-stranded break repair [15] and nucleotide excision repair [16]. csn mutants show auto-degradation of a CRL substrate receptor, which is regulated in cell-type-specific manner [17].
CSN is not only involved in developmental processes [11] but also plays significant role in cell cycle progression [18]. csn mutants show delay in S-phase progression in yeast [19], defective S phase progression in mouse thymocytes [20], and G2 phase arrest in Arabidopsis roots [18].
Enzymatically, CSN is a metalloprotease which cleaves neural precursor cell expressed, developmentally downregulated 8 (NEDD8) from the cullin subunit of CRLs by a process called deneddylation [7,21]. This catalytic activity is located in the JAB1/MPN/Mov34 metalloenzyme (JAMM) motif of CSN5 subunit [22].
Complete loss of any CSN subunit leads to seedling lethality in early stage, which obstructs further analysis of the role of subunits in plant growth and development [4]. However, in Arabidopsis, subunit CSN5 is encoded by two partially redundant genes [23], which allows mutant plants to grow to adulthood [24]. Subsequent discovery of viable hypomorphic mutants of other subunits enabled to study the role of CSN subunits in the adult and reproductive stages. These hypomorphic mutants can be broadly classified into two categories: 1. Mutants compromised in cullin deneddylation and auxin/3-indoleacetic acid (AUX/IAA) degradation (e.g., csn1-10, csn2-5, csn4-2035, csn5a-1, and csn5a-2) and 2. Mutants with normal cullin deneddylation and AUX/IAA degradation activity (e.g., csn3-3, csn5b-1, csn6a-1, and csn6b-1) [5]. csn5a-1 exhibits hyper-neddylation of CUL1, CUL3, and CUL4, whereas csn5b-1 shows normal cullin neddylation similar to wild type [25]. Studies have shown that, while CSN5A is crucial for seed germination, CSN1 plays a prominent role in seed maturation. The seed germination phenotype of csn1-10 is due to over-accumulation of RGL2; however, the germination phenotype of csn5a-1 is not only because of RGL2 but also ABI5. Thus, ABI5 is especially affected in csn5a-1 but not in csn1-10 [26].
In this study, we employed viable hypomorphic csn mutants to study the role of CSN in response to abiotic stress. We found that, while these mutants are hypersensitive to salinity and UV-C, growth of csn5a-1 was enhanced after heat stress. This enhanced growth is likely a result of numerous parameters including increased photosynthetic output and increase in CUL1 deneddylation and auxin activity. Thus, CSN5A is required to buffer plants during heat by maintaining auxin homeostasis.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
All the Arabidopsis lines used in this work were of Columbia-0 (Col-0) background. The transgenic lines were described earlier: csn5a-1 and csn5b-1 [24], csn1-10 [12], spa1-3-4 [27], cop1-4 [28], DR5::N7-VENUS [29], and DII-VENUS [30]. Sterile seeds were sown on petri plates containing 1× Murashige and Skoog salts (MS) [31], 0.8% agar, 1% sucrose, and 0.05% MES (2-(N-morpholino) ethanesulfonic acid) at pH 5.7. After 2 days of cold stratification (4 °C in dark) plates were transferred to the growth chamber at 21 °C under long day condition (16 h white light at 100 µmol m2s−1 and 8 h darkness) at 70% relative humidity; 10 days after sowing (DAS), seedlings were transferred to the soil, and stress treatment was given at 14 DAS. For root and confocal microscopy studies, seedlings were grown in liquid MS.
2.2. Stress Treatments
Fourteen DAS, seedlings in soil were treated with either 44 °C or 28 °C for 2 h a day, starting at 11:00, for 7 d. Relative humidity of the chamber was maintained at 55–70% during heat treatment. Other conditions assayed starting at 14 DAS include 200 mM NaCl for 21 d, drought (21 d), UV-C (5000 erg, 1 min), flood (7 d), and cold (4 °C overnight, 7 d). Phenotypes were analyzed either 30, 60 or 70 DAS according to the experiment. For lateral root study, seedlings 10 DAS were grown in liquid MS and treated with heat (2 h, 44 °C, 7 d) and lateral roots were analyzed at 17 DAS.
2.3. Phenomics
Rosette area and photosynthetic parameters were examined by Plant ScreenTM Phenotyping System (Photon Systems Instruments (PSI), Drasov, Czech Republic). Seedlings 10 DAS grown in MS were transferred to PSI standard pots (one seedling per pot), and imaging was done on control and heat-treated seedlings at 14, 18, 21, 25, 28, 32 and 35 DAS.
2.4. Analysis of Cell Size, Cell Number and Stomatal Density
Cell size and cell number studies were performed as described in literature [32]. Sixth true leaf of 30 DAS seedlings was fixed, and chlorophyll was cleared with 70% ethanol. Images of cells were taken under differential interference contrast (DIC) of an Olympus DP71 microscope. Cell size and cell number were calculated using ImageJ software (http://imagej.-nih.gov/ij/) similar to Reference [33].
2.5. Ploidy Analysis
Ploidy analysis was performed as described in literature [34] protocol with slight modification; 20 mg of seedling at 30 DAS control and heat-treated were taken in test tubes containing 1 mL of ice cold Galbraith’s buffer (45 mM MgCl2, 20 mM MOPS (3-(N-morpholino)propanesulfonic acid), 30 mM sodium citrate, and 0.1% TritonX-100 at pH 7) and lysed by tissue lyser (1.5 min at 25 Hz). Homogenate was filtered through 40 µm nylon mesh. The filtrate was centrifuged at 200 g for 5 min to sediment the nuclei, and the pellet was suspended in 200 µL of Galbraith’s buffer. The samples were stained with 50 µg/mL propidium iodide containing 50 µg/mL RNase. After staining, samples were analyzed using Stratedigm S1000Exi flow cytometer (San Jose, CA, USA). Data analysis was done by FlowJo software (Version10) (Tree Star, Inc. Ashland, OR, USA).
2.6. qRT-PCR Analysis
RNA was extracted from 21-DAS control and heat-treated seedlings 3 h after the treatment using TriReagent (MRC) method according to the manufacture instruction. cDNA was synthesized using qScript cDNA Synthetic Kit (Quantabio, Beverly, MA, USA), and qRT-PCR was performed using PerfectCT SYBR Green FastMix. Actin was used as a normalization gene. The experiments were performed in three to five biological replicates with three technical repetitions for each biological sample. List of primers and genes used for qRT-PCR is given in Table S1. For statistical analysis, unpaired Student’s test was used.
2.7. Western Blot
For immunoblots, proteins were extracted 3 h following heat treatment (2 h, 44 °C, 14–21 DAS) from heat-treated and control seedlings. Seedlings were frozen in liquid nitrogen, homogenized in extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 5 mM EDTA (ethylenediaminetetraacetic acid), 5% glycerol, 1 mM PMSF (phenylmethylsulfonyl fluoride), 1 mM DTT (dithiothreitol), and 1X complete cocktail (Roche 1836145)), and centrifuged at 14,000 rpm for 30 min at 4 °C. Protein concentration was determined by Bradford protein assay, and equal amounts of protein were loaded for western blot with specific antibodies anti-CSN5 (Abcam, ab195635, Cambridge, UK), anti-CUL3 (Enzo, BML-PW0470, Enzo Life Sciences, NY, USA), and anti-CUL1 (Enzo, BML-PW0190).
2.8. Confocal Microscopy
Seedlings 6 DAS, from the DR5::N7-VENUS and DII-VENUS lines, grown on liquid MS were treated with heat (2 h, 44 °C, 7 d); roots were stained with propidium iodide (2 µg/mL) and imaged using Zeiss LSM780 confocal microscope 3 h and 3 d after the heat treatment. For the DR5::N7-VENUS line, fluorescence emission was collected between 520–570 nm (band pass) for yellow fluorescent protein (YFP). For the DII-VENUS line, confocal imaging was done with a ChS1 detector for YFP signals, and the fluorescence emission was collected between 508–543 nm (band pass). For propidium iodide, fluorescence emission was collected between 595–695 nm (band pass). YFP fluorescence was quantified using ImageJ software.
3. Results
3.1. Heat Treatment (44 °C, 2 h, 7 d) Enhances the Growth and Lateral Roots of csn5a-1
To study the role of CSN in mediating responses to environmental stress, we studied two hypomorphic mutants, csn1-10 and csn5a-1, under various growth conditions as detailed in the Methods section. We chose these strains since, opposed to loss-of-function csn mutants which are seedling lethal, these strains are viable, reaching seed sets.
At the stage of adult plants, both mutants and WT (wild type) showed slower growth after salt and drought stress, though csn5a-1 was comparatively less affected than csn1-10 or the WT, with csn5a-1 leaves remaining green whereas the leaves of WT and csn1-10 were mostly dried (Figure 1a–c). UV-C reduces the growth of all plants, with csn5a-1 and csn1-10 being hyper-sensitive. Flood and cold stress in the conditions assayed did not show any prominent effect on the growth of WT, csn1-10, as well as csn5a-1 (Figure 1d–f).

Figure 1.
Phenotypic characterization of COP9 (constitutive photomorphogenesis 9) signalosome (CSN) hypomorphic strains (csn5a-1 and csn1-10) under various environmental stress: (a–c) Phenotype of plants 70 days after sowing (DAS): Col-0, csn1-10, and csn5a-1 plants respectively under salt (200 mM NaCl, 21 d), drought (21 d), and heat (2 h, 44 °C, 7 d) stress. White arrows indicate enhanced growth of csn5a-1 under heat stress (2 h, 44 °C, 7 d) in comparison to the control condition. (d–f) Phenotype of plants 60 DAS: Col-0, csn1-10, and csn5a-1 respectively under UV-C (5000 erg, 1 min), flood (7 d), and cold (4 °C overnight, 7 d) stress. (g) Phenotype of plants 30 DAS: csn1-10, Col-0, and csn5a-1 seedlings underwent temperature stresses of 28 °C (2 h, 14–21 DAS) and 44 °C (2 h, 14–21 DAS). Scale bars, 5 cm.
Most interestingly, the growth of csn5a-1 was enhanced after the transient heat regimen of two hours a day at 44 °C for seven days (Figure 1c), whereas these conditions did not affect the growth of WT or csn1-10. Similar experiments but with a milder heat stress of two hours a day at 28 °C showed that the growth of csn5a-1 increased linearly with the highest growth observed in csn5a-1 leaf rosette area at 44 °C (Figure 1g).
To assay if the heat stress-induced growth is specific to csn5a-1 or common for other photo-morphogenesis mutants, we studied additional CSN hypomorphic mutants (csn5b-1 and csn1-10), as well as cop1-4 and a SPA (suppressor of phyA-105) triple mutant (spa1, spa3 and spa4). We observed a significant increase in the growth of csn5a-1 after heat stress, with the leaf rosette area doubling by 28 DAS (Figure S1a). However, this same heat stress did not affect growth of Col-0 (Figure S1b) or any of the other mutants assayed (Figure S1c–f), indicating the specificity of heat stress-induced growth for csn5a-1. Further, we observed an increase in the number of lateral roots following heat stress in csn5a-1 mutants grown in liquid media (Figure S2a–c).
3.2. Enhanced Growth of csn5a-1 after Heat Treatment Correlates with an Increase in Photosynthetic Capacity
To understand how the heat stress may change physiology, leading to the enhanced growth of csn5a-1, we measured several photosynthetic parameters using a high-throughput phenotyping robot. We collected data for chlorophyll fluorescence decline ratio in steady state (Rfd_Lss), which is an indicator of potential photosynthetic activity of leaf and directly correlates to net photosynthetic CO2 assimilation rate [35]. We detected a significant increase in Rfd_Lss after 7 d heat stress in csn5a-1, which remains high up to 11 d following heat stress before declining to the level of csn5a-1 control (Figure 2a). However, the increased Rfd_Lss in Col-0 (Figure 2b), csn1-10 (Figure 2c), and csn5b-1 (Figure 2d) disappeared by the fourth day following the last heat stress. This result led us to suggest that CSN5A provides buffering activity during heat stress, as it appears necessary for plants to return to ground photosynthetic rates following the stress. Rfd_Lss did not change after heat stress in cop1-4 and spa1-3-4, further suggesting that the heat response detected is specific for the mutation in CSN5A and not a general response of perturbation in photomorphogenesis (Figure 2e,f). Further, we observed a different pattern of leaf greenness in WT and csn5a-1 mutant following the heat treatment (Figure S3), where the variation in relative hue abundance indicates the change in the level of chlorophyll content after heat treatment. The hue for pale green (RGB 90, 98, 58) changes more dramatically in csn5a-1 after 7 d of heat treatment. The percentage of pale green increased almost two-fold (58% in csn5a-1 heat versus 30% in csn5a-1 control) following heat treatment of csn5a-1 mutant in 21-DAS seedling. However, such drastic change in hue abundance was not observed in WT after heat treatment. Change in the hue abundance in csn5a-1 mutants after heat treatment suggests a change in chlorophyll content, which improves photosynthesis. In summary, heat stress leads to an increase in photosynthetic activity, which increases net photosynthetic CO2 assimilation rate in csn5a-1 and correlates with an increase in plant size.
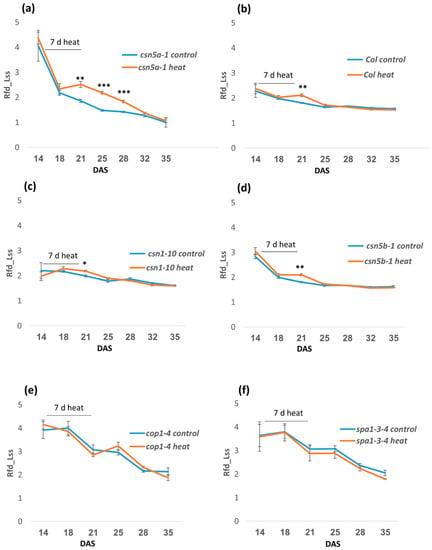
Figure 2.
Phenomics study showing time series of relative fluorescence decline ratio in steady-state (Rfd_Lss) for the photo-morphogenesis repressor mutants following heat stress (2 h, 44 °C, 14–21 DAS). (a) Rfd_Lss increases significantly in csn5a-1 after 7 d heat stress and continues to be higher for 11D before getting equal to csn5a-1 control. (b) Rfd_Lss increases in Col-0 after 7 d heat stress but becomes equal to Col-0 control after 4D. (c) Rfd_Lss increases in csn1-10 after 7 d heat stress but becomes equal to csn1-10 control after 4D. (d) Rfd_Lss increases in csn5b-1 after 7 d heat stress but becomes equal to csn5b-1 control after 4D. (e) Rfd_Lss does not change in cop1-4 after 7 d heat stress. (f) Rfd_Lss does not change in cop1-4 after 7 d heat stress. Error bars represent SEM of biological replicates (n = 4). Student’s t test * p < 0.05; ** p < 0.01; *** p < 0.001.
3.3. Heat Treatment Increases the Cell Size and Ploidy of csn5a-1
To further study the influence of heat stress leading to the increased growth in csn5a-1, we measured cell size and cell numbers on the abaxial side of the 6th true leaf of seedlings 30 DAS. Differential interference contrast (DIC) images showed an increase in the abaxial epidermal cell size of csn5a-1 after heat stress, whereas Col-0 and csn1-10 did not show any significant change in cell size following heat stress (Figure 3a,b). Similarly, an increase in palisade mesophyll cell size was also detected following heat treatment in csn5a-1 (Figure S4). The number of abaxial cells did not change in 6th true leaf of Col-0, csn1-10, and csn5a-1 following heat stress (Figure 3c), indicating that the increase in leaf size of csn5a-1 (Figure 3d) is primarily a result of increased cell size.
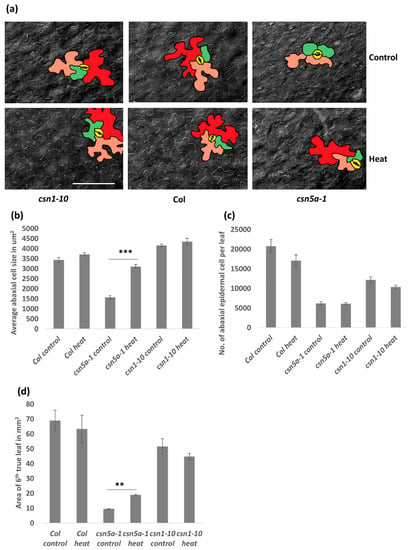
Figure 3.
Image and graph of abaxial epidermal cell size in 6th true leaf of plants 30 DAS having undergone 7 d heat treatment: Cell size increases in csn5a-1 after 7 d heat treatment. (a) Differential interference contrast (DIC) image of abaxial epidermal cell after 7 d heat treatment of 30 DAS csn1-10 (left), Col-0 (middle), and csn5a-1 (right). Cells colored in different color are most likely representing, 2n (green), 4n (orange), 8n (red), and stomata (yellow). Scale bar, 100 µm. (b) Graph displaying average abaxial cell size in µm2. (c) Graph displaying the number of abaxial cells per 6th true leaf. (d) Graph displaying area of 6th true leaf. Error bars represent SEM of biological replicates (n = 2–4). Student’s t test ** p < 0.01; *** p < 0.001.
Increased cell size is often a result of endo-reduplication [36]. Different colors were assigned to represent the predicted ploidy of abaxial epidermal cells (based on Reference [37]), 2n (green), 4n (orange), and 8n (red), which clearly showed the presence of larger cells (8n) in csn5a-1 after heat treatment but not in csn5a-1 control plants. To confirm the level of ploidy in csn5a-1 following heat treatment, we employed flow cytometry. As anticipated, for csn5a-1, we observed an increase in 8n nuclei (12.5% to 22.4%) and decrease in 2n nuclei (45.8% to 31.4%) (Figure 4a,b) after heat treatment. Col-0 and csn1-10 did not show any significant change in the ploidy level after heat stress. Thus, heat stress plays a crucial role in inducing endo-reduplication in csn5a-1, which we predict results in the increased cell size, increased photosynthetic capability, and increased plant size.
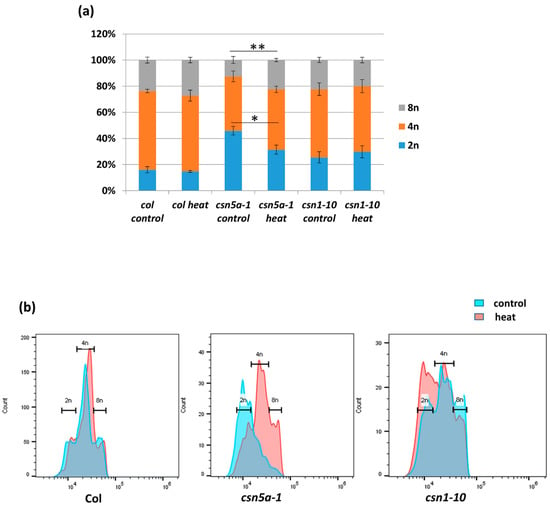
Figure 4.
Difference in the percentage of nuclear ploidy after 7 d heat treatment in csn5a-1 30 DAS. (a) Percentage of diploid, tetraploid, and octaploid nuclei in 30 DAS Col-0, csn5a-1, and csn1-10 seedlings following 7 d heat treatment showing increase in the ploidy of csn5a-1 following heat treatment. (b) Graph showing increase in the level of higher ploidy (4n and 8n) of csn5a-1 (middle) following heat treatment whereas Col-0 (left) and csn1-10 (right) does not show increase in higher ploidy (4n and 8n). Error bars represent SEM of biological replicates (n = 3). Student’s t test * p < 0.05; ** p < 0.01.
3.4. Deneddylation of CUL1, But Not CUL3, is Enhanced in csn5a-1 after Heat Treatment
To determine if the heat stress conditions influence deneddylation activity of the mutant CSN in csn5a-1 seedlings, we employed immunoblot analysis to determine the levels of CSN5 protein and the relative neddylation state of CUL3 and CUL1. As seen in Figure 5a, the levels of CSN5 remained stable following heat shock. Similarly, we could detect no difference in neddylation state of CUL3 following the heat treatment (Figure 5b). However, we detected an increase in deneddylated CUL1 after heat stress in csn5a-1 (Figure 5c). Densitometry analysis using ImageJ revealed a 5-fold increase of CUL1/CUL1Nedd ratio in csn5a-1 following heat treatment, whereas the WT showed a small increase of only 0.19-fold (Table S2). Thus, the phenotypic changes detected in csn5a-1 can possibly be explained by a rescue of CSN-dependent CUL1 deneddylation activity.
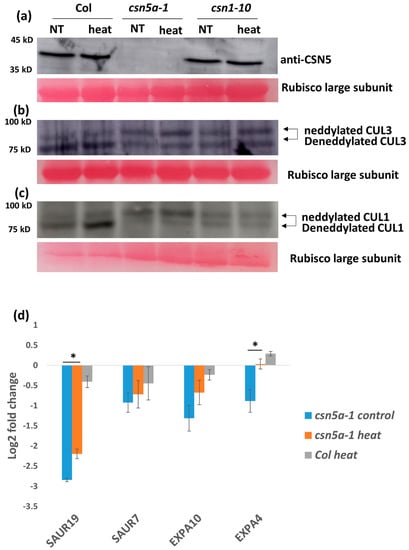
Figure 5.
Increased growth of csn5a-1 following 7 d heat treatment is due to an increase in deneddylation activity (against CUL1) and auxin response. (a) Immunoblot using equal protein concentrations with CSN5 antibodies did not show any change in the protein band of csn5a-1 after heat treatment. (b) Immunoblot using equal protein concentrations with CUL3 antibodies did not show any change in cullin neddylation/deneddylation (93/85 kDa) ratio in either in csn5a-1 or Col-0 after heat treatment. (c) Immunoblot using equal protein concentrations with CUL1 showed increase in deneddylation activity of csn5a-1 and Col-0. Rubisco large subunit stained with ponceau is used as loading control. NT, non-treated. (d) Expression of auxin responsive genes (SAUR19 (Small auxin-up RNA 19) and EXPA4 (Expansin A4) of which the expression was downregulated in csn5a-1 control plants compared to WT (Col-0) control plants increases after heat treatment. Expression of WT (Col-0) control plant is taken as the baseline (0) in the log2 fold change. Error bars represent SEM of biological replicates (n = 3–5). Student’s t test * p < 0.05; ** p < 0.01.
3.5. Auxin Activity Increases in csn5a-1 Following Heat Treatment
csn5a-1 is defective in the auxin-response pathway [24]. Thus, to understand the role of auxin in the enhanced growth of csn5a-1 following heat stress, we analyzed expression levels of the auxin-induced genes SAUR7 (Small auxin-up RNA 7), SAUR19, EXPA4 (Expansin A4), and EXPA10. We observed 1–3 log2 fold downregulations in these genes in csn5a-1 seedlings compared to Col-0, confirming the reduced auxin-response in csn5a-1. The expression of SAUR19 and EXPA4 increases 3 h after heat treatment in csn5a-1 seedlings but not in heat-treated Col-0 (Figure 5d).
To further test the increased activity of CSN-dependent deneddylation of CUL1 containing CRL, we assayed the activity of a known SCF target—the auxin-induced activity of SCFTIR1-mediated protein degradation. We crossed csn5a-1 with the auxin reporter lines DR5::N7-VENUS and DII-VENUS. The DR5::N7-VENUS signal is under the regulation of several ARF (auxin response factor) binding sites [29,38] and thus is directly related to auxin levels, whereas in DII-VENUS, VENUS protein is attached to auxin interaction domain II of AUX/IAA [30] and thus the VENUS signal is inversely related to auxin levels. We generated two homozygous mutant lines: csn5a-1 × DR5::N7-VENUS and csn5a-1 × DII-VENUS. We confirmed the integration and homozygosity by PCR in the F2 population and used the seeds for further study.
Seeds were grown on liquid media and confocal imaging was done 3 h as well as 3 d after the heat treatment (2 h a day at 44 °C, for 6–13 d). We observed significant increases in the levels of VENUS signal in csn5a-1 × DR5::N7-VENUS 3 h after the final heat treatment compared to untreated plants, while no such increase was found in the WT (DR5) background (Figure 6a). Three days following the final heat treatment, the level of DR5::N7-VENUS fluorescence reverted to control levels. However, the fluorescence level of heat treated csn5a-1 × DR5::N7-VENUS remained high compared to the control even after three days (Figure 6b), indicating an increase in auxin activity after heat treatment in csn5a-1.
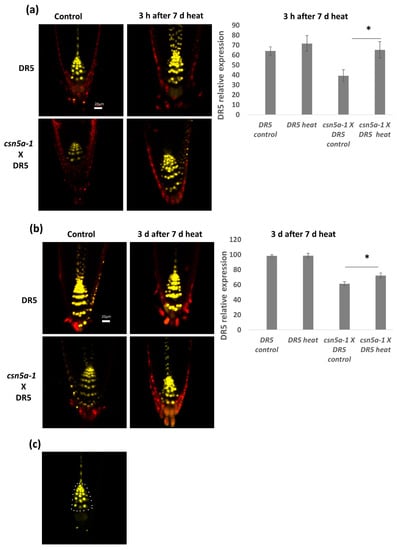
Figure 6.
Auxin activity increases in csn5a-1 following 7 d heat treatment: (a) Relative integrated density in the roots of DR5::N7-VENUS and csn5a-1 × DR5::N7-VENUS, 3 h after 7 d heat treatment. (b) Relative integrated density of DR5::N7-VENUS and csn5a-1 × DR5::N7-VENUS, 3 d after 7 d heat treatment. Cell wall was stained with propidium iodide. (c) White dots represent the area used to quantify DR5 expression levels. Scale bars, 20 µm. Error bars represent SEM of biological replicates (n = 4–7). Student’s t test * p < 0.05.
To determine if the increase in auxin activity is due to a decrease in auxin repressor activity, we assayed the homozygous line csn5a-1 × DII-VENUS. In this line, higher VENUS levels indicate higher levels of the auxin repressor and lower levels of auxin activity. We observed a decrease of the DII-VENUS fluorescent level in both DII-VENUS and csn5a-1 × DII-VENUS lines 3 h after the final heat treatment, suggesting that heat enhances the degradation of the auxin repressor (Figure 7a). Three days after the final heat treatment, DII-VENUS fluorescence levels in the control line returned to baseline values (e.g., pre-heat treatment). On the other hand, in the csn5a-1 × DII-VENUS line, DII-VENUS fluorescence levels remained low for three days following the final heat treatment (Figure 7b), indicating enhanced degradation and maintained auxin response. These results correlate with the increased deneddylation of CUL1 in csn5a-1 following heat stress. Thus, heat stress increases CUL1 deneddylation, which mediates degradation of AUX/IAA repressor to increase the auxin signaling and growth of csn5a-1.
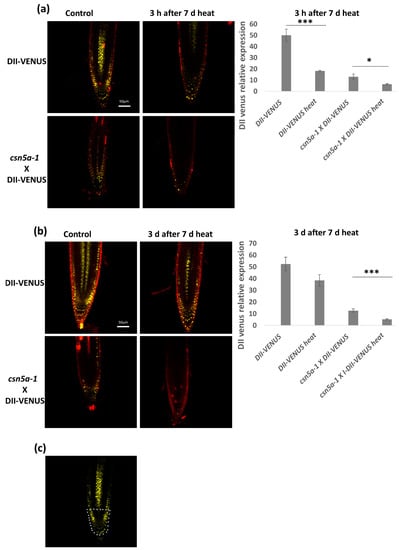
Figure 7.
Auxin repressor activity remains low even after 3 d of 7 d heat treatment. (a) Relative integrated density in the roots of DII-VENUS and csn5a-1 × DII-VENUS, 3 h after 7 d heat treatment. (b) Relative integrated density of DII-VENUS and csn5a-1 × DII-VENUS, 3 days after 7 d heat treatment. Cell wall was stained with propidium iodide. (c) White dots represent the area used to quantify VENUS expression levels. Scale bars, 50 µm. Error bars represent SEM of biological replicates (4–7). Student’s t test * p < 0.05; *** p < 0.001.
4. Discussion
Numerous studies of the COP9 signalosome have highlighted its role in regulating plant responses to diverse signals [6,7,8,9,10,11,13,14,18]. Here, we showed that exposure to heat stress partially rescues the dwarf phenotype of the csn5a-1 mutant. This rescue manifested at numerous levels. At the level of growth morphology, a phenomic analysis revealed a gradual, significant increase in rosette size following heat treatment in csn5a-1 but not in the WT (Col-0). The increased growth detected was also seen at the cellular level, with both leaf epidermal and mesophyll cells showing increased size in csn5a-1 following the heat treatment. This increased cell size correlated with a rescue of ploidy in epidermal cells.
Physiologically, we detected an increase of net photosynthetic CO2 assimilation rate following heat stress. In WT and csn5b-1, the net photosynthetic CO2 assimilation rate remained elevated for four days following the heat regimen. For csn5a-1, the net photosynthetic CO2 assimilation rate remained elevated for 11 days following heat treatment (Figure 2a). Physiologically, leaf growth is regulated by carbon availability, hydraulic properties, and wall extensibility, which promotes cell division, cell expansion, and increase in ploidy [39]. Thus, the increased CO2 assimilation rate in csn5a-1 indicates an increase in photosynthesis rate and is likely the main cause for the increases in growth of the mutant.
When first confronted with the heat-enhanced growth phenotype of csn5a-1, we considered that this heat-enhanced growth phenotype may be a characteristic of other photomorphogenic mutants. However, it appears to be specific for the mutation in csn5a-1, as the growth of csn1-10, csn5b-1, cop1-4, and spa1-3-4 was not enhanced following the same heat treatments.
A straightforward explanation for the improved growth of csn5a-1 following heat is that CSN activity could be enhanced under elevated temperatures. Indeed, we detected enhanced deneddylation of CUL1 in csn5a-1 following heat conditions (Figure 5c). In the absence of CSN5A, the deneddylation activity in this mutant suggests the contribution of CSN complexes containing CSN5B [40]. Thus, it appears that the catalytic activity of this specific subset of CSN complexes is heat enhanced. On the other hand, we could not detect a change in CUL3 deneddylation status in csn5a-1 following the same heat conditions (Figure 5b). While CUL3 forms the E3 complex with RBX1 and BTB proteins and is involved in ethylene synthesis [41], CUL1 coordinates SCF E3 complexes together with SKP1 and RBX1 and has a major role in auxin signaling [7]. Thus, the enhanced growth of the csn5a-1 correlates with increased deneddylation activity of CUL1, which increases auxin signaling following heat treatment.
We propose that, overall, CSN5A functions in buffering a plant’s response to heat, partially through regulating the auxin response to heat. Auxin is of course essential for plant growth, including regulation of cell division, cell expansion, and differentiation [42,43,44]. Accordingly, auxin-resistant mutants are impaired in cell division, cell expansion, and lateral root formation [45,46]. csn5a-1 also shows similar phenotypes which implicated CSN5 and the CSN-mediated degradation of AUX/IAA in regulating auxin responses such as cell division and expansion [7].
Indeed, we found that auxin signaling is enhanced in csn5a-1 following heat stress. In control lines, heat induces an increase in auxin signaling, which reverts to baseline levels three days following the heat treatment. In csn5a-1, this heat-induced increase in auxin signaling was maintained even after 3 d of stress (Figure 6b), suggesting a role of CSN5A in auxin homeostasis following recurrent heat treatment.
These results are in line with earlier transcriptomic analyses carried out in our lab which suggested that heat stress influences auxin signaling. In the short duration of 3 h of heat stress, we found a change in the auxin response pathway (Figure S5). Network analysis revealed that the heat-induced upregulated auxin pathway genes fall mostly in the GH3 and Small auxin-up RNA (SAUR) families, forming two major networks. The first network connects SAUR55, SAUR16, GH3.1, and GH3.17 while the second connects SAUR14, SAUR6, and the O-fucosyltransferase family (Figure S6). Auxin mediates acid growth by loosening the wall at low pH, which elevates wall extensibility and results in fast cell elongation [47,48], and this process is controlled in part by an additional SAUR, SAUR19 [49]. Further, this acid growth requires EXPANSINS which are non-enzymatic wall-loosening proteins which break polysaccharide networks by fragmenting and loosening the connection between cellulose microfibrils and polysaccharides [31,50,51].
We propose that recurrent heat stress leads to the degradation of the AUX/IAA auxin repressor in a CUL1-deneddylation dependent manner, resulting in higher auxin activity, and that this leads to an induction of downstream genes such as SAURs (e.g., SAUR19) and EXPANSINs (e.g., EXPA4), as was shown in the expression study in Figure 5d. These genes then function in pathways, resulting in cell elongation and plant growth. CSN5A is required for AUX/IAA recovery following heat stress to maintain proper auxin homeostasis in plants.
Supplementary Materials
Figure S1: Number of lateral root increases in csn5a-1 seedlings following 7 d heat treatment, Figure S2: Number of lateral root increases in csn5a-1 seedlings following 7 d heat treatment, Figure S3: Change in the percentage of leaf greenness in csn5a-1 mutant after heat stress is different from Col-0, Figure S4: Image of mesophyll cells and size in 6th true leaves of 30 DAS seedlings having undergone 7 d heat treatment, Figure S5: Heat map showing the distribution of differentially upregulated pathways, Figure S6: Network association of genes upregulated in csn5a-1 heat compared to WT heat, Table S1: Primers used in this research work, Table S2. Fold change of deneddylated vs neddylated cullin following heat treatment.
Author Contributions
A.K.S. and D.A.C. conceived and designed the research experiments. A.K.S., B.S.Y., S.D., and M.B. performed the experiments. A.K.S., B.S.Y., A.F., and D.A.C. analyzed the data. A.K.S. and D.A.C. wrote the manuscript.
Funding
This research was funded through the Israeli Centers for Research Excellence Program of the Planning and Budgeting Committee, Israel Science Foundation Grant 757/12 (to D.A.C.) and the Israel Science Foundation Grant (060411552/18; to D.A.C.).
Acknowledgments
We thank the following for providing seeds: William Grey for csn1-10 and Ute Höcker for spa1-3-4. We thank Guilia Meshulam for assistance with the phenomics study.
Conflicts of Interest
There is no conflict of interest.
References
- Deng, X.W.; Dubiel, W.G.; Wei, N.; Hofmann, K.; Mundt, K.; Colicelli, L.; Kato, J.; Naumann, M.; Segal, D.; Seeger, M. Unified nomenclature for the COP9 signalosome and its subunits: An essential regulator of development. Trends Genet. 2000, 16, 202–203. [Google Scholar] [CrossRef]
- Chamovitz, D.A.; Wei, N.; Osterlund, M.T.; von Arnim, A.G.; Staub, J.M.; Matsui, M.; Deng, X.W. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 1996, 86, 115–121. [Google Scholar] [CrossRef]
- Wei, N.; Chamovitz, D.A.; Deng, X.W. Arabidopsis Cop9 Is a Component of a Novel Signaling Complex Mediating Light Control of Development. Cell 1994, 78, 117–124. [Google Scholar] [CrossRef]
- Serino, G.; Su, H.W.; Peng, Z.H.; Tsuge, T.; Wei, N.; Gu, H.Y.; Deng, X.W. Characterization of the last subunit of the Arabidopsis COP9 signalosome: Implications for the overall structure and origin of the complex. Plant Cell 2003, 15, 719–731. [Google Scholar] [CrossRef]
- Singh, A.K.; Chamovitz, D.A. Role of Cop9 Signalosome Subunits in the Environmental and Hormonal Balance of Plant. Biomolecules 2019, 9. [Google Scholar] [CrossRef]
- Franciosini, A.; Lombardi, B.; Iafrate, S.; Pecce, V.; Mele, G.; Lupacchini, L.; Rinaldi, G.; Kondou, Y.; Gusmaroli, G.; Aki, S. The Arabidopsis COP9 Signalosome Interacting F-Box Kelch 1 Protein Forms an SCF Ubiquitin Ligase and Regulates Hypocotyl Elongation. Mol. Plant 2013, 6, 1616–1629. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Serino, G.; Callis, J.; Crosby, W.L.; Lyapina, S.; Deshaies, R.J.; Gray, W.M.; Estelle, M.; Deng, X.W. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 2001, 292, 1379–1382. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.H.; Ma, L.G.; Wang, X.P.; Xie, D.X.; Dinesh-Kumar, S.P.; Wei, N.; Deng, X.W. The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 2003, 15, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Feng, S.H.; Nakayama, N.; Crosby, W.L.; Irish, V.; Deng, X.W.; Wei, N. The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell 2003, 15, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Dohmann, E.M.N.; Nill, C.; Schwechheimer, C. DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur. J. Cell Biol. 2010, 89, 163–168. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Figueroa, P.; Serino, G.; Deng, X.W. Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis cullin3-based E3 ligases. Plant Cell 2007, 19, 564–581. [Google Scholar] [CrossRef]
- Zhang, W.J.; Ito, H.; Quint, M.; Huang, H.; Noel, L.D.; Gray, W.M. Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. P. Natl. Acad. Sci. USA 2008, 105, 8470–8475. [Google Scholar] [CrossRef]
- Liu, Y.L.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef]
- Hind, S.R.; Pulliam, S.E.; Veronese, P.; Shantharaj, D.; Nazir, A.; Jacobs, N.S.; Stratmann, J.W. The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J. 2011, 65, 480–491. [Google Scholar] [CrossRef]
- Meir, M.; Galanty, Y.; Kashani, L.; Blank, M.; Khosravi, R.; Fernandez-Avila, M.J.; Cruz-Garcia, A.; Star, A.; Shochot, L.; Thomas, Y. The COP9 signalosome is vital for timely repair of DNA double-strand breaks. Nucleic Acids Res. 2015, 43, 4517–4530. [Google Scholar] [CrossRef] [PubMed]
- Groisman, R.; Polanowska, J.; Kuraoka, I.; Sawada, J.; Saijo, M.; Drapkin, R.; Kisselev, A.F.; Tanaka, K.; Nakatani, Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 2003, 113, 357–367. [Google Scholar] [CrossRef]
- Mayor-Ruiz, C.; Jaeger, M.G.; Bauer, S.; Brand, M.; Sin, C.; Hanzl, A.; Mueller, A.C.; Menche, J.; Winter, G.E. Plasticity of the Cullin-RING Ligase Repertoire Shapes Sensitivity to Ligand-Induced Protein Degradation. Mol. Cell 2019, 75, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Dohmann, E.M.N.; Levesque, M.P.; De Veylder, L.; Reichardt, I.; Juergens, G.; Schmid, M.; Schwechheimer, C. The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 2008, 135, 2013–2022. [Google Scholar] [CrossRef]
- Mundt, K.E.; Porte, J.; Murray, J.M.; Brikos, C.; Christensen, P.U.; Caspari, T.; Hagan, I.M.; Millar, J.B.A.; Simanis, V.; Hofmann, K. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 1999, 9, 1427–1430. [Google Scholar] [CrossRef]
- Panattoni, M.; Sanvito, F.; Basso, V.; Doglioni, C.; Casorati, G.; Montini, E.; Bender, J.R.; Mondino, A.; Pardi, R. Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J. Exp. Med. 2008, 205, 465–477. [Google Scholar] [CrossRef]
- Lyapina, S.; Cope, G.; Shevchenko, A.; Serino, G.; Tsuge, T.; Zhou, C.S.; Wolf, D.A.; Wei, N.; Shevchenko, A.; Deshaies, R.J. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 2001, 292, 1382–1385. [Google Scholar] [CrossRef] [PubMed]
- Cope, G.A.; Suh, G.S.B.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Kwok, S.F.; Solano, R.; Tsuge, T.; Chamovitz, D.A.; Ecker, J.R.; Matsui, M.; Deng, X.W. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 1998, 10, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Dohmann, E.M.N.; Kuhnle, C.; Schwechheimer, C. Loss of the constitutive photomorphogenic9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 2005, 17, 1967–1978. [Google Scholar] [CrossRef]
- Jin, D.; Li, B.S.; Deng, X.W.; Wei, N. Plant COP9 Signalosome subunit 5, CSN5. Plant. Sci. 2014, 224, 54–61. [Google Scholar] [CrossRef]
- Jin, D.; Wu, M.; Li, B.S.; Bucker, B.; Keil, P.; Zhang, S.M.; Li, J.G.; Kang, D.M.; Liu, J.; Dong, J. The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLoS Genet. 2018, 14, e1007237. [Google Scholar] [CrossRef]
- Fittinghoff, K.; Laubinger, S.; Nixdorf, M.; Fackendahl, P.; Baumgardt, R.L.; Batschauer, A.; Hoecker, U. Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 2006, 47, 577–590. [Google Scholar] [CrossRef]
- Mcnellis, T.W.; Vonarnim, A.G.; Araki, T.; Komeda, Y.; Misera, S.; Deng, X.W. Genetic and Molecular Analysis of an Allelic Series of Cop1 Mutants Suggests Functional Roles for the Multiple Protein Domains. Plant Cell 1994, 6, 487–500. [Google Scholar]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef]
- Brunoud, G.; Wells, D.M.; Oliva, M.; Larrieu, A.; Mirabet, V.; Burrow, A.H.; Beeckman, T.; Kepinski, S.; Traas, J.; Bennett, M.J. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 2012, 482, 103. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, M.S.; Krishnamachari, V.; Payton, P.; Sun, Y.; Grimson, M.; Farag, M.A.; Ryu, C.M.; Allen, R.; Melo, I.S. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 2007, 226, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Cao, L.; Wang, S.; Li, Y.P.; Shi, X.Z.; Liu, H.; Li, L.X.; Zhang, Z.L.; Fowke, L.C.; Wang, H. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 2013, 75, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bashir, T.; Sailer, C.; Gurumoorthy, V.; Ramakrishnan, A.M.; Dhanapal, S.; Grossniklaus, U.; Baskar, R. Parental Age Affects Somatic Mutation Rates in the Progeny of Flowering Plants. Plant Physiol. 2015, 168, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Dolezel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Miehe, J.A. Fluorescence imaging as a diagnostic tool for plant stress. Trends Plant Sci. 1997, 2, 316–320. [Google Scholar] [CrossRef]
- Sugimoto-Shirasu, K.; Roberts, K. “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 2003, 6, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Boudolf, V.; Barroco, R.; Engler, J.D.; Verkest, A.; Beeckman, T.; Naudts, M.; Inze, D.; De Veylder, L. B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 2004, 16, 945–955. [Google Scholar] [CrossRef]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar]
- Massonnet, C.; Tisné, S.; Radziejwoski, A.; Vile, D.; De Veylder, L.; Dauzat, M.; Granier, C. New insights into the control of endoreduplication: Endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol. 2011, 157, 2044–2055. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Feng, S.H.; Deng, X.W. The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 2004, 16, 2984–3001. [Google Scholar] [CrossRef]
- Thomann, A.; Lechner, E.; Hansen, M.; Dumbliauskas, E.; Parmentier, Y.; Kieber, J.; Scheres, B.; Genschik, P. Arabidopsis CULLIN3 Genes Regulate Primary Root Growth and Patterning by Ethylene-Dependent and -Independent Mechanisms. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Rayle, D.L.; Cleland, R.E. Evidence that auxin-induced growth of soybean hypocotyls involves proton excretion. Plant Physiol. 1980, 66, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y. Auxin-induced cell elongation and cell wall changes. Bot. Mag. Shokubutsu-gaku-zasshi 1990, 103, 345. [Google Scholar] [CrossRef]
- Velasquez, S.M.; Barbez, E.; Kleine-Vehn, J.; Estevez, J.M. Auxin and cellular elongation. Plant Physiol. 2016, 170, 1206–1215. [Google Scholar] [CrossRef]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]
- Perrot-Rechenmann, C. Cellular Responses to Auxin: Division versus Expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Karcz, W.; Stolarek, J.; Pietruszka, M.; Malkowski, E. The Dose-Response Curves for Iaa Induced Elongation Growth and Acidification of the Incubation Medium of Zea-Mays Coleoptile Segments. Physiol. Plant 1990, 80, 257–261. [Google Scholar] [CrossRef]
- Arsuffi, G.; Braybrook, S.A. Acid growth: An ongoing trip. J. Exp. Bot. 2018, 69, 137–146. [Google Scholar] [CrossRef]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef]
- Wang, L.; Ruan, Y.L. Regulation of cell division and expansion by sugar and auxin signaling. Front. Plant Sci. 2013, 4, 163. [Google Scholar] [CrossRef]
- Kuluev, B.R.; Knyazev, A.B.; Lebedev, Y.P.; Chemeris, A.V. Morphological and physiological characteristics of transgenic tobacco plants expressing expansin genes: AtEXP10 from Arabidopsis and PnEXPA1 from poplar. Russ. J. Plant Physiol. 2012, 59, 97–104. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).