Sustained Neurotrophin Release from Protein Nanoparticles Mediated by Matrix Metalloproteinases Induces the Alignment and Differentiation of Nerve Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Assays for MMPs
2.2. RT-PCR and qPCR
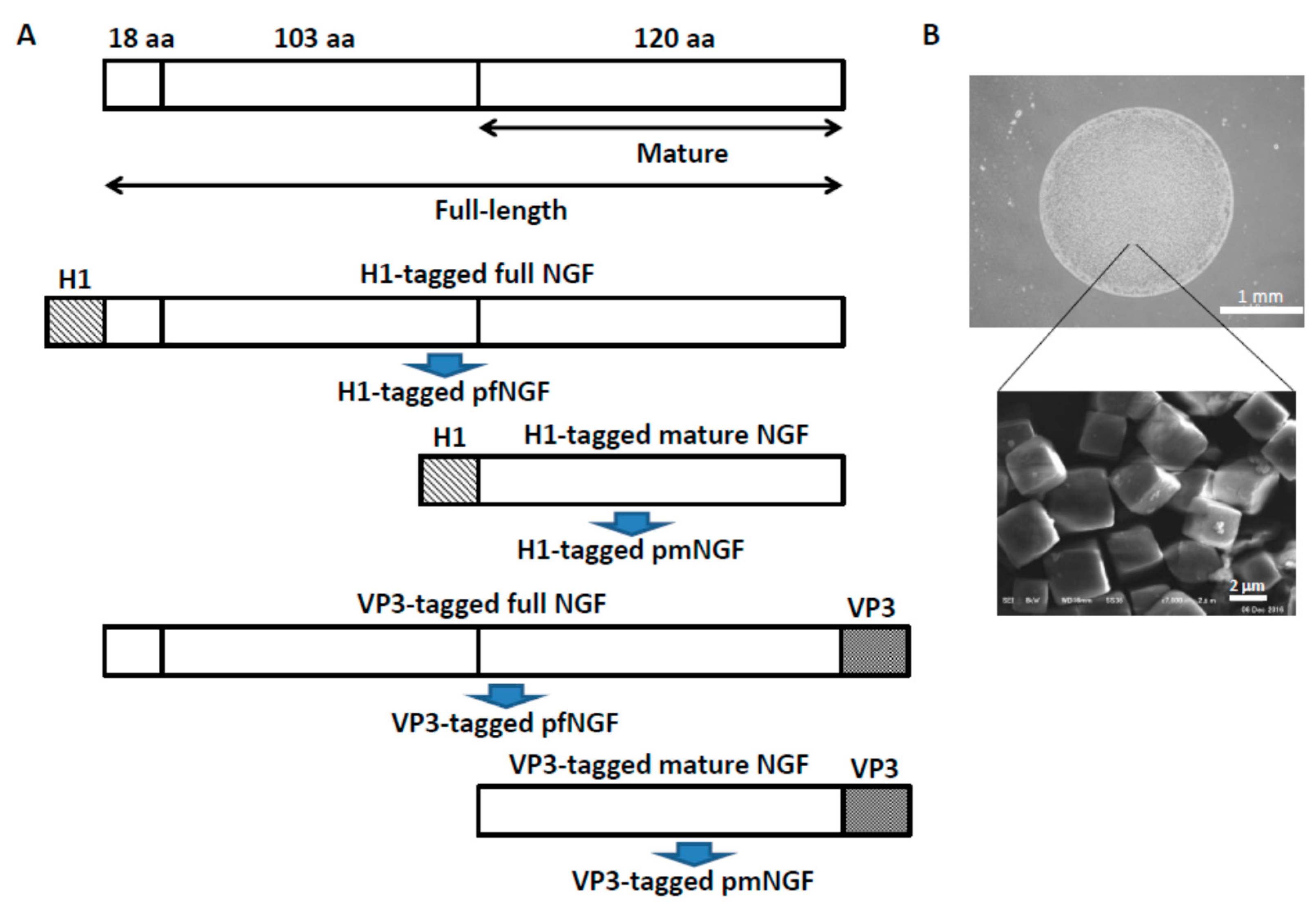
2.3. Construction of Expression Vectors for pNGF
2.4. Expression and Purification of pNGF
2.5. Cover Slip Coating
2.6. PC12 Cell Culture
2.7. Preparation of Cells for SEM Imaging
2.8. Immunocytochemistry
3. Results
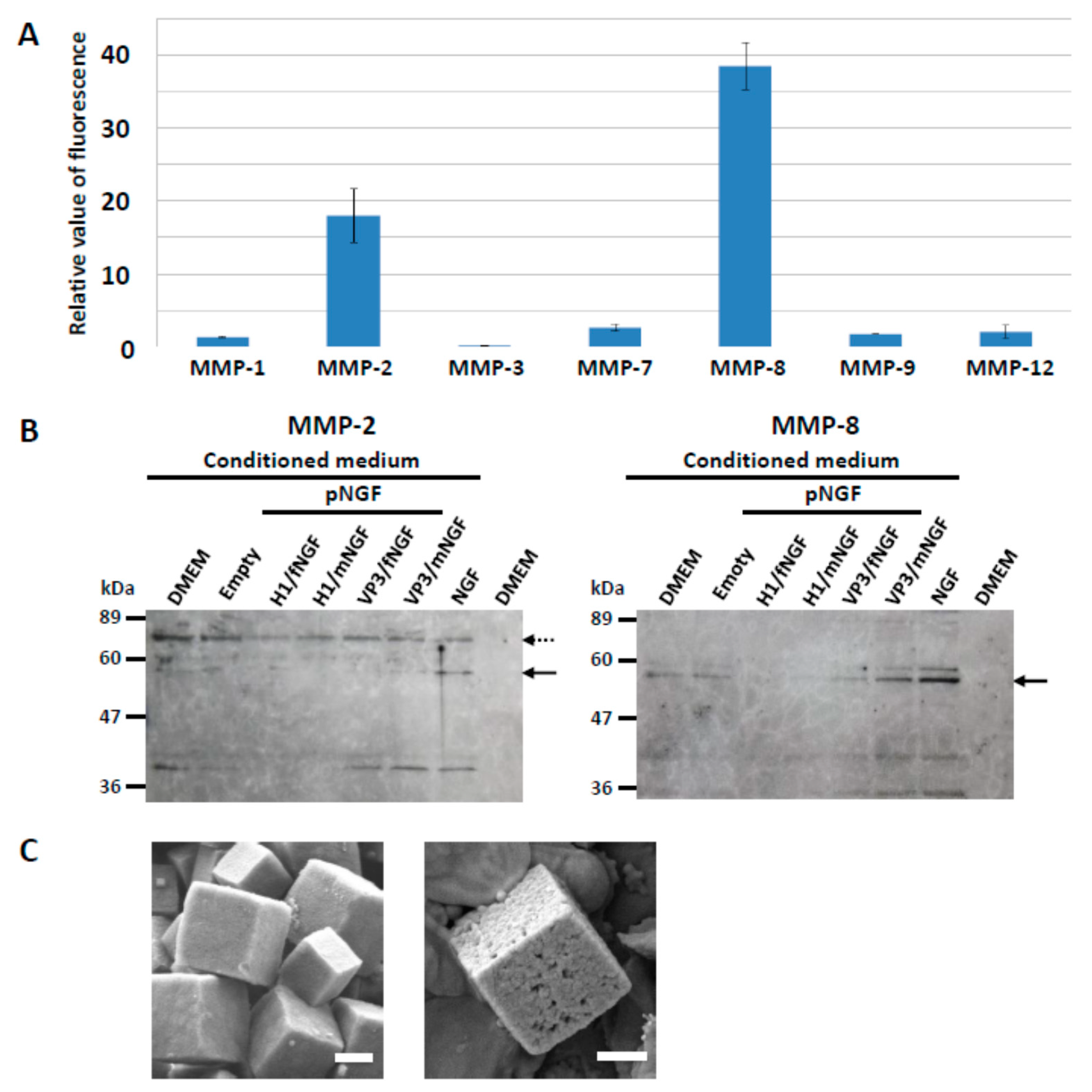
3.1. Release of Cargo Proteins from PODS Crystals
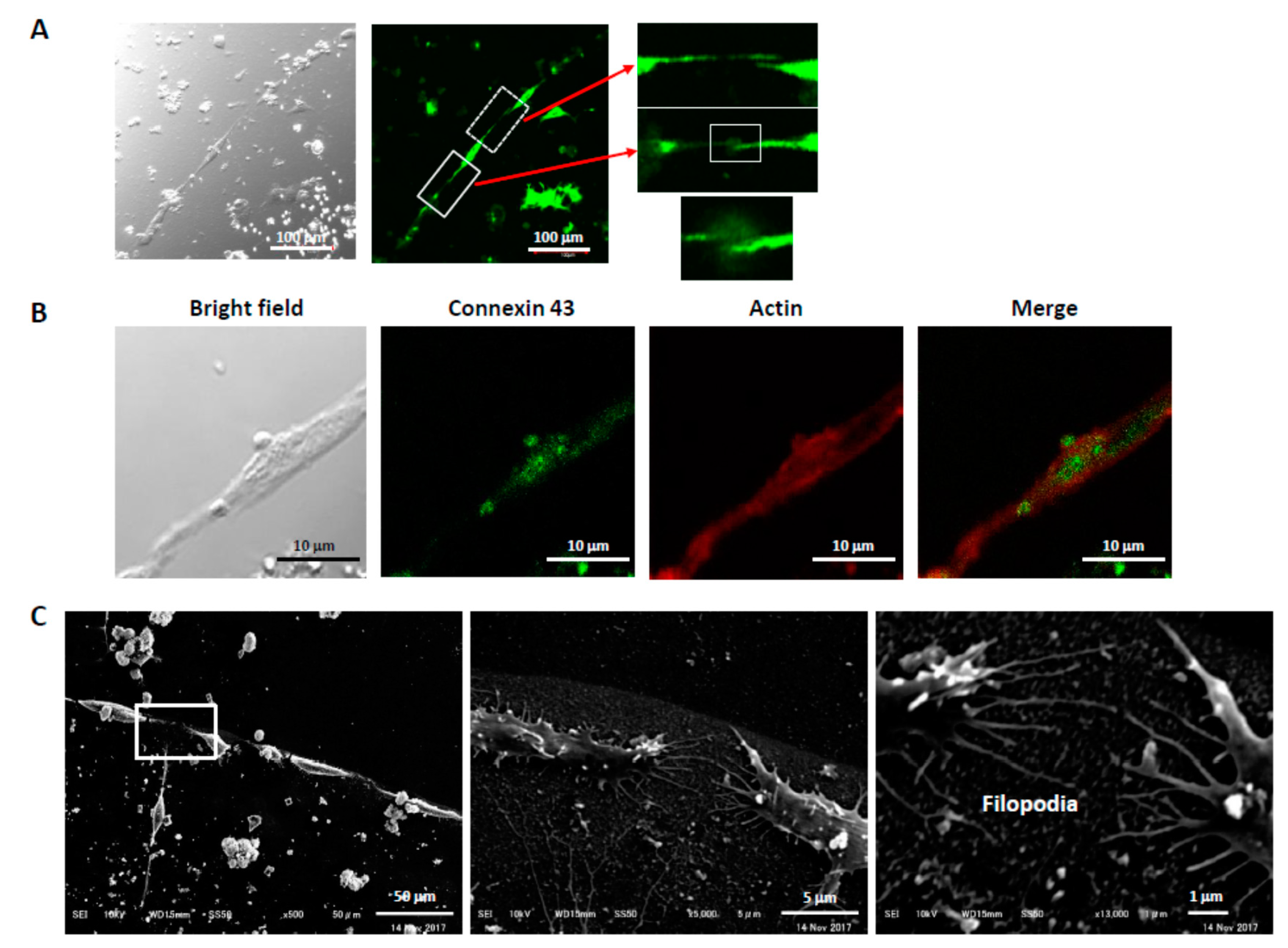
3.2. Alignment of PC12 Cells by pNGF
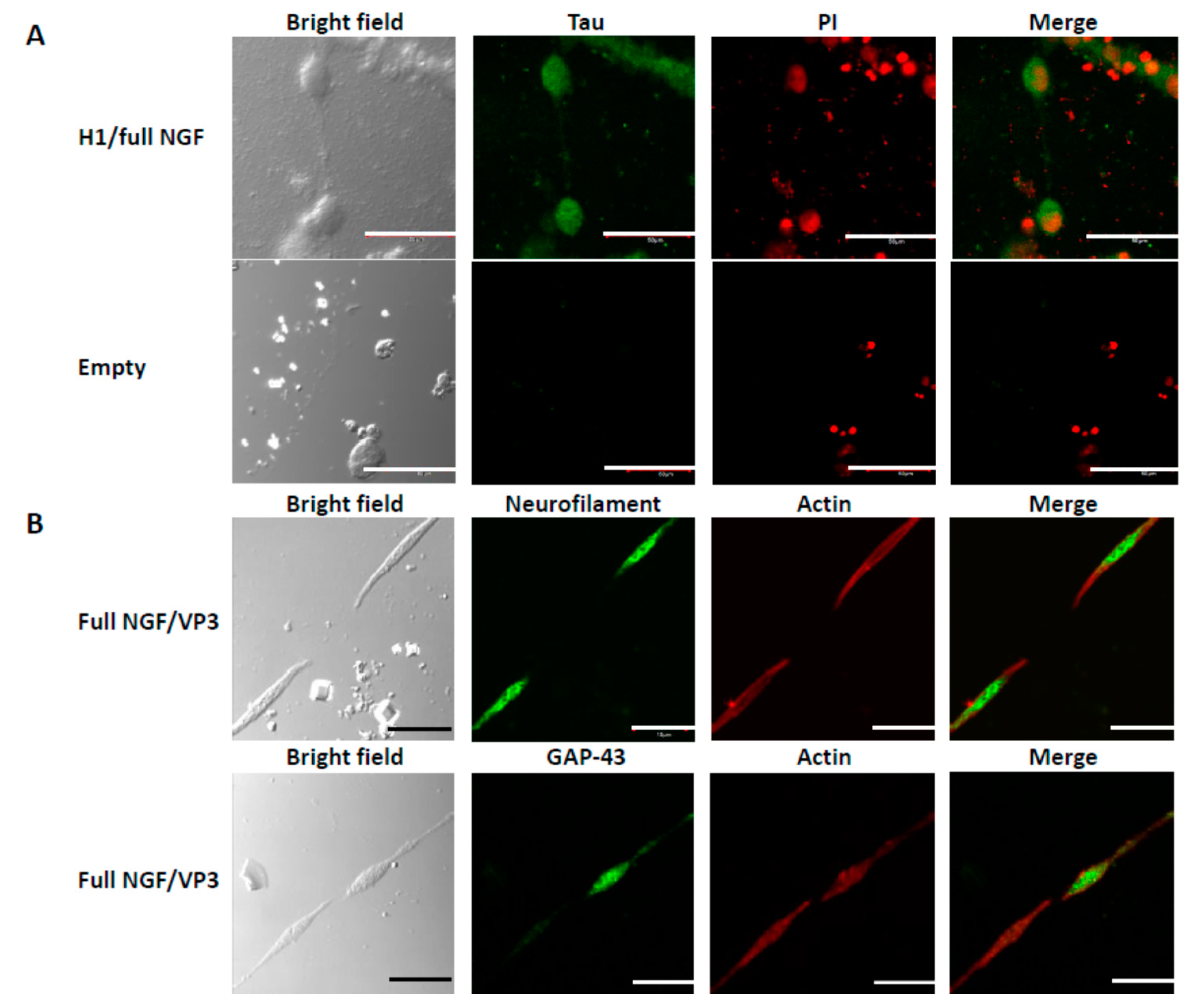
3.3. Differentiation of PC12 Cells
3.4. Connections Between Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barthes, J.; Ozcelik, H.; Hindie, M.; Ndreu-Halili, A.; Hasan, A.; Vrana, N.E. Cell microenvironment engineering and monitoring for tissue engineering and regenerative medicine: The recent advances. Biomed. Res. Int 2014, 2014, 921905. [Google Scholar] [CrossRef] [PubMed]
- Bourget, J.M.; Guillemette, M.; Veres, T.; Auger, F.A.; Germain, L. Alignment of Cells and Extracellular Matrix within Tissue- Engineered Substitutes. In Advances in Biomaterials Science and Biomedical Applications; Pignatello, R., Ed.; Intech Open: London, UK, 2013. [Google Scholar]
- Gattazzo, F.; Urciuolo, A.; Bonaldo, P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys Acta 2014, 1840, 2506–2519. [Google Scholar] [CrossRef] [PubMed]
- Muiznieks, L.D.; Keeley, F.W. Molecular assembly and mechanical properties of the extracellular matrix: A fibrous protein perspective. Biochim. Biophys Acta 2013, 1832, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Surguchev, A.A.; Emamzadeh, F.N.; Surguchov, A. Cell responses to extracellular α-synuclein. Molecules 2019, 24, 305. [Google Scholar] [CrossRef] [PubMed]
- Loffek, S.; Schilling, O.; Franzke, C.W. Series “Matrix metalloproteinases in lung health and disease”: Biological role of matrix metalloproteinases: A critical balance. Eur Respir J. 2011, 38, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, N.; Folch, A. Large-scale microfluidic gradient arrays reveal axon guidance behaviors in hippocampal neurons. Microsyst. Nanoeng. 2017, 3, 17003. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, M.; Golding, J.P.; Loughlin, A.J.; Kingham, P.J.; Phillips, J.B. Engineered neural tissue with aligned, differentiated adipose-derived stem cells promotes peripheral nerve regeneration across a critical sized defect in rat sciatic nerve. Biomaterials 2015, 37, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Gnavi, S.; Fornasari, B.E.; Tonda-Turo, C.; Laurano, R.; Zanetti, M.; Ciardelli, G.; Geuna, S. The Effect of Electrospun Gelatin Fibers Alignment on Schwann Cell and Axon Behavior and Organization in the Perspective of Artificial Nerve Design. Int J. Mol. Sci. 2015, 16, 12925–12942. [Google Scholar] [CrossRef]
- Han, D.; Cheung, K.C. Biodegradable Cell-Seeded Nanofiber Scaffolds for Neural Repair. Polymers 2011, 3, 1684–1733. [Google Scholar] [CrossRef]
- Wang, W.; Itoh, S.; Konno, K.; Kikkawa, T.; Ichinose, S.; Sakai, K.; Ohkuma, T.; Watabe, K. Effects of Schwann cell alignment along the oriented electrospun chitosan nanofibers on nerve regeneration. J. Biomed. Mater. Res. A 2009, 91, 994–1005. [Google Scholar] [CrossRef]
- Skaper, S.D. The neurotrophin family of neurotrophic factors: An overview. Methods Mol. Biol. 2012, 846, 1–12. [Google Scholar]
- Sofroniew, M.V.; Howe, C.L.; Mobley, W.C. Nerve growth factor signaling, neuroprotection, and neural repair. Annu. Rev. Neurosci. 2001, 24, 1217–1281. [Google Scholar] [CrossRef]
- Mingorance-Le Meur, A.; Mohebiany, A.N.; O’Connor, T.P. Varicones and growth cones: Two neurite terminals in PC12 cells. PLoS ONE 2009, 4, e4334. [Google Scholar] [CrossRef]
- Mori, H.; Metcalf, P. Cypoviruses. In Insect Virology; Asgari, S., Johnson, K.N., Eds.; Caister Academic Press: British, UK, 2010; pp. 307–324. [Google Scholar]
- Ijiri, H.; Coulibaly, F.; Nishimura, G.; Nakai, D.; Chiu, E.; Takenaka, C.; Ikeda, K.; Nakazawa, H.; Hamada, N.; Kotani, E.; et al. Structure-based targeting of bioactive proteins into cypovirus polyhedra and application to immobilized cytokines for mammalian cell culture. Biomaterials 2009, 30, 4297–4308. [Google Scholar] [CrossRef]
- Mori, H.; Shukunami, C.; Furuyama, A.; Notsu, H.; Nishizaki, Y.; Hiraki, Y. Immobilization of bioactive fibroblast growth factor-2 into cubic proteinous microcrystals (Bombyx mori cypovirus polyhedra) that are insoluble in a physiological cellular environment. J. Biol. Chem. 2007, 282, 17289–17296. [Google Scholar] [CrossRef]
- Nishishita, N.; Ijiri, H.; Takenaka, C.; Kobayashi, K.; Goto, K.; Kotani, E.; Itoh, T.; Mori, H.; Kawamata, S. The use of leukemia inhibitory factor immobilized on virus-derived polyhedra to support the proliferation of mouse embryonic and induced pluripotent stem cells. Biomaterials 2011, 32, 3555–3563. [Google Scholar] [CrossRef]
- Peng, W.J.; Yan, J.W.; Wan, Y.N.; Wang, B.X.; Tao, J.H.; Yang, G.J.; Pan, H.F.; Wang, J. Matrix metalloproteinases: A review of their structure and role in systemic sclerosis. J. Clin. Immunol. 2012, 32, 1409–1414. [Google Scholar]
- Surgucheva, I.; Chidambaram, K.; Willoughby, D.A.; Surguchov, A. Matrix metalloproteinase 9 expression: New regulatory elements. J. Ocul. Biol. Dis. Infor. 2010, 3, 41–52. [Google Scholar] [CrossRef]
- Wang, H.; Parry, S.; Macones, G.; Sammel, M.D.; Ferrand, P.E.; Kuivaniemi, H.; Tromp, G.; Halder, I.; Shriver, M.D.; Romero, R.; et al. Functionally significant SNP MMP8 promoter haplotypes and preterm premature rupture of membranes (PPROM). Hum. Mol. Genet 2004, 13, 2659–2669. [Google Scholar] [CrossRef]
- Saarialho-Kere, U.K.; Crouch, E.C.; Parks, W.C. Matrix metalloproteinase matrilysin is constitutively expressed in adult human exocrine epithelium. J. Investig. Dermatol. 1995, 105, 190–196. [Google Scholar] [CrossRef]
- Fontanet, P.; Irala, D.; Alsina, F.C.; Paratcha, G.; Ledda, F. Pea3 transcription factor family members Etv4 and Etv5 mediate retrograde signaling and axonal growth of DRG sensory neurons in response to NGF. J. Neurosci. 2013, 33, 15940–15951. [Google Scholar] [CrossRef]
- Nordstrom, L.A.; Lochner, J.; Yeung, W.; Ciment, G. The metalloproteinase stromelysin-1 (transin) mediates PC12 cell growth cone invasiveness through basal laminae. Mol. Cell Neurosci. 1995, 6, 56–68. [Google Scholar] [CrossRef]
- Pollock, J.D.; Krempin, M.; Rudy, B. Differential effects of NGF, FGF, EGF, cAMP, and dexamethasone on neurite outgrowth and sodium channel expression in PC12 cells. J. Neurosci. 1990, 10, 2626–2637. [Google Scholar] [CrossRef]
- Drubin, D.G.; Feinstein, S.C.; Shooter, E.M.; Kirschner, M.W. Nerve growth factor-induced neurite outgrowth in PC12 cells involves the coordinate induction of microtubule assembly and assembly-promoting factors. J. Cell Biol. 1985, 101, 1799–1807. [Google Scholar] [CrossRef]
- Matsumoto, G.; Ueda, T.; Shimoyama, J.; Ijiri, H.; Omi, Y.; Yube, H.; Sugita, Y.; Kubo, K.; Maeda, H.; Kinoshita, Y.; et al. Bone regeneration by polyhedral microcrystals from silkworm virus. Sci. Rep. 2012, 2, 935. [Google Scholar] [CrossRef]
- Matsumoto, G.; Hirohata, R.; Hayashi, K.; Sugimoto, Y.; Kotani, E.; Shimabukuro, J.; Hirano, T.; Nakajima, Y.; Kawamata, S.; Mori, H. Control of angiogenesis by VEGF and endostatin-encapsulated protein microcrystals and inhibition of tumor angiogenesis. Biomaterials 2014, 35, 1326–1333. [Google Scholar] [CrossRef]
- Matsushima, K.; Suyama, T.; Takenaka, C.; Nishishita, N.; Ikeda, K.; Ikada, Y.; Sawa, Y.; Jakt, L.M.; Mori, H.; Kawamata, S. Secreted frizzled related protein 4 reduces fibrosis scar size and ameliorates cardiac function after ischemic injury. Tissue Eng. Part. A 2010, 16, 3329–3341. [Google Scholar] [CrossRef]
- Shimizu, T.; Ishikawa, T.; Iwai, S.; Ueki, A.; Sugihara, E.; Onishi, N.; Kuninaka, S.; Miyamoto, T.; Toyama, Y.; Ijiri, H.; et al. Fibroblast growth factor-2 is an important factor that maintains cellular immaturity and contributes to aggressiveness of osteosarcoma. Mol. Cancer Res. 2012, 10, 454–468. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzaki, Y.; Maruta, R.; Takaki, K.; Kotani, E.; Kato, Y.; Yoshimura, R.; Endo, Y.; Whitty, C.; Pernstich, C.; Gandhi, R.; et al. Sustained Neurotrophin Release from Protein Nanoparticles Mediated by Matrix Metalloproteinases Induces the Alignment and Differentiation of Nerve Cells. Biomolecules 2019, 9, 510. https://doi.org/10.3390/biom9100510
Matsuzaki Y, Maruta R, Takaki K, Kotani E, Kato Y, Yoshimura R, Endo Y, Whitty C, Pernstich C, Gandhi R, et al. Sustained Neurotrophin Release from Protein Nanoparticles Mediated by Matrix Metalloproteinases Induces the Alignment and Differentiation of Nerve Cells. Biomolecules. 2019; 9(10):510. https://doi.org/10.3390/biom9100510
Chicago/Turabian StyleMatsuzaki, Yuka, Rina Maruta, Keiko Takaki, Eiji Kotani, Yasuko Kato, Ryoichi Yoshimura, Yasuhisa Endo, Ciara Whitty, Christian Pernstich, Raj Gandhi, and et al. 2019. "Sustained Neurotrophin Release from Protein Nanoparticles Mediated by Matrix Metalloproteinases Induces the Alignment and Differentiation of Nerve Cells" Biomolecules 9, no. 10: 510. https://doi.org/10.3390/biom9100510
APA StyleMatsuzaki, Y., Maruta, R., Takaki, K., Kotani, E., Kato, Y., Yoshimura, R., Endo, Y., Whitty, C., Pernstich, C., Gandhi, R., Jones, M., & Mori, H. (2019). Sustained Neurotrophin Release from Protein Nanoparticles Mediated by Matrix Metalloproteinases Induces the Alignment and Differentiation of Nerve Cells. Biomolecules, 9(10), 510. https://doi.org/10.3390/biom9100510




