Gene Expression Signature-Based Approach Identifies Antifungal Drug Ciclopirox As a Novel Inhibitor of HMGA2 in Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Antibodies
2.2. Computational Prediction and Assessment of Novel HMGA2 Inhibitor
2.3. Cell Cultures
2.4. Cell Viability Assay
2.5. Focus Formation Assays
2.6. Flow Cytometry Analysis
2.7. Western Blotting
2.8. In Vivo Analysis of Effect of CPX
2.9. Immunohistochemistry
2.10. In-Silico Modeling Analysis
2.11. Statistical Analysis
3. Results
3.1. Identification of a Potential Inhibitor of HMGA2
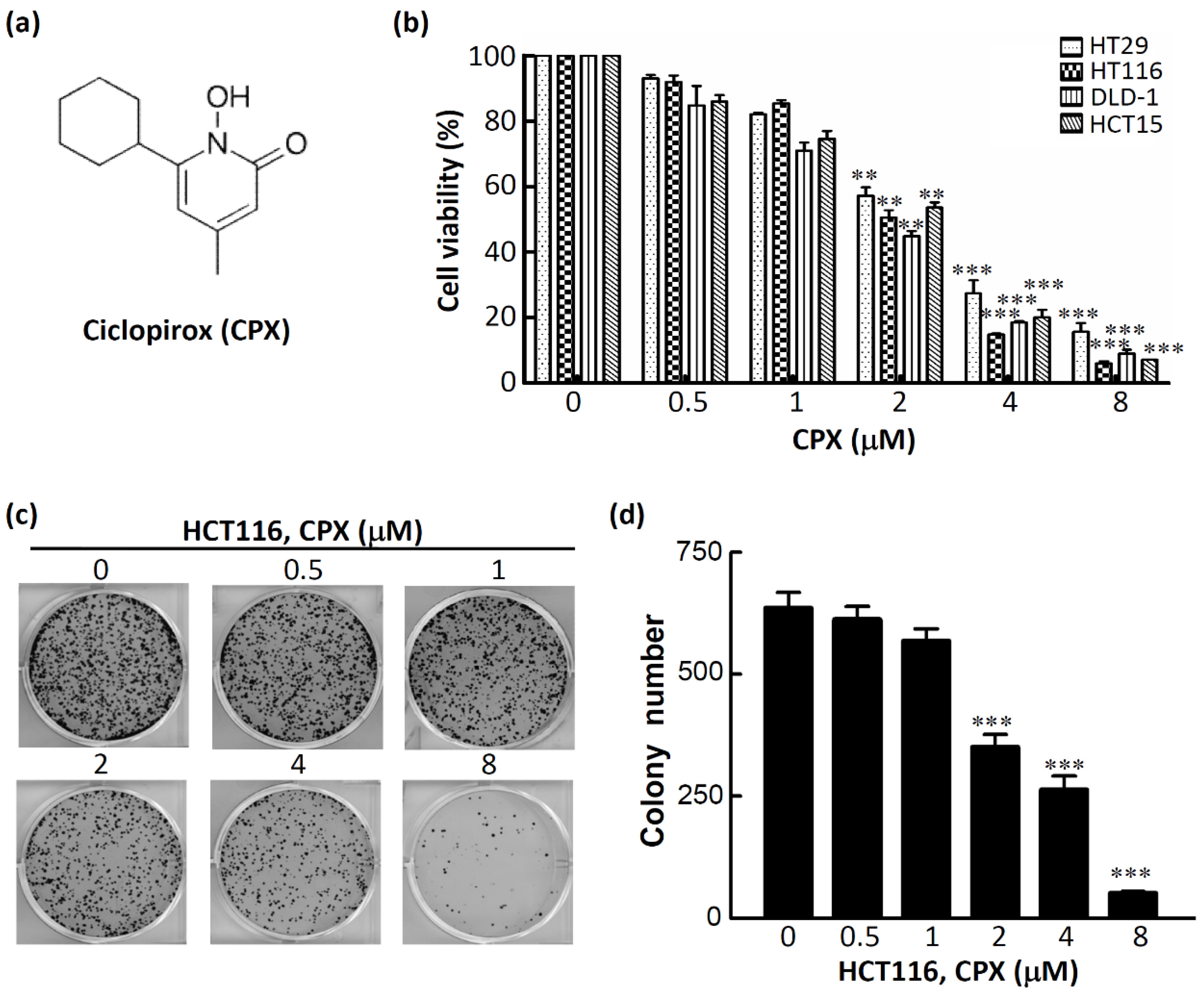
3.2. CPX Inhibits Cell Proliferation in CRC Cells
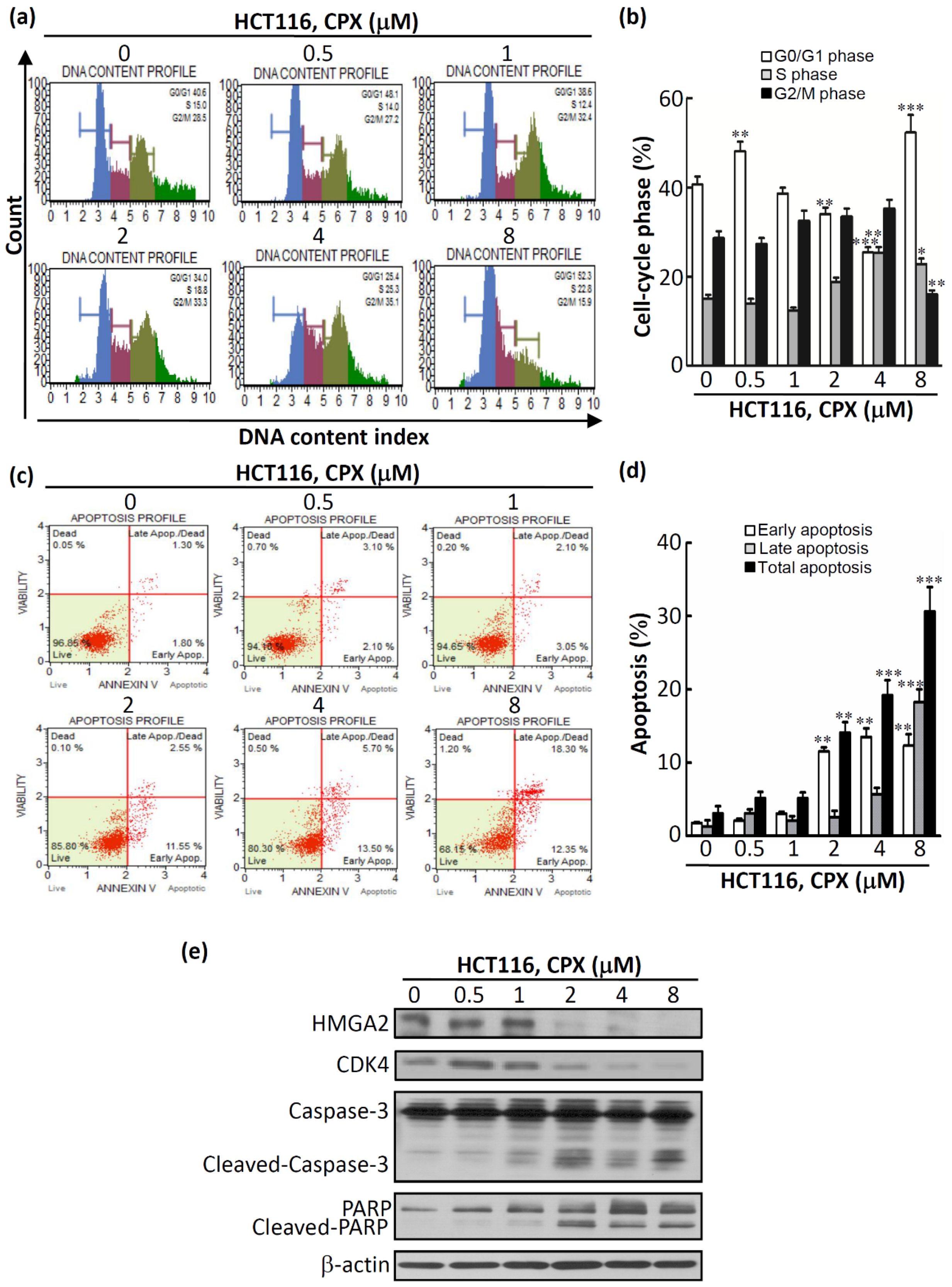
3.3. The Therapeutic Efficacy of CPX in CRC Cells
3.4. In Vivo Efficacy of CPX in Human CRCHCT116 Xenograft Mice Models
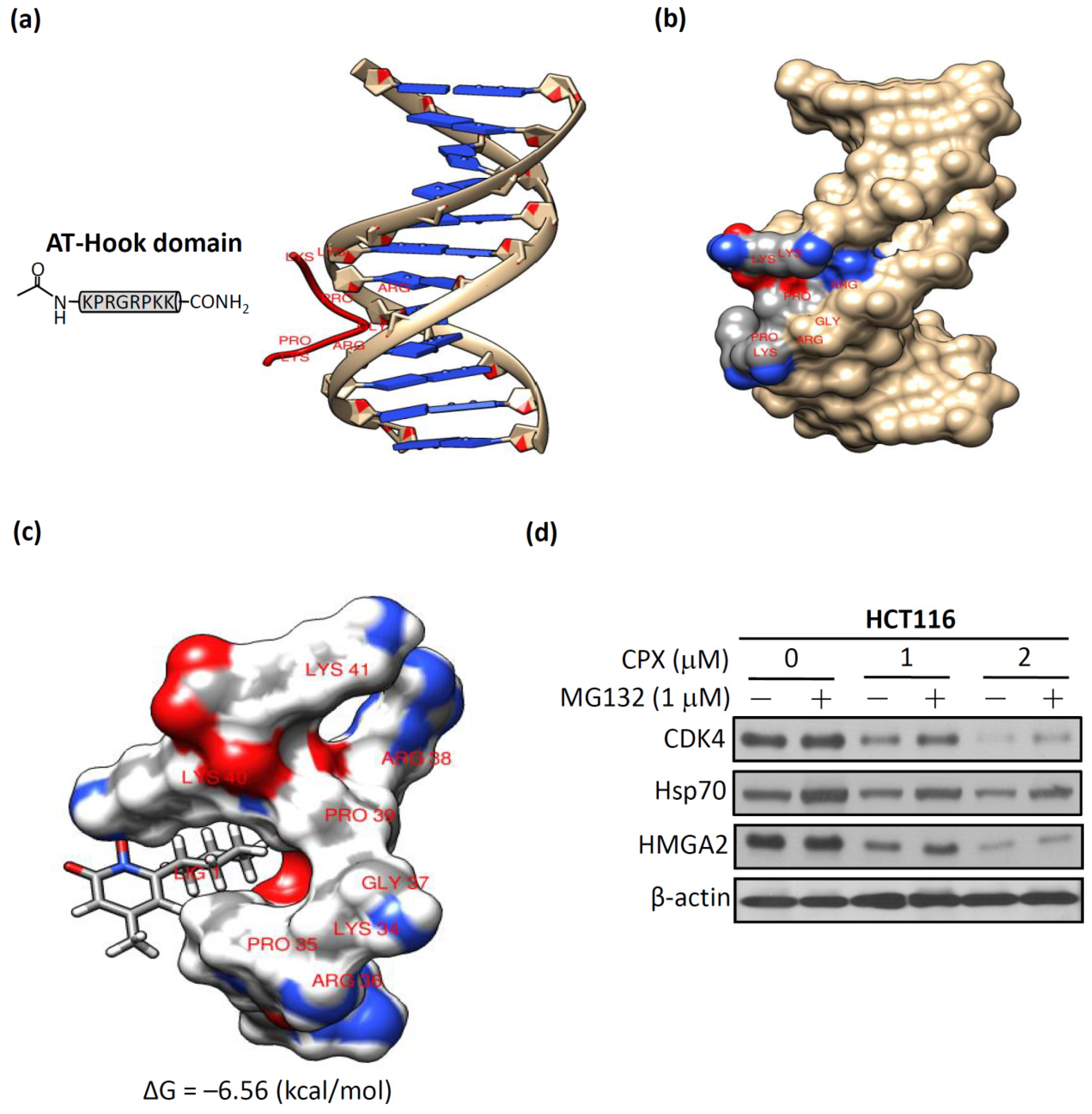
3.5. Molecular Docking of CPX on A-T Hook Motif of HMGA2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Boil. 2014, 2, 5. [Google Scholar]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Di Cello, F.; Hillion, J.; Hristov, A.; Wood, L.J.; Mukherjee, M.; Schuldenfrei, A.; Kowalski, J.; Bhattacharya, R.; Ashfaq, R.; Resar, L.M.S. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 2008, 6, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Dai, M.; Li, Q.; Wang, Z.; Lu, Y.; Song, Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac. Cancer 2017, 8, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Malek, A.; Bakhidze, E.; Noske, A.; Sers, C.; Aigner, A.; Schäfer, R.; Tchernitsa, O. HMGA2 gene is a promising target for ovarian cancer silencing therapy. Int. J. Cancer 2008, 123, 348–356. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, S.; Shan, J.; Hu, Z.; Liu, X.; Chen, L.; Ren, X.; Yao, L.; Sheng, H.; Li, L.; et al. Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett. 2016, 376, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Sakata, J.; Hirosue, A.; Yoshida, R.; Kawahara, K.; Matsuoka, Y.; Yamamoto, T.; Nakamoto, M.; Hirayama, M.; Takahashi, N.; Nakamura, T.; et al. HMGA2 Contributes to Distant Metastasis and Poor Prognosis by Promoting Angiogenesis in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 2473. [Google Scholar] [CrossRef]
- Chiou, S.-H.; Dorsch, M.; Kusch, E.; Naranjo, S.; Kozak, M.M.; Koong, A.C.; Winslow, M.M.; Grüner, B.M. Hmga2 is dispensable for pancreatic cancer development, metastasis, and therapy resistance. Sci. Rep. 2018, 8, 14008. [Google Scholar] [CrossRef]
- Kao, C.-Y.; Yang, P.-M.; Wu, M.-H.; Huang, C.-C.; Lee, Y.-C. Heat shock protein 90 is involved in the regulation of HMGA2-driven growth and epithelial-to-mesenchymal transition of colorectal cancer cells. PeerJ 2016, 4, e1683. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Li, A.Y.-J.; Chen, L.; Lai, L.; Lin, H.H.; Hu, S.; Yao, L.; Peng, J.; Loera, S.; et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 2011, 17, 2570–2580. [Google Scholar] [CrossRef]
- Califano, D.; Pignata, S.; Losito, N.S.; Ottaiano, A.; Greggi, S.; De Simone, V.; Cecere, S.; Aiello, C.; Esposito, F.; Fusco, A.; et al. High HMGA2 expression and high body mass index negatively affect the prognosis of patients with ovarian cancer. J. Cell Physiol. 2014, 229, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Hao, M.; Takeda, T.; Bryant, S.H.; Wang, Y. Large-Scale Prediction of Drug-Target Interaction: A Data-Centric Review. AAPS J. 2017, 19, 1264–1275. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Setola, V.; Irwin, J.J.; Laggner, C.; Abbas, A.I.; Hufeisen, S.J.; Jensen, N.H.; Kuijer, M.B.; Matos, R.C.; Tran, T.B.; et al. Predicting new molecular targets for known drugs. Nature 2009, 462, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Guo, X.; Duan, C.; Ma, W.; Zhang, Y.; Xu, P.; Gao, Z.; Wang, Z.; Yan, H.; Zhang, Y.; et al. Comparative analysis of gene expression profiles between the normal human cartilage and the one with endemic osteoarthritis. Osteoarthr. Cartil. 2009, 17, 83–90. [Google Scholar] [CrossRef]
- Hu, G.; Agarwal, P. Human Disease-Drug Network Based on Genomic Expression Profiles. PLoS ONE 2009, 4, e6536. [Google Scholar] [CrossRef]
- Del Rio, M.; Molina, F.; Bascoul-Mollevi, C.; Copois, V.; Bibeau, F.; Chalbos, P.; Bareil, C.; Kramar, A.; Salvetat, N.; Fraslon, C.; et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J. Clin. Oncol. 2007, 25, 773–780. [Google Scholar] [CrossRef]
- Holleman, A.; Cheok, M.H.; Boer, M.L.D.; Yang, W.; Veerman, A.J.; Kazemier, K.M.; Pei, D.; Cheng, C.; Pui, C.-H.; Relling, M.V.; et al. Gene-Expression Patterns in Drug-Resistant Acute Lymphoblastic Leukemia Cells and Response to Treatment. New Engl. J. Med. 2004, 351, 533–542. [Google Scholar] [CrossRef]
- Robert, J.; Vekris, A.; Pourquier, P.; Bonnet, J. Predicting drug response based on gene expression. Crit. Rev. Oncol. 2004, 51, 205–227. [Google Scholar] [CrossRef]
- Sessa, C.; Shapiro, G.I.; Bhalla, K.N.; Britten, C.; Jacks, K.S.; Mita, M.; Papadimitrakopoulou, V.; Pluard, T.; Samuel, T.A.; Akimov, M.; et al. First-in-Human Phase I Dose-Escalation Study of the HSP90 Inhibitor AUY922 in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2013, 19, 3671–3680. [Google Scholar] [CrossRef]
- Duan, Q.; Flynn, C.; Niepel, M.; Hafner, M.; Muhlich, J.L.; Fernandez, N.F.; Rouillard, A.D.; Tan, C.M.; Chen, E.Y.; Golub, T.R.; et al. LINCS Canvas Browser: Interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014, 42, W449–W460. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Marshall, K.A.; et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009, 37, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Tang, W.C.; Cheng, Y.W.; Sia, P.; Huang, C.C.; Lee, Y.C.; Jiang, H.Y.; Wu, M.H.; Lai, I.L.; Lee, J.W.; et al. Targeting of multiple oncogenic signaling pathways by Hsp90 inhibitor alone or in combination with berberine for treatment of colorectal cancer. Biochim Biophys Acta 2015, 1853, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-C.; Lee, P.-T.; Wu, M.-H.; Huang, C.-C.; Ko, C.-Y.; Lee, Y.-C.; Lin, D.-Y.; Cheng, Y.-W.; Lee, K.-H. Distinct roles and differential expression levels of Wnt5a mRNA isoforms in colorectal cancer cells. PLoS ONE 2017, 12, e0181034. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.W.; Chou, C.-J.; Huang, T.-C.; Yang, P.-M. An Integrated Bioinformatics Analysis Repurposes an Antihelminthic Drug Niclosamide for Treating HMGA2-Overexpressing Human Colorectal Cancer. Cancers 2019, 11, 1482. [Google Scholar] [CrossRef]
- Venkatesan, N.; Krishnakumar, S.; Deepa, P.R.; Deepa, M.; Khetan, V.; Reddy, M.A. Molecular deregulation induced by silencing of the high mobility group protein A2 gene in retinoblastoma cells. Mol. Vis. 2012, 18, 2420–2437. [Google Scholar]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Reeves, R. Nuclear functions of the HMG proteins. Biochim Biophys Acta 2010, 1799, 3–14. [Google Scholar] [CrossRef]
- Aravind, L. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998, 26, 4413–4421. [Google Scholar] [CrossRef] [PubMed]
- Cattaruzzi, G.; Altamura, S.; Tessari, M.A.; Rustighi, A.; Giancotti, V.; Pucillo, C.; Manfioletti, G. The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res. 2007, 35, 1751–1760. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Filarsky, M.; Zillner, K.; Araya, I.; Villar-Garea, A.; Merkl, R.; Längst, G.; Németh, A. The extended AT-hook is a novel RNA binding motif. RNA Boil. 2015, 12, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 2001, 277, 63–81. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Shen, T.; Huang, S. Repositioning the Old Fungicide Ciclopirox for New Medical Uses. Curr. Pharm. Des. 2016, 22, 4443–4450. [Google Scholar] [CrossRef]
- Subissi, A.; Monti, D.; Togni, G.; Mailland, F. Ciclopirox: Recent nonclinical and clinical data relevant to its use as a topical antimycotic agent. Drugs 2010, 70, 2133–2152. [Google Scholar] [CrossRef]
- Kellner, H.M.; Arnold, C.E.; Christ, O.; Eckert, H.G.; Herok, J.; Hornke, I.; Rupp, W. [Pharmacokinetics and biotransformation of the antimycotic drug ciclopiroxolamine in animals and man after topical and systemic administration]. Arzneimittelforschung 1981, 31, 1337–1353. [Google Scholar]
- Luo, Y.; Zhou, H.; Liu, L.; Shen, T.; Chen, W.; Xu, B.; Han, X.; Zhang, F.; Scott, R.S.; Alexander, J.S.; et al. The fungicide ciclopirox inhibits lymphatic endothelial cell tube formation by suppressing VEGFR-3-mediated ERK signaling pathway. Oncogene 2011, 30, 2098–2107. [Google Scholar] [CrossRef]
- Zhou, H.; Shen, T.; Luo, Y.; Liu, L.; Chen, W.; Xu, B.; Han, X.; Pang, J.; Rivera, C.A.; Huang, S. The antitumor activity of the fungicide ciclopirox. Int. J. Cancer 2010, 127, 2467–2477. [Google Scholar] [CrossRef]
- Clement, P.M.; Hanauske-Abel, H.M.; Wolff, E.C.; Kleinman, H.K.; Park, M.H. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro. Int. J. Cancer 2002, 100, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Minden, M.D.; Hogge, D.E.; Weir, S.J.; Kasper, J.; Webster, D.A.; Patton, L.; Jitkova, Y.; Hurren, R.; Gronda, M.; Goard, C.A.; et al. Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. Am. J. Hematol. 2014, 89, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Kinnings, S.L.; Liu, N.; Buchmeier, N.; Tonge, P.J.; Xie, L.; Bourne, P.E. Drug Discovery Using Chemical Systems Biology: Repositioning the Safe Medicine Comtan to Treat Multi-Drug and Extensively Drug Resistant Tuberculosis. PLoS Comput. Boil. 2009, 5, e1000423. [Google Scholar] [CrossRef] [PubMed]
- Polamreddy, P.; Gattu, N. The drug repurposing landscape from 2012 to 2017: Evolution, challenges, and possible solutions. Drug Discov. Today 2019, 24, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Mestres, J. Drug Repurposing: Far Beyond New Targets for Old Drugs. AAPS J. 2012, 14, 759–763. [Google Scholar] [CrossRef]
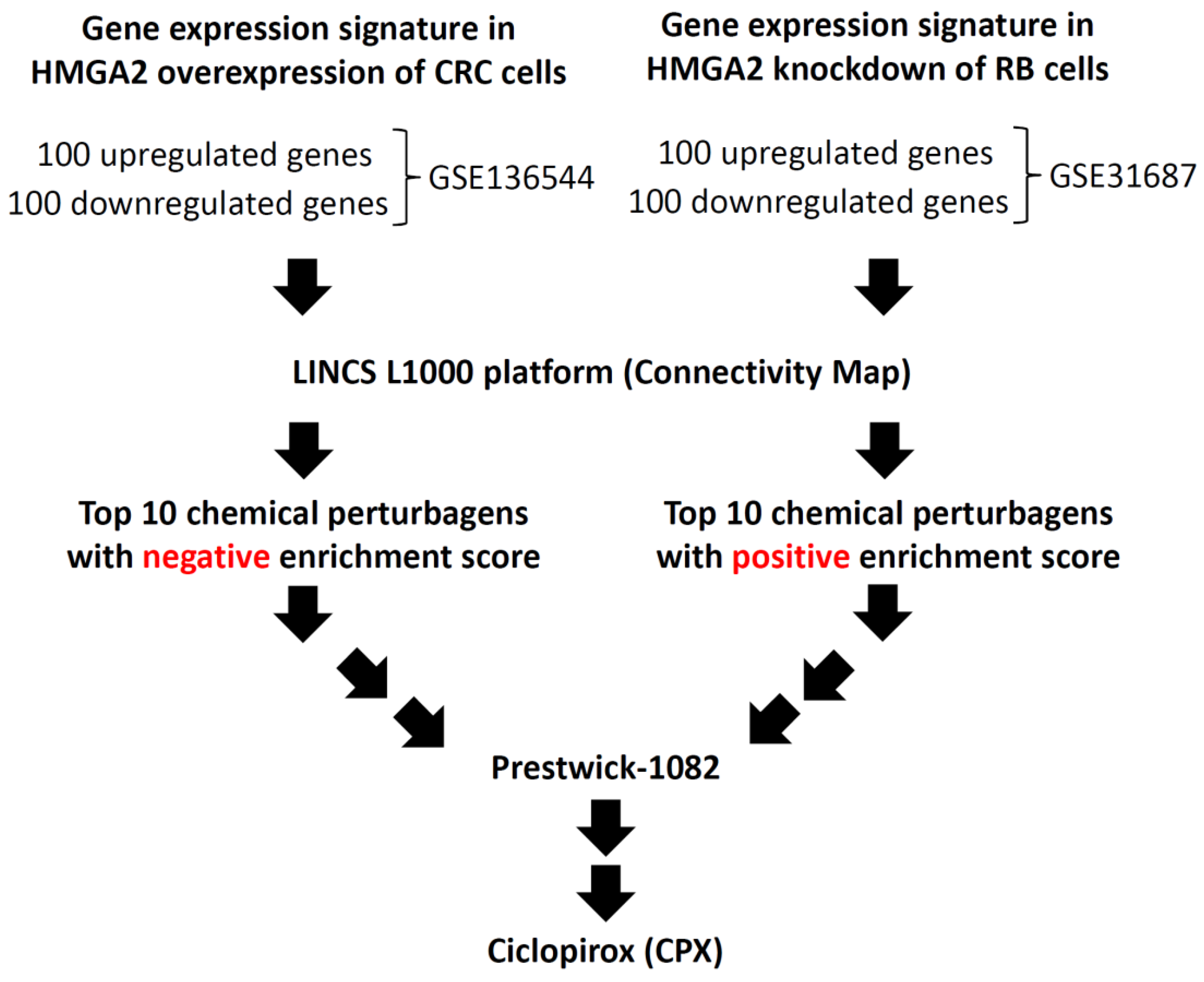

| Rank | Drug | Description | Enrichment Score |
|---|---|---|---|
| 1 | Ciclopirox | A synthetic antifungal agent for topical dermatologic treatment of superficial mycoses | 0.805 |
| 2 | Tetrandrine | A calcium channel blocker with anti-inflammatory, immunologic, and antiallergenic effects | 0.773 |
| 3 | Viomycin | A group of nonribosomal peptide antibiotics exhibiting anti-tuberculosis properties | 0.766 |
| 4 | Prestwick-692 | A small-molecule perturbation from the Connectivity Map (CMAP) resource | 0.764 |
| 5 | Oxyphenbutazone | A nonsteroidal anti-inflammatory drug | 0.758 |
| 6 | Perphenazine | A typical antipsychotic drug | 0.753 |
| 7 | Clotrimazole | A broad spectrum antimycotic or antifungal agent | 0.740 |
| 8 | Metixene | An anticholinergic used as an antiparkinsonian agent | 0.739 |
| 9 | Iloprost | A drug used to treat pulmonary arterial hypertension | 0.733 |
| 10 | Astemizole | A second-generation antihistamine drug | 0.732 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-M.; Cheng, C.-H.; Pan, S.-L.; Yang, P.-M.; Lin, D.-Y.; Lee, K.-H. Gene Expression Signature-Based Approach Identifies Antifungal Drug Ciclopirox As a Novel Inhibitor of HMGA2 in Colorectal Cancer. Biomolecules 2019, 9, 688. https://doi.org/10.3390/biom9110688
Huang Y-M, Cheng C-H, Pan S-L, Yang P-M, Lin D-Y, Lee K-H. Gene Expression Signature-Based Approach Identifies Antifungal Drug Ciclopirox As a Novel Inhibitor of HMGA2 in Colorectal Cancer. Biomolecules. 2019; 9(11):688. https://doi.org/10.3390/biom9110688
Chicago/Turabian StyleHuang, Yu-Min, Chia-Hsiung Cheng, Shiow-Lin Pan, Pei-Ming Yang, Ding-Yen Lin, and Kuen-Haur Lee. 2019. "Gene Expression Signature-Based Approach Identifies Antifungal Drug Ciclopirox As a Novel Inhibitor of HMGA2 in Colorectal Cancer" Biomolecules 9, no. 11: 688. https://doi.org/10.3390/biom9110688
APA StyleHuang, Y.-M., Cheng, C.-H., Pan, S.-L., Yang, P.-M., Lin, D.-Y., & Lee, K.-H. (2019). Gene Expression Signature-Based Approach Identifies Antifungal Drug Ciclopirox As a Novel Inhibitor of HMGA2 in Colorectal Cancer. Biomolecules, 9(11), 688. https://doi.org/10.3390/biom9110688







