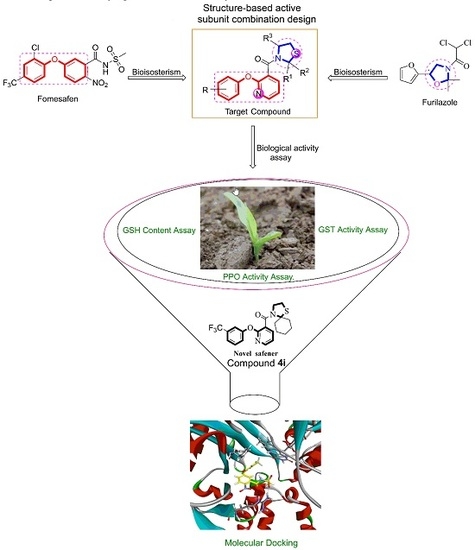
Novel Thiazole Phenoxypyridine Derivatives Protect Maize from Residual Pesticide Injury Caused by PPO-Inhibitor Fomesafen
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Synthetic Chemistry
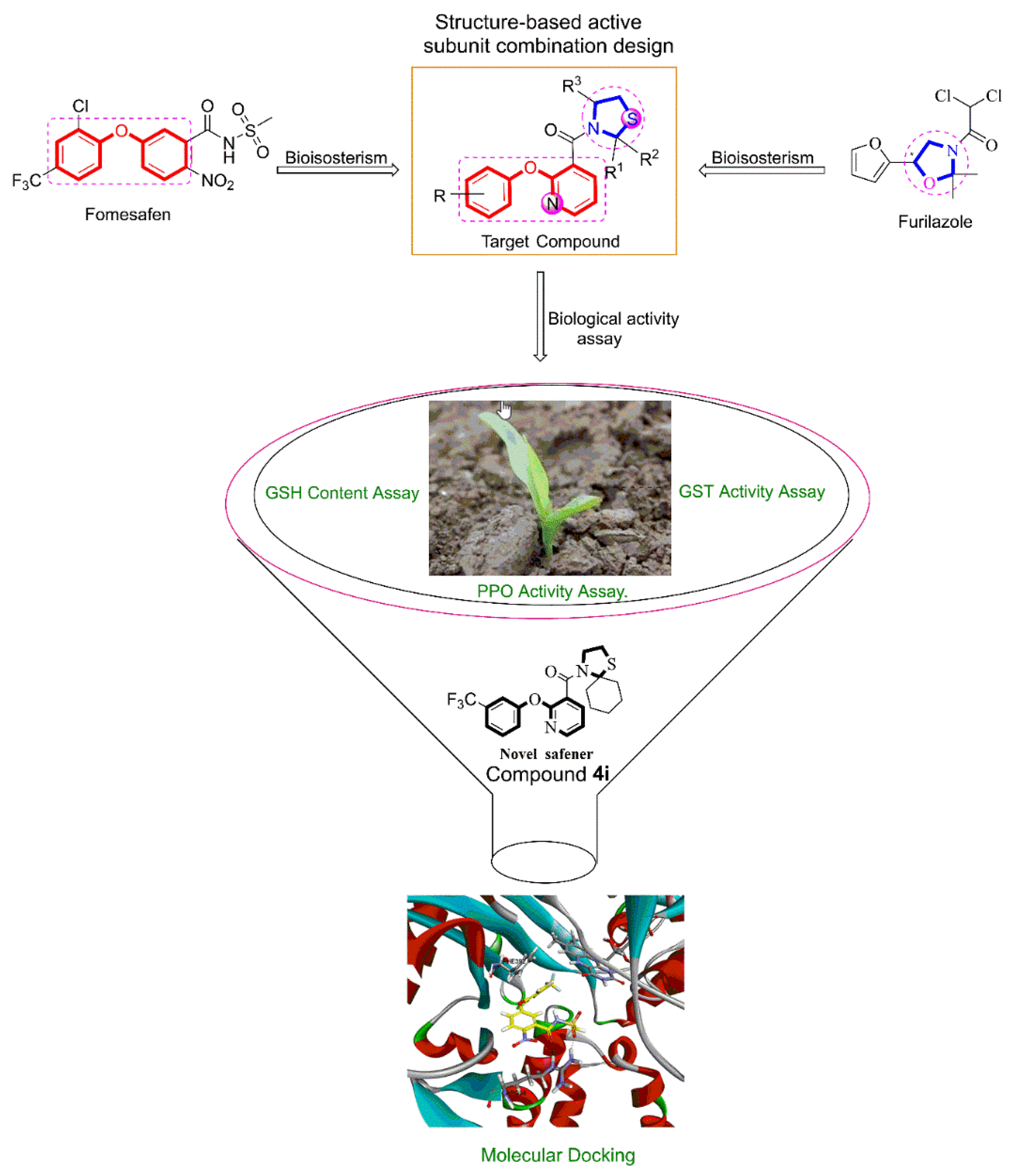
2.2.1. General Method for Preparation of 2-Phenoxynicotinic Acid 1
2.2.2. General Method for Preparation of 2-phenoxynicotinoyl Chloride 2
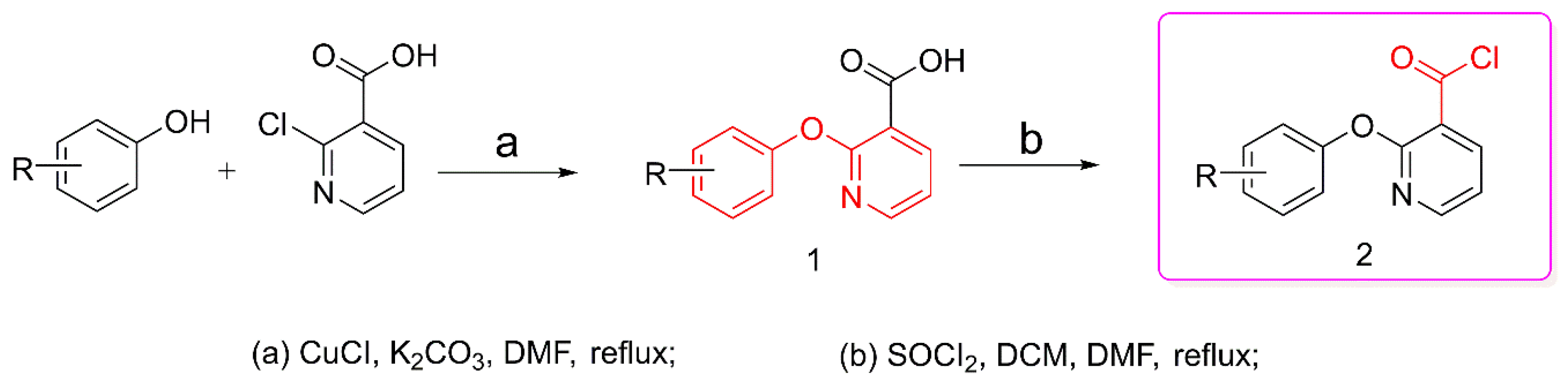
2.2.3. General Method for Preparation of Thiazolidines 3
2.2.4. General Method for Preparation of Thiazole Phenoxypyridine 4
2.3. X-ray Diffraction
2.4. Plant Material and Growth Conditions
2.5. GSH Content Assay
2.6. GST Activity Assay
2.7. PPO Activity Assay
2.8. Molecular Docking
3. Results and Discussion
3.1. Synthesis
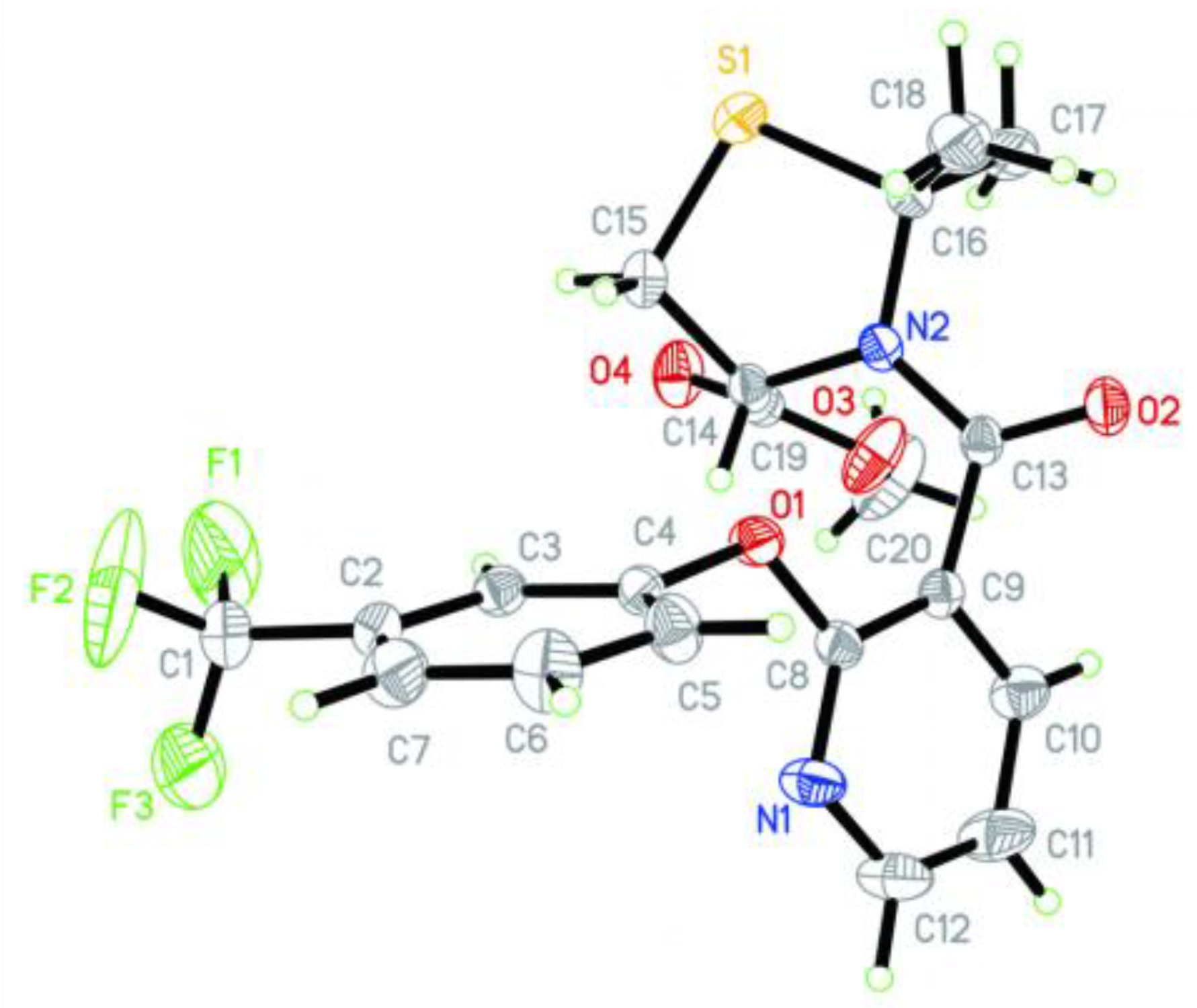
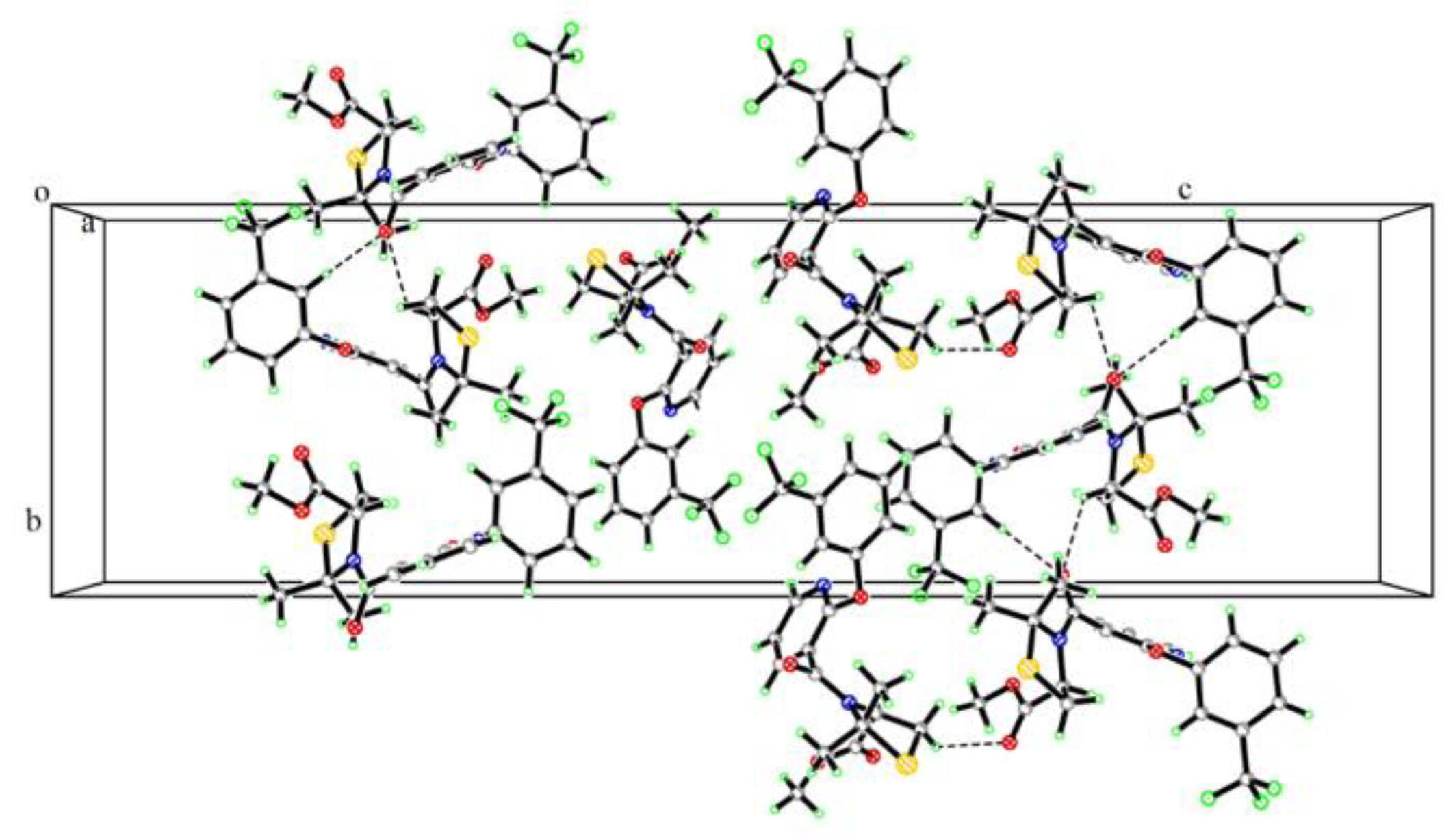
3.2. Crystallographic Data for Compound 4a
3.3. Maize Growth
3.4. GSH Content, GST Activity, and PPO Activity
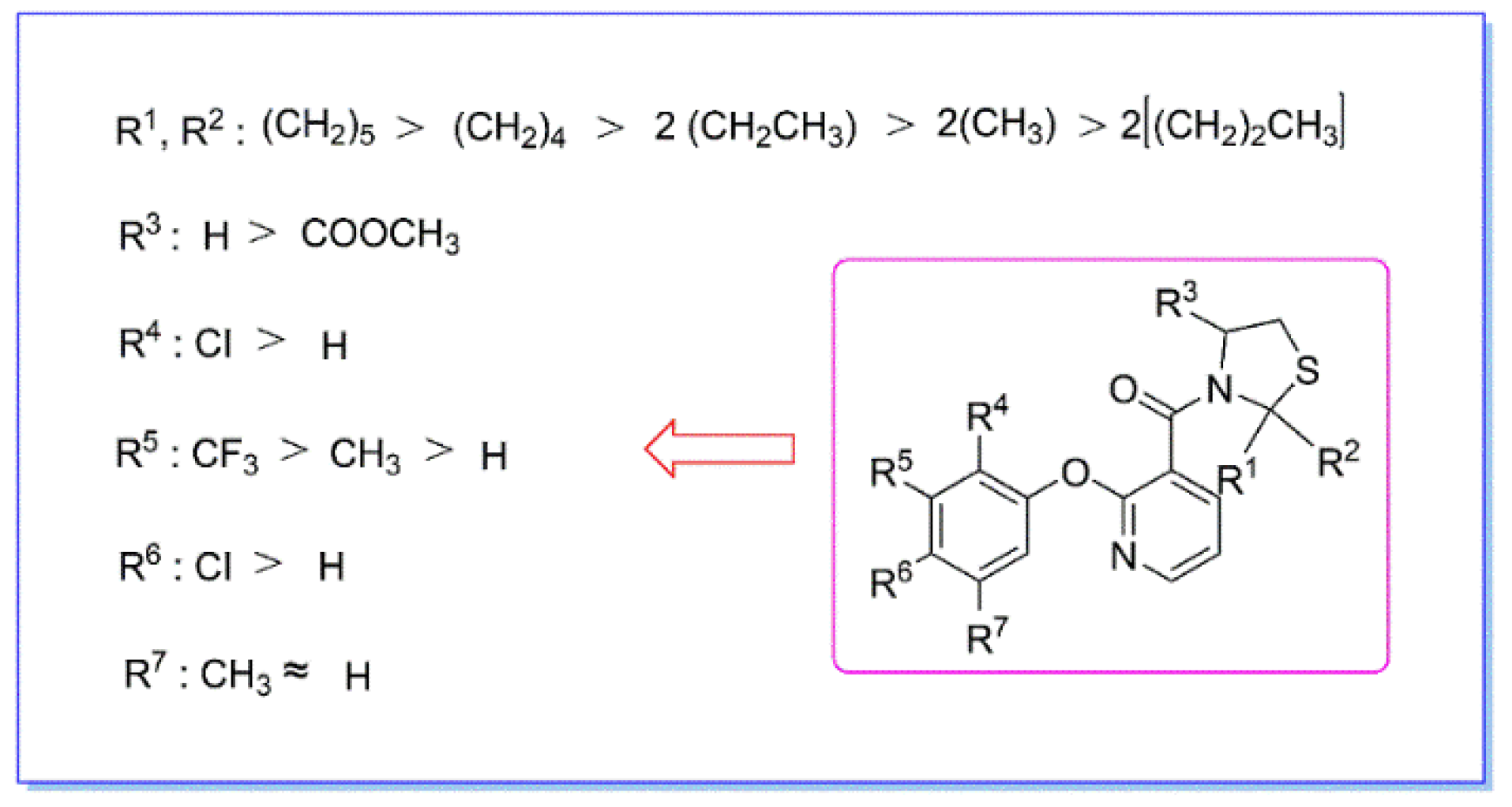
3.5. Structure–Activity Relationship
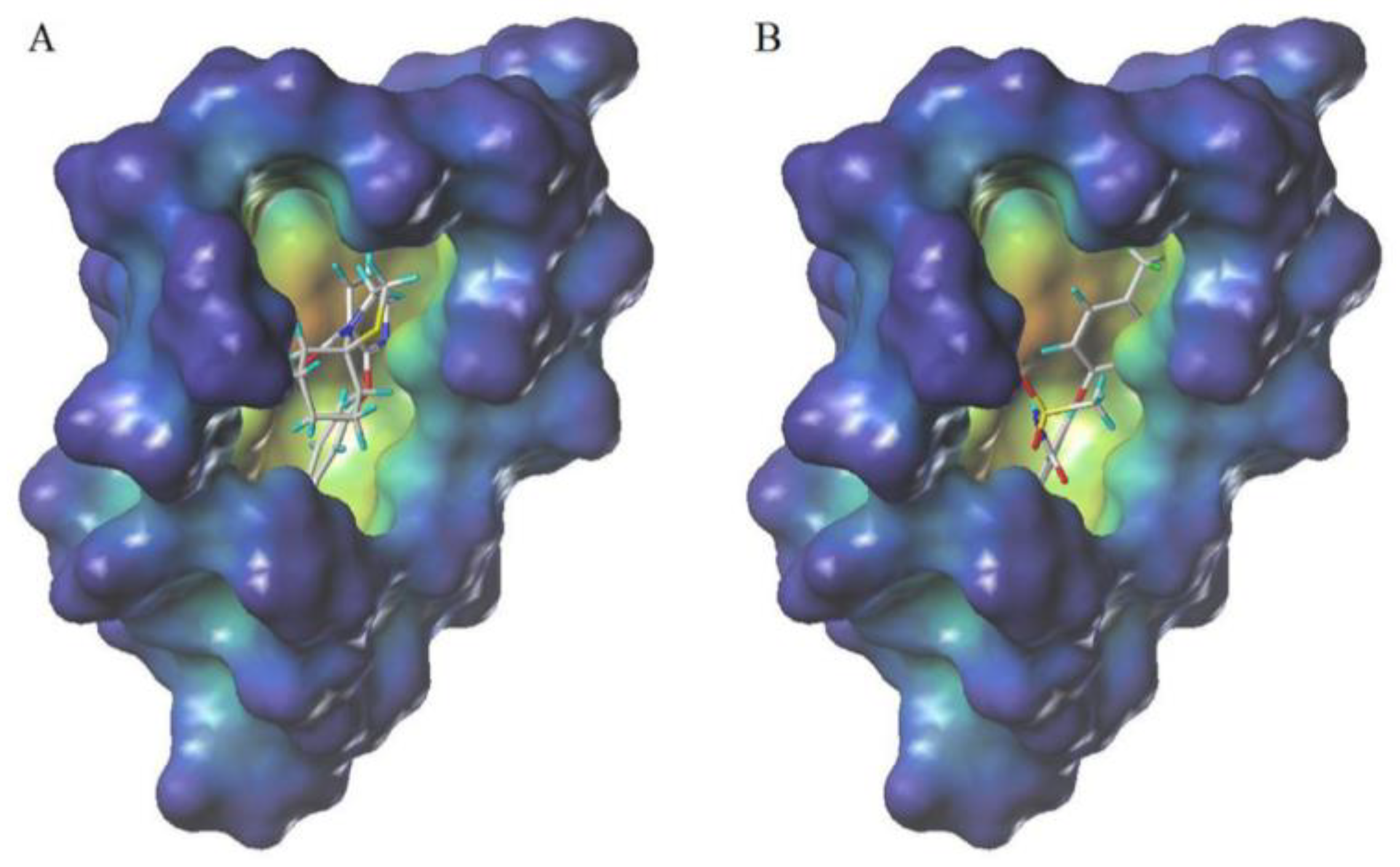
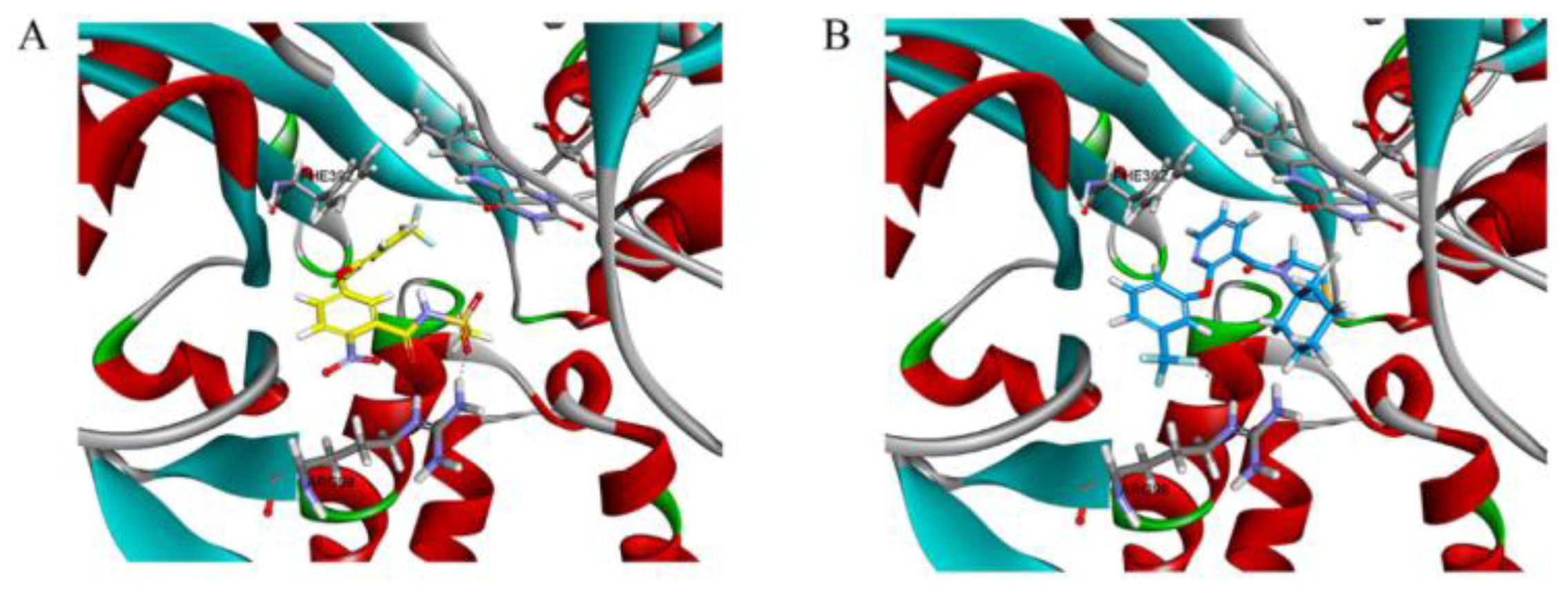
3.6. Molecular Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Layer, G.; Reichelt, J.; Jahn, D.; Heinz, D.W. Structure and function of enzymes in heme biosynthesis. Protein Sci. 2010, 19, 1137–1161. [Google Scholar] [CrossRef]
- Wang, B.F.; Wen, X.; Xi, Z. Molecular Simulations Bring New Insights into Protoporphyrinogen IX Oxidase/Protoporphyrinogen IX Interaction Modes. Mol. Inf. 2016, 35, 475–482. [Google Scholar] [CrossRef]
- Silva, F.M.; Kostygov, A.Y.; Spodareva, V.V.; Butenko, A.; Tossou, R.; Lukes, J.; Yurchenko, V.; Alves, J.M.P. The reduced genome of Candidatus Kinetoplastibacterium sorsogonicusi, the endosymbiont of Kentomonas sorsogonicus (Trypanosomatidae): Loss of the haem-synthesis pathway. Parasitology 2018, 145, 1287–1293. [Google Scholar] [CrossRef]
- Figueiredo, M.R.A.; Leibhart, L.J.; Reicher, Z.J.; Tranel, P.J.; Nissen, S.J.; Westra, P.; Bernards, M.L.; Kruger, G.R.; Gaines, T.A.; Jugulam, M. Metabolism of 2,4-dichlorophenoxyacetic acid contributes to resistance in a common waterhemp (Amaranthus tuberculatus) population. Pest Manag. Sci. 2018, 74, 2356–2362. [Google Scholar] [CrossRef]
- Trezzi, M.M.; Vidal, R.A.; Kruse, N.D.; Gustman, M.S.; Xavier, E.; Rosin, D.; Dedordi, G.F. Eletrolite Leakage as a Technique to Diagnose Euphorbia Heterophylla Biotypes Resistant to Ppo-Inhibitors Herbicides. Planta Daninha 2011, 29, 655–662. [Google Scholar] [CrossRef]
- Sikkema, P.H.; Shropshire, C.; Soltani, N. Response of dry bean to pre-plant incorporated and pre-emergence applications of S-metolachlor and fomesafen. Crop Prot. 2009, 28, 744–748. [Google Scholar] [CrossRef]
- Cobucci, T.; Prates, H.T.; Falcão, C.L.M.; Rezende, M.M.V. Effect of Imazamox, Fomesafen, and Acifluorfen Soil Residue on Rotational Crops. Weed Sci. 1998, 46, 258–263. [Google Scholar] [CrossRef]
- Rauch, B.J.; Bellinder, R.R.; Brainard, D.C. Dissipation of Fomesafen in New York State Soils and Potential to Cause Carryover Injury to Sweet Corn. Weed Technol. 2007, 21, 206–212. [Google Scholar] [CrossRef]
- Khorram, M.S.; Zheng, Y.; Lin, D.L.; Zhang, Q.; Fang, H.; Yu, Y.L. Dissipation of fomesafen in biochar-amended soil and its availability to corn (Zea mays L.) and earthworm (Eisenia fetida). J. Soils Sediments 2016, 16, 2439–2448. [Google Scholar] [CrossRef]
- Oliveira, M.C.; Gaines, T.A.; Dayan, F.E.; Patterson, E.L.; Jhala, A.J.; Knezevic, S.Z. Reversing resistance to tembotrione in an Amaranthus tuberculatus (var. rudis) population from Nebraska, USA with cytochrome P450 inhibitors. Pest Manag. Sci. 2018, 74, 2296–2305. [Google Scholar] [CrossRef]
- Moss, S.R.; Perryman, S.A.M.; Tatnell, L.V. Managing herbicide-resistant blackgrass (Alopecurus myosuroides): Theory and practice. Weed Technol. 2007, 21, 300–309. [Google Scholar] [CrossRef]
- Ercoli, L.; Mariotti, M.; Masoni, A.; Arduini, I. Growth responses of sorghum plants to chilling temperature and duration of exposure. Eur. J. Agron. 2004, 21, 93–103. [Google Scholar] [CrossRef]
- Shen, X.; Gao, X.; Eneji, A.E.; Chen, Y. Chemical control of weedy rice in precise hill-direct-seeded rice in South China. Weed Biol. Manag. 2013, 13, 39–43. [Google Scholar] [CrossRef]
- Lucini, L.; Molinari, G.P. Residues of the herbicide fenoxaprop-P-ethyl, its agronomic safener isoxadifen-ethyl and their metabolites in rice after field application. Pest Manag. Sci. 2010, 66, 621–626. [Google Scholar] [CrossRef]
- Ye, F.; Zhai, Y.; Kang, T.; Wu, S.L.; Li, J.J.; Gao, S.; Zhao, L.X.; Fu, Y. Rational design, synthesis and structure-activity relationship of novel substituted oxazole isoxazole carboxamides as herbicide safener. Pest. Biochem. Physiol. 2019, 157, 60–68. [Google Scholar] [CrossRef]
- Liu, C.L.; Guan, A.Y.; Yang, J.D.; Chai, B.S.; Li, M.; Li, H.C.; Yang, J.C.; Xie, Y. Efficient Approach To Discover Novel Agrochemical Candidates: Intermediate Derivatization Method. J. Agric. Food Chem. 2016, 64, 45–51. [Google Scholar] [CrossRef]
- Guan, A.Y.; Liu, C.L.; Chen, W.; Yang, F.; Xie, Y.; Zhang, J.B.; Li, Z.N.; Wang, M.A. Design, Synthesis, and Structure-Activity Relationship of New Pyrimidinamine Derivatives Containing an Aryloxy Pyridine Moiety. J. Agric. Food Chem. 2017, 65, 1272–1280. [Google Scholar] [CrossRef]
- Ye, F.; Ma, P.; Zhang, Y.-Y.; Li, P.; Yang, F.; Fu, Y. Herbicidal Activity and Molecular Docking Study of Novel ACCase Inhibitors. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Xie, Y.; Chi, H.W.; Guan, A.Y.; Liu, C.L.; Ma, H.J.; Cui, D.L. Design, Synthesis, and Herbicidal Activity of Novel Substituted 3-(Pyridin-2-yl)benzenesulfonamide Derivatives. J. Agric. Food Chem. 2014, 62, 12491–12496. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, B.; Gou, Z.P.; Li, Y.; Zhang, X.; Wang, Y.Q.; Yu, S.J.; Li, Y.H.; Sun, D.Q. Design of novel CSA analogues as potential safeners and fungicides. Bioorg. Med. Chem. Lett. 2015, 25, 791–794. [Google Scholar] [CrossRef]
- Chen, J.L.; Tang, W.; Che, J.Y.; Chen, K.; Yan, G.; Gu, Y.C.; Shi, D.Q. Synthesis and Biological Activity Evaluation of Novel alpha-Amino Phosphonate Derivatives Containing a Pyrimidinyl Moiety as Potential Herbicidal Agents. J. Agric. Food Chem. 2015, 63, 7219–7229. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, K.; Wang, P.; Kang, J.X.; Gao, S.; Zhao, L.X.; Ye, F. Design, Synthesis, and Herbicidal Activity Evaluation of Novel Aryl-Naphthyl Methanone Derivatives. Front. Chem. 2019, 7. [Google Scholar] [CrossRef]
- Matringe, M. Protoporphyrinogen oxidase as a molecular target for diphenylether herbicides. Biochem. J. 1989, 260, 231–235. [Google Scholar] [CrossRef]
- Orr, G.L.; Hess, F.D. Mechanism of Action of the Diphenyl Ether Herbicide Acifluorfen-Methyl in Excised Cucumber (Cucumis sativus L.) Cotyledons: LIGHT ACTIVATION AND THE SUBSEQUENT FORMATION OF LIPOPHILIC FREE RADICALS. Plant Physiol. 1982, 69, 502–507. [Google Scholar] [CrossRef]
- Fujikawa, K.I.; Kondo, K.; Yokomichi, I.; Kimura, F.; Haga, T.; Nishiyama, R. Studies on Herbicidal Activities of Phenoxypyridines. Agric. Biol. Chem. 1970, 34, 12. [Google Scholar] [CrossRef]
- Tajik, H.; Dadras, A. Synthesis and herbicidal activity of novel 5-chloro-3-fluoro-2-phenoxypyridines with a 1,3,4-oxadiazole ring. J. Pestic. Sci. 2011, 36, 27–32. [Google Scholar] [CrossRef]
- Nelson, E.A.; Penner, D. Leaching of isoxaflutole and the herbicide safeners R-29148 and furilazole. Weed Technol. 2007, 21, 106–109. [Google Scholar] [CrossRef]
- Liu, Y.J.; Zhang, J.A.; Zhang, L.J.; Zou, X.Z.; Li, N.; Li, Y. Study on pesticide activities of four ligands and their transition metal complexes with 8-mercaptoquinoline and pyridine terminal groups. Inorg. Chem. Commun. 2015, 57, 40–43. [Google Scholar] [CrossRef]
- Abu-Qare, A.W.; Duncan, H.J. Herbicide safeners: Uses, limitations, metabolism, and mechanisms of action. Chemosphere 2002, 48, 970–974. [Google Scholar] [CrossRef]
- Braga, A.L.; Milani, P.; Vargas, F.; Paixao, M.W.; Sehnem, J.A. Modular chiral thiazolidine catalysts in asymmetric aryl transfer reactions. Tetrahedron Asymmetry 2006, 17, 2793–2797. [Google Scholar] [CrossRef]
- Gronwald, J.W.; Fuerst, E.P.; Eberlein, C.V.; Egli, M.A. Effect of herbicide antidotes on glutathione content and glutathione s-transferase activity of sorghum shoots. Pestic. Biochem. Phys. 1987, 29, 66–76. [Google Scholar] [CrossRef]
- Scarponi, L.; Quagliarini, E.; Del Buono, D. Induction of wheat and maize glutathione S-transferase by some herbicide safeners and their effect on enzyme activity against butachlor and terbuthylazine. Pest Manag. Sci. 2006, 62, 927–932. [Google Scholar] [CrossRef]
- Green, J.M.; Owen, M.D.K. Herbicide-Resistant Crops: Utilities and Limitations for Herbicide-Resistant Weed Management. J. Agric. Food Chem. 2011, 59, 5819–5829. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Evans, K.M.; Cunningham, O.D.; Hodgson, D.R.W.; Steel, P.G.; Edwards, R. Catabolism of glutathione conjugates in Arabidopsis thaliana—Role in metabolic reactivation of the herbicide safener fenclorim. J. Biol. Chem. 2008, 283, 21102–21112. [Google Scholar] [CrossRef]
- Koch, M.; Breithaupt, C.; Kiefersauer, R.; Freigang, J.; Huber, R.; Messerschmidt, A. Crystal structure of protoporphyrinogen IX oxidase: A key enzyme in haem and chlorophyll biosynthesis. EMBO J. 2004, 23, 1720–1728. [Google Scholar] [CrossRef]
- Roy, K.; Paul, S. Docking and 3D QSAR studies of protoporphyrinogen oxidase inhibitor 3H-pyrazolo[3,4-d][1,2,3]triazin-4-one derivatives. J. Mol. Model. 2010, 16, 137–153. [Google Scholar] [CrossRef]
- Yang, S.G.; Hao, G.F.; Dayan, F.E.; Tranel, P.J.; Yang, G.F. Insight into the Structural Requirements of Protoporphyrinogen Oxidase Inhibitors: Molecular Docking and CoMFA of Diphenyl Ether, Isoxazole Phenyl, and Pyrazole Phenyl Ether. Chin. J. Chem. 2013, 31, 1153–1158. [Google Scholar] [CrossRef]
- Hao, G.F.; Zuo, Y.; Yang, S.G.; Yang, G.F. Protoporphyrinogen Oxidase Inhibitor: An Ideal Target for Herbicide Discovery. CHIMIA 2011, 65, 961–969. [Google Scholar] [CrossRef]

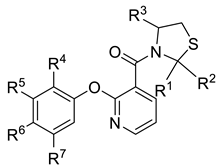
| Compd | R1 | R2 | R3 | R4 | R5 | R6 | R7 | Yield (%) |
|---|---|---|---|---|---|---|---|---|
| 4a | CH3 | CH3 | COOCH3 | H | CF3 | H | H | 85 |
| 4b | (CH2)2CH3 | (CH2)2CH3 | COOCH3 | H | CF3 | H | H | 70 |
| 4c | CH3 | CH3 | H | H | CF3 | H | H | 74 |
| 4d | (CH2)5 | COOCH3 | H | CF3 | H | H | 87 | |
| 4e | CH2CH3 | CH2CH3 | H | H | CF3 | H | H | 67 |
| 4f | CH2CH3 | CH2CH3 | COOCH3 | H | CF3 | H | H | 73 |
| 4g | (CH2)2CH3 | CH3 | H | H | H | Cl | H | 51 |
| 4h | (CH2)4 | H | H | CF3 | H | H | 76 | |
| 4i | (CH2)5 | H | H | CF3 | H | H | 84 | |
| 4j | (CH2)2CH3 | CH3 | H | H | CF3 | H | H | 62 |
| 4k | (CH2)5 | H | H | CH3 | Cl | CH3 | 93 | |
| 4l | CH2CH3 | CH2CH3 | H | H | CH3 | Cl | CH3 | 64 |
| 4m | (CH2)2CH3 | (CH2)2CH3 | H | H | CH3 | Cl | CH3 | 57 |
| 4n | CH3 | CH3 | H | H | CH3 | Cl | CH3 | 87 |
| 4o | CH3 | C(CH3)3 | H | H | CH3 | Cl | CH3 | 39 |
| 4p | (CH2)4 | H | H | H | Cl | H | 43 | |
| 4q | (CH2)5 | H | H | H | Cl | H | 49 | |
| 4r | (CH2)2CH3 | (CH2)2CH3 | H | H | H | Cl | H | 31 |
| 4s | CH3 | CH3 | H | H | H | Cl | H | 46 |
| 4t | (CH2)2CH3 | (CH2)2CH3 | H | H | CF3 | H | H | 43 |
| 4u | CH3 | C(CH3)3 | H | H | H | Cl | H | 21 |
| 4v | CH3 | CH3 | H | Cl | H | Cl | H | 38 |
| 4w | CH2CH3 | CH2CH3 | H | Cl | H | Cl | H | 35 |
| 4x | (CH2)4 | H | Cl | H | Cl | H | 38 | |
| 4y | (CH2)2CH3 | (CH2)2CH3 | H | Cl | H | Cl | H | 23 |
| 4z | (CH2)4 | H | H | CH3 | Cl | CH3 | 42 | |
| Treatment | Recovery of Plant Height (%) | Recovery of Root Length (%) | Recovery of Plant Weight (%) | Recovery of Root Weight (%) |
|---|---|---|---|---|
| 4a | 30.20 ± 1.11 c | 32.35 ± 0.79 b | 33.36 ± 1.46 abcd | 36.35 ±2.86 bc |
| 4b | 40.75 ± 1.70 e | 35.52 ± 1.48 bc | 38.28 ± 0.68 defg | 44.11 ± 3.85 f |
| 4c | 54.02 ± 0.92 g | 52.31 ± 4.04 gh | 40.40 ± 2.47 deg | 33.62 ± 1.30 b |
| 4d | 56.17 ± 1.36 g | 27.39 ± 2.3 2a | 41.62 ± 0.55 fgh | 54.81 ± 1.05 gh |
| 4e | 40.88 ± 0.66 e | 42.37 ± 2.91 de | 29.09 ± 1.22 ab | 17.85 ± 1.28 a |
| 4f | 27.26 ± 0.99 b | 42.57 ± 2.49 de | 35.91 ± 1.89 de | 42.16 ± 0.37 def |
| 4g | 29.59 ± 1.75 bc | 47.78 ± 2.65 fg | 37.35 ± 1.11 def | 42.79 ± 2.31 cef |
| 4h | 50.37 ± 1.80 f | 57.13 ± 4.98 ij | 63.75 ± 1.06 kl | 66.48 ± 2.83 j |
| 4i | 72.41 ± 1.49 l | 73.34 ± 0.76 l | 77.33 ± 6.08 m | 81.96 ± 3.91 l |
| 4j | 65.81 ± 1.13 j | 60.35 ± 2.20 j | 64.54 ± 1.25 kl | 55.02 ± 1.33 gh |
| 4k | 61.05 ± 1.37 i | 50.33 ± 2.51 fgh | 61.43 ± 1.8 1kl | 62.31 ± 0.56 ij |
| 4l | 58.65 ± 0.65 h | 68.38 ± 2.05 k | 62.76 ± 1.52 kl | 65.95 ± 1.63 j |
| 4m | 40.40 ± 1.46 m | 59.24 ± 2.81 j | 49.04 ± 3.89 ij | 56.46 ± 0.33 g |
| 4n | 29.07 ± 1.05 bc | 38.92 ± 2.94 cd | 34.26 ± 2.63 bcd | 17.38 ± 8.10 a |
| 4o | 69.04 ± 1.32 k | 67.78 ± 1.90 k | 67.03 ± 3.24 k | 67.18 ± 2.03 i |
| 4p | 40.68 ± 2.93 e | 50.77 ± 2.65fgh | 66.55 ± 3.69 k | 62.86 ± 0.49 ij |
| 4q | 27.82 ± 0.95 bc | 39.95 ± 6.26 cd | 28.15 ± 2.29 a | 39.86 ± 5.55 def |
| 4r | 55.11 ± 1.19 g | 42.77 ± 2.26 de | 63.22 ± 7.56 kl | 51.190 ± 3.68 g |
| 4s | 40.17 ± 0.96 e | 46.12 ± 0.49 ef | 42.73 ± 2.43 gh | 52.07 ± 3.34 g |
| 4t | 37.29 ± 1.39 d | 38.38 ± 3.58 cd | 46.08 ± 3.30 hi | 38.70 ± 0.63 bcde |
| 4u | 54.32s ± 1.89 g | 51.11 ± 0.87 gh | 59.22 ± 3.30 k | 60.04 ± 1.57 hi |
| 4v | 51.54 ± 2.40 f | 67.78 ± 1.02 k | 52.99 ± 0.89 j | 63.24 ± 0.62 ij |
| 4w | 22.75 ± 0.81 a | 40.22 ± 0.51 cd | 33.86 ± 2.26 bcd | 37.41 ± 3.35 bcd |
| 4x | 64.93 ± 1.57 j | 53.57 ± 1.69 hi | 65.95 ± 2.91 k | 73.27 ± 3.89 k |
| 4y | 41.70 ± 1.87 e | 33.69 ± 1.88 b | 30.894 ± 3.82 abc | 34.93 ± 0.69 bc |
| 4z | 69.39 ± 0.22 l | 65.93 ± 1.44 k | 65.94 ± 0.63 l | 73.43 ± 2.04 k |
| Treatment | GSH Content (μg·g−1) | GST Activity (nmol ·min−1·mg−1 Protein) | PPO Activity (U/L) |
|---|---|---|---|
| Fomesafen | 4.5 ± 0.2 fg | 6.4± 0.2 cde | 79.3 ± 0.4 b |
| Control | 3.2 ± 0.3 abcd | 5.9 ± 0.2 abc | 162.4 ± 0.9 p |
| 4a | 3.5 ± 0.5 bcde | 8.1 ± 0.6 hi | 99.6 ± 3.5 fg |
| 4b | 3.9 ± 0.4 def | 5.7 ± 0.3 ab | 97.4 ± 0.5 f |
| 4c | 3.8 ± 0.2 de | 9.4 ± 0.2 k | 104.3 ± 1.4 gh |
| 4d | 3.6 ± 0.2 cde | 9.0 ± 0.2 jk | 107.3 ± 4.5 hi |
| 4e | 2.5 ± 0.3 a | 8.5 ± 0.2 ij | 113.1 ± 0.3 ij |
| 4f | 3.9 ± 0.4 ef | 8.6 ± 0.4 ij | 86.2 ± 3.2 cd |
| 4g | 4.1 ± 0.4 ef | 8.4 ± 0.1 ij | 115.9 ± 0.7 jk |
| 4h | 6.4 ± 0.6 i | 6.3 ± 0.2 bcd | 153.8 ± 6.6 o |
| 4i | 8.4 ± 0.4 k | 10.4 ± 0.8 l | 154.7 ± 4.5 o |
| 4j | 5.1 ± 0.2 gh | 6.3 ± 0.1 bcde | 143.9 ± 0.3 n |
| 4k | 5.4 ± 0.2 h | 6.5 ± 0.2 de | 126.5 ± 1.6 m |
| 4l | 6.8 ± 0.2 i | 6.9 ± 0.4 ef | 123.0 ± 0.2 lm |
| 4m | 5.1 ± 0.4 gh | 9.5 ± 0.4 k | 85.3 ± 0.2 cd |
| 4n | 3.8 ± 0.2 cde | 8.6 ± 0.1 ij | 88.4 ± 3.9 de |
| 4o | 6.8 ± 0.7 i | 9.3 ± 0.2 k | 155.5 ± 11.2 o |
| 4p | 3.0 ± 0.3 abc | 6.7 ± 0.4 de | 93.6 ± 2.3 ef |
| 4q | 3.8 ± 0.2 cde | 5.5 ± 0.2 a | 111.1 ± 2.1 ij |
| 4r | 3.9 ± 0.3 ef | 5.5 ± 0.3 a | 81.6 ± 0.2 bc |
| 4s | 4.1 ± 0.3 ef | 6.4 ± 0.1 cde | 66.9 ± 0.7 a |
| 4t | 3.1 ± 0.2 abc | 7.3 ± 0.1 fg | 97.2 ± 0.8 f |
| 4u | 3.8 ± 0.2 cde | 5.8 ± 0.3 abc | 113.1 ± 0.2 ij |
| 4v | 4.9 ± 0.2 gh | 6.2 ± 0.3 bcd | 120.0 ± 0.7 kl |
| 4w | 3.1 ± 0.6 abc | 7.7 ± 0.2 gh | 86.8 ± 2.4 cd |
| 4x | 5.1 ± 0.3 gh | 6.8 ± 0.4 def | 128.8 ± 4.7 m |
| 4y | 2.8 ± 0.1 ab | 5.9 ± 0.4 abc | 94.5 ± 0.6 f |
| 4z | 7.8 ± 0.6 j | 9.2 ± 0.2 k | 151.4 ± 6.2 o |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.-X.; Yin, M.-L.; Wang, Q.-R.; Zou, Y.-L.; Ren, T.; Gao, S.; Fu, Y.; Ye, F. Novel Thiazole Phenoxypyridine Derivatives Protect Maize from Residual Pesticide Injury Caused by PPO-Inhibitor Fomesafen. Biomolecules 2019, 9, 514. https://doi.org/10.3390/biom9100514
Zhao L-X, Yin M-L, Wang Q-R, Zou Y-L, Ren T, Gao S, Fu Y, Ye F. Novel Thiazole Phenoxypyridine Derivatives Protect Maize from Residual Pesticide Injury Caused by PPO-Inhibitor Fomesafen. Biomolecules. 2019; 9(10):514. https://doi.org/10.3390/biom9100514
Chicago/Turabian StyleZhao, Li-Xia, Min-Lei Yin, Qing-Rui Wang, Yue-Li Zou, Tao Ren, Shuang Gao, Ying Fu, and Fei Ye. 2019. "Novel Thiazole Phenoxypyridine Derivatives Protect Maize from Residual Pesticide Injury Caused by PPO-Inhibitor Fomesafen" Biomolecules 9, no. 10: 514. https://doi.org/10.3390/biom9100514
APA StyleZhao, L.-X., Yin, M.-L., Wang, Q.-R., Zou, Y.-L., Ren, T., Gao, S., Fu, Y., & Ye, F. (2019). Novel Thiazole Phenoxypyridine Derivatives Protect Maize from Residual Pesticide Injury Caused by PPO-Inhibitor Fomesafen. Biomolecules, 9(10), 514. https://doi.org/10.3390/biom9100514