Association of Genetic Variation at AQP4 Locus with Vascular Depression
Abstract
1. Introduction
2. Materials and Methods
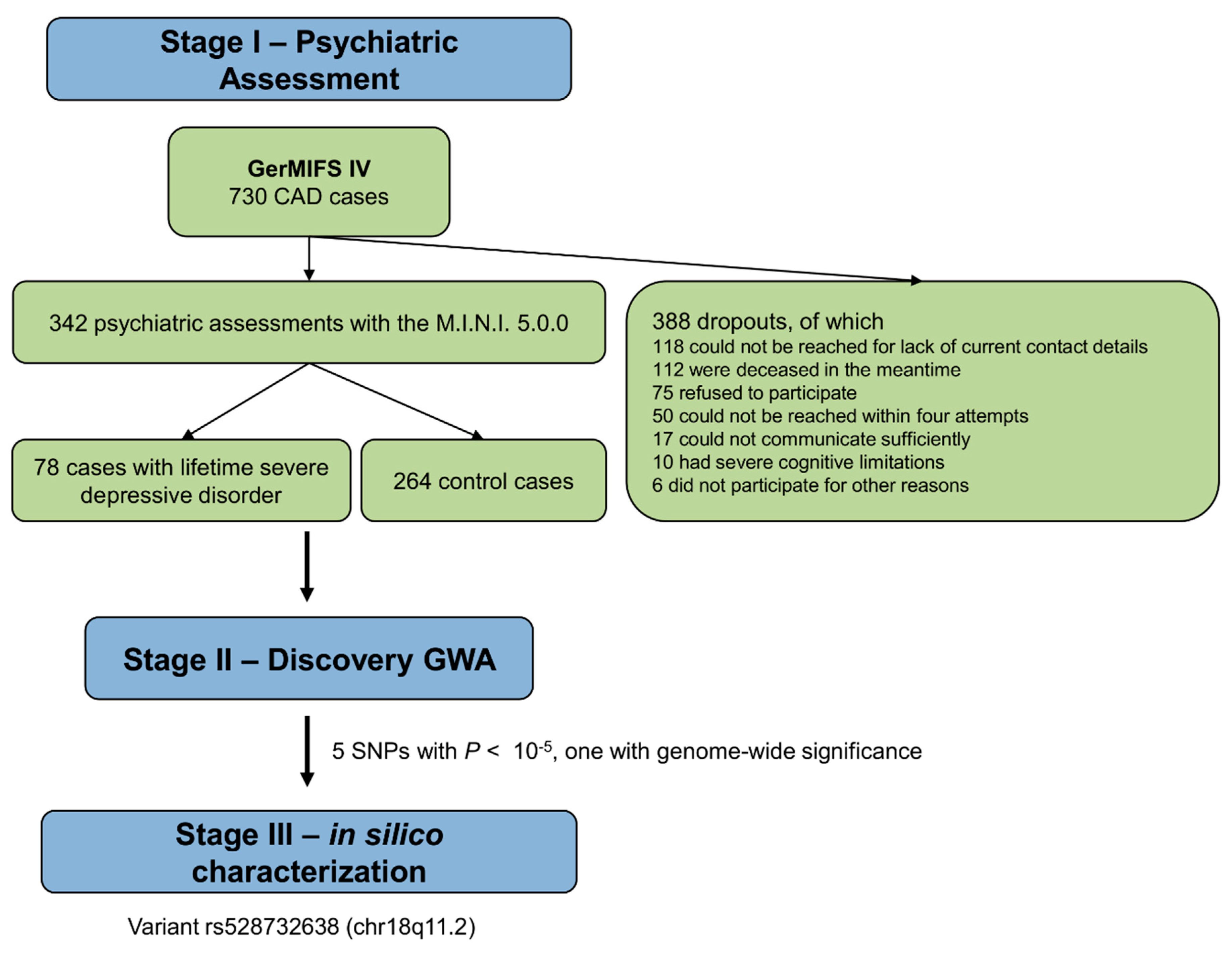
2.1. Study Protocol and Study Subjects
2.2. Assessment of Depression
2.3. DNA Isolation and Genotyping
2.4. Pre/Post-Processing and Test of Genome-Wide Associations
2.5. Functional Annotation and Software
3. Results
3.1. Demographic and Clinical Characteristics
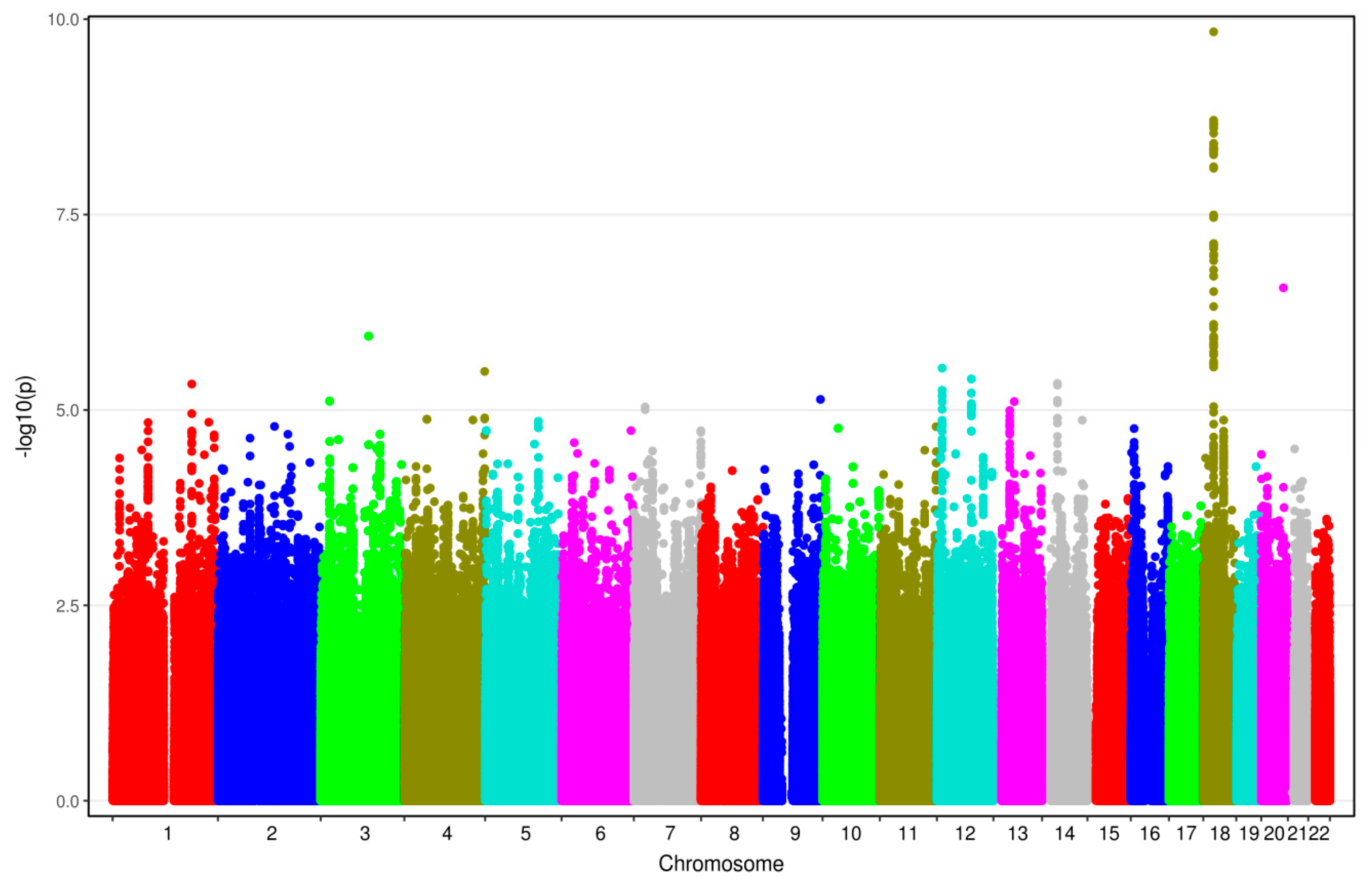
3.2. Results from GWAS
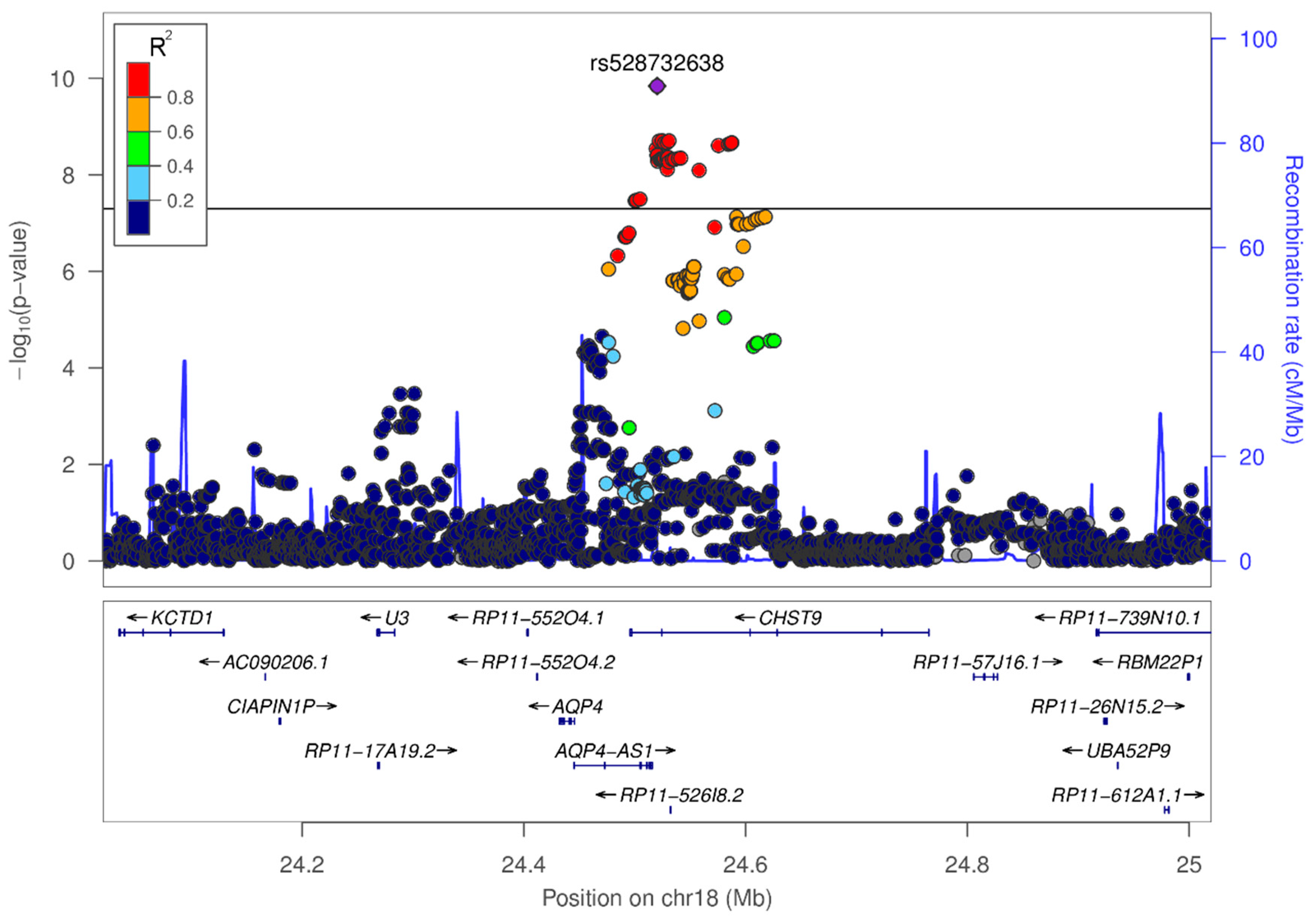
3.3. In-Silico Characterization of Variant rs528732638
4. Discussion
4.1. Variant rs528732638 on Chromosome 18q11.2 Represents the First Vascular Depression Locus
4.2. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mathers, C.; Fat, D.M.; Boerma, J.T. The Global Burden of Disease: 2004 Update; World Health Organization: Geneva, Switzerland, 2008; ISBN 9241563710. [Google Scholar]
- Geschwind, D.H.; Flint, J. Genetics and genomics of psychiatric disease. Science 2015, 349, 1489–1494. [Google Scholar] [CrossRef]
- Bosker, F.; Hartman, C.; Nolte, I.; Prins, B.; Terpstra, P.; Posthuma, D.; Van Veen, T.; Willemsen, G.; DeRijk, R.; De Geus, E. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol. Psychiatry 2011, 16, 516–532. [Google Scholar] [CrossRef]
- Otte, C.; Gold, S.M.; Penninx, B.W.; Pariante, C.M.; Etkin, A.; Fava, M.; Mohr, D.C.; Schatzberg, A.F. Major depressive disorder. Nat. Rev. Dis. Primers 2016, 2, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Ripke, S.; Wray, N.R.; Lewis, C.M.; Hamilton, S.P.; Weissman, M.M.; Breen, G.; Byrne, E.M.; Blackwood, D.H.; Boomsma, D.I.; Cichon, S. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 2013, 18, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Flint, J.; Kendler, K.S. The genetics of major depression. Neuron 2014, 81, 484–503. [Google Scholar] [CrossRef]
- Alexopoulos, G.S.; Meyers, B.S.; Young, R.C.; Campbell, S.; Silbersweig, D.; Charlson, M. The ‘vascular depression’ hypothesis. Arch. Gen. Psychiatry 1997, 54, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, K.; Hays, J.C.; Blazer, D.G. MRI-defined vascular depression. Am. J. Psychiatry 1997, 154, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulos, G.S.; Kiosses, D.N.; Klimstra, S.; Kalayam, B.; Bruce, M.L. Clinical presentation of the “depression–executive dysfunction syndrome” of late life. Am. J. Geriatr. Psychiatry 2002, 10, 98–106. [Google Scholar]
- Taylor, W.D.; Aizenstein, H.J.; Alexopoulos, G.S. The vascular depression hypothesis: Mechanisms linking vascular disease with depression. Mol. Psychiatry 2013, 18, 963–974. [Google Scholar] [CrossRef]
- Alexopoulos, G.S.; Schultz, S.K.; Lebowitz, B.D. Late-life depression: A model for medical classification. Biol. Psychiatry 2005, 58, 283–289. [Google Scholar] [CrossRef]
- Köhler, S.; Thomas, A.; Barnett, N.; O’Brien, J. The pattern and course of cognitive impairment in late-life depression. Psychol. Med. 2010, 40, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Paranthaman, R.; Burns, A.S.; Cruickshank, J.K.; Jackson, A.; Scott, M.L.; Baldwin, R.C. Age at onset and vascular pathology in late-life depression. Am. J. Geriatr. Psychiatry 2012, 20, 524–532. [Google Scholar] [CrossRef]
- Feng, C.; Fang, M.; Xu, Y.; Hua, T.; Liu, X.-Y. Microbleeds in late-life depression: Comparison of early-and late-onset depression. Biomed. Res. Int. 2014, 2014, 682092. [Google Scholar] [CrossRef]
- Herrmann, L.L.; Le Masurier, M.; Ebmeier, K.P. White matter hyperintensities in late life depression: A systematic review. J. Neurol. Neurosurg. Psychiatry 2008, 79, 619–624. [Google Scholar] [CrossRef] [PubMed]
- van Sloten, T.T.; Sigurdsson, S.; van Buchem, M.A.; Phillips, C.L.; Jonsson, P.V.; Ding, J.; Schram, M.T.; Harris, T.B.; Gudnason, V.; Launer, L.J. Cerebral small vessel disease and association with higher incidence of depressive symptoms in a general elderly population: The AGES-Reykjavik Study. Am. J. Psychiatry 2015, 172, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Sexton, C.E.; Le Masurier, M.; Allan, C.L.; Jenkinson, M.; McDermott, L.; Kalu, U.G.; Herrmann, L.L.; Bradley, K.M.; Mackay, C.E.; Ebmeier, K.P. Magnetic resonance imaging in late-life depression: Vascular and glucocorticoid cascade hypotheses. Br. J. Psychiatry 2012, 201, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Andreescu, C.; Tudorascu, D.L.; Butters, M.A.; Tamburo, E.; Patel, M.; Price, J.; Karp, J.F.; Reynolds, C.F.; Aizenstein, H. Resting state functional connectivity and treatment response in late-life depression. Psychiatry Res. Neuroimaging 2013, 214, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Hou, Z.; Wang, X.; Sui, Y.; Yuan, Y. Association between altered resting-state cortico-cerebellar functional connectivity networks and mood/cognition dysfunction in late-onset depression. J. Neural Transm. 2015, 122, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Valkanova, V.; Ebmeier, K.P. Vascular risk factors and depression in later life: A systematic review and meta-analysis. Biol. Psychiatry 2013, 73, 406–413. [Google Scholar] [CrossRef]
- Saran, R.; Puri, A.; Agarwal, M. Depression and the heart. Indian Heart J. 2012, 64, 397–401. [Google Scholar] [CrossRef]
- Robinson, R.G.; Spalletta, G. Poststroke depression: A review. Can. J. Psychiatry 2010, 55, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Saczynski, J.S.; Beiser, A.; Seshadri, S.; Auerbach, S.; Wolf, P.; Au, R. Depressive symptoms and risk of dementia The Framingham Heart Study. Neurology 2010, 75, 35–41. [Google Scholar] [CrossRef] [PubMed]
- González, H.M.; Tarraf, W.; Whitfield, K.; Gallo, J.J. Vascular depression prevalence and epidemiology in the United States. J. Psychiatr. Res. 2012, 46, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, S.B.; Lee, J.J.; Yoon, J.C.; Han, J.W.; Kim, T.H.; Jeong, H.-G.; Newhouse, P.A.; Taylor, W.D.; Kim, J.H. Epidemiology of MRI-defined vascular depression: A longitudinal, community-based study in Korean elders. J. Affect. Disord. 2015, 180, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Seripa, D.; Panza, F.; D’Onofrio, G.; Paroni, G.; Bizzarro, A.; Fontana, A.; Paris, F.; Cascavilla, L.; Copetti, M.; Masullo, C. The serotonin transporter gene locus in late-life major depressive disorder. Am. J. Geriatr. Psychiatry 2013, 21, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Lee, S.-H.; Kim, C.K.; Ryu, W.-S.; Kwon, H.-M.; Choi, S.-Y.; Yoon, B.-W. Advanced coronary artery calcification and cerebral small vessel diseases in the healthy elderly. Circ. J. 2011, 75, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Bos, D.; Ikram, M.A.; Elias-Smale, S.E.; Krestin, G.P.; Hofman, A.; Witteman, J.C.; van der Lugt, A.; Vernooij, M.W. Calcification in major vessel beds relates to vascular brain disease. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2331–2337. [Google Scholar] [CrossRef] [PubMed]
- Nikpay, M.; Goel, A.; Won, H.-H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Ackenheil, M.; Stotz-Ingenlath, G.; Dietz-Bauer, R.; Vossen, A. MINI (Mini International Neuropsychiatric Interview), German Version 5.0.0 DSM IV; Psychiatric University Clinic Munich: Munich, Germany, 1999. [Google Scholar]
- Schaich, A.; Westermair, A.L.; Munz, M.; Nitsche, S.; Willenborg, B.; Willenborg, C.; Schunkert, H.; Erdmann, J.; Schweiger, U. Mental health and psychosocial functioning over the lifespan of German patients undergoing cardiac catheterization for coronary artery disease. Front. Psychiatry 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Marchini, J.; Howie, B.; Myers, S.; McVean, G.; Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007, 39, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Munz, M.; Tönnies, S.; Balke, W.-T.; Simon, E. Multidimensional gene search with Genehopper. Nucleic Acids Res. 2015, 43, W98–W103. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Sammeth, M.; Friedländer, M.R.; AC’t Hoen, P.; Monlong, J.; Rivas, M.A.; Gonzalez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G.; et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Shabalin, A.A.; Huang, S.; Madar, V.; Zhou, Y.-H.; Wang, W.; Zou, F.; Sun, W.; Sullivan, P.F.; Wright, F.A. seeQTL: A searchable database for human eQTLs. Bioinformatics 2011, 28, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Dudbridge, F.; Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008, 32, 227–234. [Google Scholar] [CrossRef]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2013, 42, D1001–D1006. [Google Scholar] [CrossRef]
- Iacovetta, C.; Rudloff, E.; Kirby, R. The role of aquaporin 4 in the brain. Vet. Clin. Pathol. 2012, 41, 32–44. [Google Scholar] [CrossRef]
- Suzuki, Y.; Nakamura, Y.; Yamada, K.; Huber, V.J.; Tsujita, M.; Nakada, T. Aquaporin-4 Positron Emission Tomography Imaging of the Human Brain: First Report. J. Neuroimaging 2013, 23, 219–223. [Google Scholar] [CrossRef]
- Venero, J.L.; Vizuete, M.A.L.; Machado, A.; Cano, J. Aquaporins in the central nervous system. Prog. Neurobiol. 2001, 63, 321–336. [Google Scholar] [CrossRef]
- Cavazzin, C.; Ferrari, D.; Facchetti, F.; Russignan, A.; Vescovi, A.L.; La Porta, C.A.; Gritti, A. Unique expression and localization of aquaporin-4 and aquaporin-9 in murine and human neural stem cells and in their glial progeny. Glia 2006, 53, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Nakada, T.; Kwee, I.L.; Igarashi, H.; Suzuki, Y. Aquaporin-4 functionality and Virchow-Robin space water dynamics: Physiological model for neurovascular coupling and glymphatic flow. Int. J. Mol. Sci. 2017, 18, 1798. [Google Scholar] [CrossRef] [PubMed]
- Previch, L.E.; Ma, L.; Wright, J.C.; Singh, S.; Geng, X.; Ding, Y. Progress in AQP research and new developments in therapeutic approaches to ischemic and hemorrhagic stroke. Int. J. Mol. Sci. 2016, 17, 1146. [Google Scholar] [CrossRef] [PubMed]
- Rajkowska, G.; Hughes, J.; Stockmeier, C.A.; Miguel-Hidalgo, J.J.; Maciag, D. Coverage of Blood Vessels by Astrocytic Endfeet is Reduced in Major Depressive Disorder. Biol. Psychiatry 2013, 73, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Di Benedetto, B.; Malik, V.A.; Begum, S.; Jablonowski, L.; Gómez-González, G.B.; Neumann, I.D.; Rupprecht, R. Fluoxetine requires the endfeet protein aquaporin-4 to enhance plasticity of astrocyte processes. Front. Cell. Neurosci. 2016, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Fan, Y.; Xie, J.; Ding, J.; Sha, L.; Shi, X.; Sun, X.; Hu, G. AQP4 knockout impairs proliferation, migration and neuronal differentiation of adult neural stem cells. J. Cell Sci. 2008, 121, 4029–4036. [Google Scholar] [CrossRef]
- Kong, H.; Sha, L.-L.; Fan, Y.; Xiao, M.; Ding, J.-H.; Wu, J.; Hu, G. Requirement of AQP4 for antidepressive efficiency of fluoxetine: Implication in adult hippocampal neurogenesis. Neuropsychopharmacology 2009, 34, 1263–1276. [Google Scholar] [CrossRef]
- Sheline, Y.I.; Gado, M.H.; Kraemer, H.C. Untreated depression and hippocampal volume loss. Am. J. Psychiatry 2003, 160, 1516–1518. [Google Scholar] [CrossRef]
- Malberg, J.E.; Eisch, A.J.; Nestler, E.J.; Duman, R.S. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J. Neurosci. 2000, 20, 9104–9110. [Google Scholar] [CrossRef]
- Manev, H.; Uz, T.; Smalheiser, N.R.; Manev, R. Antidepressants alter cell proliferation in the adult brain in vivo and in neural cultures in vitro. Eur. J. Pharmacol. 2001, 411, 67–70. [Google Scholar] [CrossRef]
- Malberg, J.E.; Duman, R.S. Cell proliferation in adult hippocampus is decreased by inescapable stress: Reversal by fluoxetine treatment. Neuropsychopharmacology 2003, 28, 1562. [Google Scholar] [CrossRef] [PubMed]
- Manley, G.T.; Fujimura, M.; Ma, T.; Noshita, N.; Filiz, F.; Bollen, A.W.; Chan, P.; Verkman, A. Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat. Med. 2000, 6, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Thrane, A.S.; Rappold, P.M.; Fujita, T.; Torres, A.; Bekar, L.K.; Takano, T.; Peng, W.; Wang, F.; Thrane, V.R.; Enger, R.; et al. Critical role of aquaporin-4 (AQP4) in astrocytic Ca2+ signaling events elicited by cerebral edema. Proc. Natl. Acad. Sci. USA 2011, 108, 846–851. [Google Scholar] [CrossRef]
- Fazzina, G.; Amorini, A.M.; Marmarou, C.R.; Fukui, S.; Okuno, K.; Dunbar, J.G.; Glisson, R.; Marmarou, A.; Kleindienst, A. The protein kinase C activator phorbol myristate acetate decreases brain edema by aquaporin 4 downregulation after middle cerebral artery occlusion in the rat. J. Neurotrauma 2010, 27, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bai, R.; Zhang, J.; Wang, X. Effect of progesterone intervention on the dynamic changes of AQP-4 in hypoxic-ischaemic brain damage. Int. J. Clin. Exp. Med. 2015, 8, 18831–18836. [Google Scholar] [PubMed]
- Higashida, T.; Peng, C.; Li, J.; Dornbos, D.; Teng, K.; Li, X.; Kinni, H.; Guthikonda, M.; Ding, Y. Hypoxia-inducible factor-1α contributes to brain edema after stroke by regulating aquaporins and glycerol distribution in brain. Curr. Neurovasc. Res. 2011, 8, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.S.; Fan, Y.Y.; Ye, G.; Li, J.; Feng, X.P.; Lin, K.; Dong, M.; Wang, Z. Curcumin alleviates brain edema by lowering AQP4 expression levels in a rat model of hypoxia-hypercapnia-induced brain damage. Exp. Ther. Med. 2016, 11, 709–716. [Google Scholar] [CrossRef]
- Wang, X.; An, F.; Wang, S.; An, Z.; Wang, S. Orientin Attenuates Cerebral Ischemia/Reperfusion Injury in Rat Model through the AQP-4 and TLR4/NF-κB/TNF-α Signaling Pathway. J. Stroke Cerebrovasc. Dis. 2017, 26, 2199–2214. [Google Scholar] [CrossRef]
- Vella, J.; Zammit, C.; Di Giovanni, G.; Muscat, R.; Valentino, M. The central role of aquaporins in the pathophysiology of ischemic stroke. Front. Cell. Neurosci. 2015, 9, 108. [Google Scholar] [CrossRef]
- Igarashi, H.; Huber, V.J.; Tsujita, M.; Nakada, T. Pretreatment with a novel aquaporin 4 inhibitor, TGN-020, significantly reduces ischemic cerebral edema. Neurol. Sci. 2011, 32, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Popescu, E.S.; Pirici, I.; Ciurea, R.N.; Balseanu, T.-A.; Catalin, B.; Margaritescu, C.; Mogoanta, L.; Hostiuc, S.; Pirici, D. Three-dimensional organ scanning reveals brain edema reduction in a rat model of stroke treated with an aquaporin 4 inhibitor. Rom. J. Morphol. Embryol. 2017, 58, 59–66. [Google Scholar] [PubMed]
- Amiry-Moghaddam, M.; Otsuka, T.; Hurn, P.D.; Traystman, R.J.; Haug, F.-M.; Froehner, S.C.; Adams, M.E.; Neely, J.D.; Agre, P.; Ottersen, O.P.; et al. An α-syntrophin-dependent pool of AQP4 in astroglial end-feet confers bidirectional water flow between blood and brain. Proc. Natl. Acad. Sci. USA 2003, 100, 2106–2111. [Google Scholar] [CrossRef]
- Rutkovskiy, A.; Valen, G.; Vaage, J. Cardiac aquaporins. Basic Res. Cardiol. 2013, 108, 393. [Google Scholar] [CrossRef] [PubMed]
- Warth, A.; Eckle, T.; Köhler, D.; Faigle, M.; Zug, S.; Klingel, K.; Eltzschig, H.K.; Wolburg, H. Upregulation of the water channel aquaporin-4 as a potential cause of postischemic cell swelling in a murine model of myocardial infarction. Cardiology 2007, 107, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Z.; Kim, M.H.; Lim, J.H.; Bae, H.-R. Time-dependent expression patterns of cardiac aquaporins following myocardial infarction. J. Korean Med. Sci. 2013, 28, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rutkovskiy, A.; Stensløkken, K.-O.; Mariero, L.H.; Skrbic, B.; Amiry-Moghaddam, M.; Hillestad, V.; Valen, G.; Perreault, M.-C.; Ottersen, O.P.; Gullestad, L.; et al. Aquaporin-4 in the heart: Expression, regulation and functional role in ischemia. Basic Res. Cardiol. 2012, 107, 280. [Google Scholar] [CrossRef]
- Vilahur, G.; Gutiérrez, M.; Casani, L.; Varela, L.; Capdevila, A.; Pons-Lladó, G.; Carreras, F.; Carlsson, L.; Hidalgo, A.; Badimon, L. Protective effects of ticagrelor on myocardial injury after infarction. Circulation 2016, 134, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Szpakowski, N.; Bennell, M.C.; Qiu, F.; Ko, D.T.; Tu, J.V.; Kurdyak, P.; Wijeysundera, H.C. Clinical Impact of Subsequent Depression in Patients With a New Diagnosis of Stable Angina. Circ. Cardiovasc. Qual. Outcomes 2016, 9. [Google Scholar] [CrossRef]
- Moussavi, S.; Chatterji, S.; Verdes, E.; Tandon, A.; Patel, V.; Ustun, B. Depression, chronic diseases, and decrements in health: Results from the World Health Surveys. Lancet 2007, 370, 851–858. [Google Scholar] [CrossRef]
- de Jager, T.A.; Dulfer, K.; Pieters, K.; Utens, E.M.; Daemen, J.; Lenzen, M.J.; van Domburg, R.T. The association between subjective health status and 14-year mortality in post-PCI patients. Int. J. Cardiol. 2017, 229, 108–112. [Google Scholar] [CrossRef] [PubMed]
| EAF | ||||||||
|---|---|---|---|---|---|---|---|---|
| Variant | Locus | Nearest Genes | EA | NEA | Cas | Con | OR [95% CI] | P |
| rs528732638 | 18q11.2 | AQP4-AS1, CHST9 | GAA | G | 0.34 | 0.13 | 3.62 [2.38–5.50] | 1.45 × 10−10 |
| Tissue | Gene |
|---|---|
| Blood (4.7 × 10−11), skeletal muscle (4.7 × 10−11) | AQP4 |
| Average brain (1.2 × 10−10), white matter (6.6 × 10−9), parietal lobe (3 × 10−3) | AQP4-AS1 |
| liver (1 × 10−6) | CDH12a |
| Peripheral blood monocytes (1.6 × 10−6) | CHST9 |
| Peripheral blood monocytes (1.6 × 10−6) | ZNF570a |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Westermair, A.L.; Munz, M.; Schaich, A.; Nitsche, S.; Willenborg, B.; Muñoz Venegas, L.M.; Willenborg, C.; Schunkert, H.; Schweiger, U.; Erdmann, J. Association of Genetic Variation at AQP4 Locus with Vascular Depression. Biomolecules 2018, 8, 164. https://doi.org/10.3390/biom8040164
Westermair AL, Munz M, Schaich A, Nitsche S, Willenborg B, Muñoz Venegas LM, Willenborg C, Schunkert H, Schweiger U, Erdmann J. Association of Genetic Variation at AQP4 Locus with Vascular Depression. Biomolecules. 2018; 8(4):164. https://doi.org/10.3390/biom8040164
Chicago/Turabian StyleWestermair, Anna L., Matthias Munz, Anja Schaich, Stefan Nitsche, Bastian Willenborg, Loreto M. Muñoz Venegas, Christina Willenborg, Heribert Schunkert, Ulrich Schweiger, and Jeanette Erdmann. 2018. "Association of Genetic Variation at AQP4 Locus with Vascular Depression" Biomolecules 8, no. 4: 164. https://doi.org/10.3390/biom8040164
APA StyleWestermair, A. L., Munz, M., Schaich, A., Nitsche, S., Willenborg, B., Muñoz Venegas, L. M., Willenborg, C., Schunkert, H., Schweiger, U., & Erdmann, J. (2018). Association of Genetic Variation at AQP4 Locus with Vascular Depression. Biomolecules, 8(4), 164. https://doi.org/10.3390/biom8040164






