Single Molecule FRET: A Powerful Tool to Study Intrinsically Disordered Proteins
Abstract
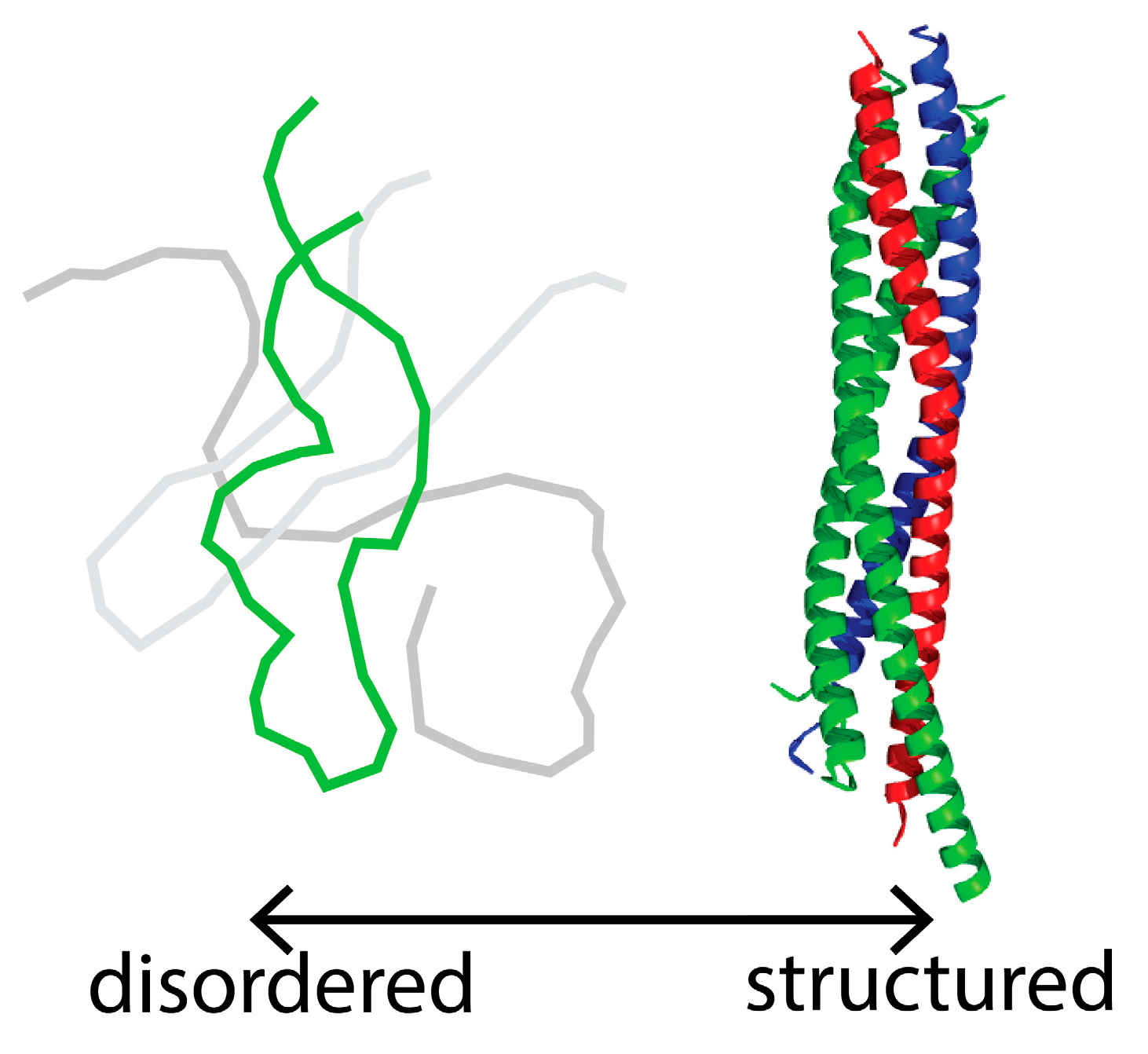
1. Introduction
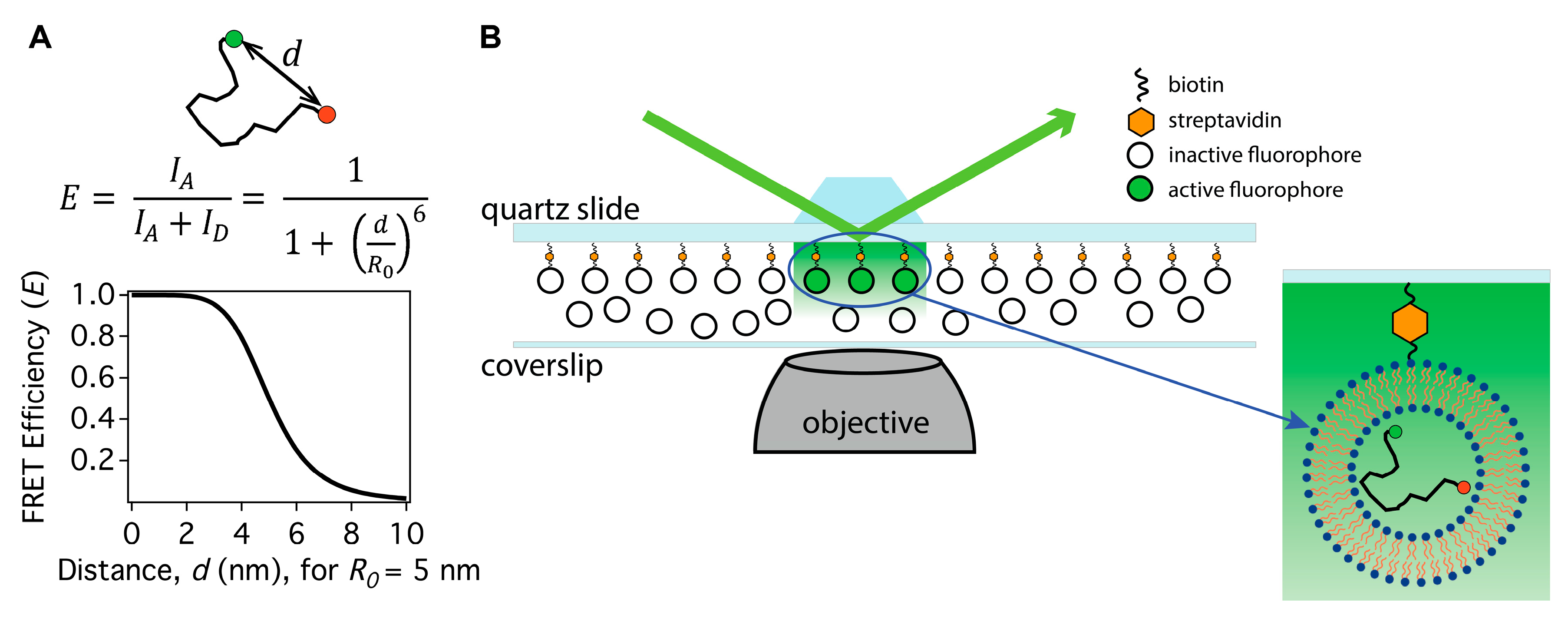
2. How to Apply Single Molecule FRET to Intrinsically Disordered Protein Studies
3. Do Single Molecule FRET and Small Angle X-ray Scattering Agree about the Size of Intrinsically Disordered Proteins?
4. Unique Phenomena Identified by Applying Single Molecule FRET to Intrinsically Disordered Proteins
4.1. Spontaneous Switching among Intrinsically Disordered Protein Ensembles
4.2. Phosphorylation Modulates Intrinsically Disordered Protein Configuration and Function
5. Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1231–1264. [Google Scholar] [CrossRef]
- Shammas, S.L. Mechanistic roles of protein disorder within transcription. Curr. Opin. Struct. Biol. 2017, 42, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in macromolecular complexes: The roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Functional advantages of dynamic protein disorder. FEBS Lett. 2015, 589, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.; Chan, J.M.; Simoncik, O.; Fojta, M.; Lane, D.P.; Hupp, T.; Vojtesek, B. Evidence for allosteric effects on p53 oligomerization induced by phosphorylation. Protein Sci. 2018, 27, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Valk, E.; Venta, R.; Ord, M.; Faustova, I.; Kõivomägi, M.; Loog, M. Multistep phosphorylation systems: Tunable components of biological signaling circuits. Mol. Biol. Cell 2014, 25, 3456–3460. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Solomon, T.L.; He, Y.; Chen, Y.; Bryan, P.N.; Orban, J. Structural metamorphism and polymorphism in proteins on the brink of thermodynamic stability. Protein Sci. 2018, 27, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Kragelj, J.; Ozenne, V.; Blackledge, M.; Jensen, M.R. Conformational propensities of intrinsically disordered proteins from NMR chemical shifts. ChemPhysChem 2013, 14, 3034–3045. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, B.; Felli, I.C.; Gil-Caballero, S.; Hošek, T.; Kümmerle, R.; Piai, A.; Pierattelli, R.; Sólyom, Z. NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: General overview and practical guidelines. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2015; Volume 870, pp. 49–122. [Google Scholar]
- Kurzbach, D.; Kontaxis, G.; Coudevylle, N.; Konrat, R. NMR spectroscopic studies of the conformational ensembles of intrinsically disordered proteins. In Advances in Experimental Medicine and Biology; Springer: Basel, Switzerland, 2015; Volume 870, pp. 149–185. [Google Scholar]
- Chen, H.; Rhoades, E. Fluorescence characterization of denatured proteins. Curr. Opin. Struct. Biol. 2008, 18, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.; Itkin, A.; Kuttner, Y.Y.; Rhoades, E.; Amir, D.; Haas, E.; Haran, G. Using fluorescence correlation spectroscopy to study conformational changes in denatured proteins. Biophys. J. 2008, 94, 4819–4827. [Google Scholar] [CrossRef] [PubMed]
- Nettels, D.; Gopich, I.V.; Hoffmann, A.; Schuler, B. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc. Natl. Acad. Sci. USA 2007, 104, 2655–2660. [Google Scholar] [CrossRef] [PubMed]
- Nettels, D.; Hoffmann, A.; Schuler, B. Unfolded protein and peptide dynamics investigated with single-molecule FRET and correlation spectroscopy from picoseconds to seconds. J. Phys. Chem. B 2008, 112, 6137–6146. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Bellaiche, M.M.J.; Kim, J.-Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly Disordered Amyloid-β Monomer Probed by Single-Molecule FRET and MD Simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B. Perspective: Chain dynamics of unfolded and intrinsically disordered proteins from nanosecond fluorescence correlation spectroscopy combined with single-molecule FRET. J. Chem. Phys. 2018, 149, 010901. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenkina, E.V.; Heyes, C.D.; Nienhaus, G.U. Single-molecule Förster resonance energy transfer study of protein dynamics under denaturing conditions. Proc. Natl. Acad. Sci. USA 2005, 102, 15471–15476. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.-N.; Gradinaru, C.C. Insights into the conformations and dynamics of intrinsically disordered proteins using single-molecule fluorescence. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.M.; Moran, C.R.; Gambin, Y.; Deniz, A.A. Single-Molecule Fluorescence Studies of Intrinsically Disordered Proteins. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 472, pp. 179–204. [Google Scholar]
- Lee, T.; Moran-Gutierrez, C.R.; Deniz, A.A. Probing protein disorder and complexity at single-molecule resolution. Semin. Cell Dev. Biol. 2015, 37, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.; Tinnefeld, P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 2012, 63, 595–617. [Google Scholar] [CrossRef] [PubMed]
- Brustad, E.M.; Lemke, E.A.; Schultz, P.G.; Deniz, A.A. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J. Am. Chem. Soc. 2008, 130, 17664–17665. [Google Scholar] [CrossRef] [PubMed]
- DeRocco, V.; Anderson, T.; Piehler, J.; Erie, D.A.; Weninger, K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. Biotechniques 2010, 49, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.C.; Moran, C.R.; Cistrone, P.A.; Dawson, P.E.; Deniz, A.A. Site-specific three-color labeling of α-synuclein via conjugation to uniquely reactive cysteines during assembly by native chemical ligation. Cell Chem. Biol. 2018, 25, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Ragunathan, K.; Joo, C.; Ha, T.; Hohng, S. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 2010, 49, 9922–9925. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.A.; Van Der Kleij, A.A.; Sonnemans, M.A.; Burbach, J.P.; van Leeuwen, F.W. Frameshift mutations at two hotspots in vasopressin transcripts in post-mitotic neurons. Proc. Natl. Acad. Sci. USA 1994, 91, 6059. [Google Scholar] [CrossRef] [PubMed]
- Hohng, S.; Joo, C.; Ha, T. Single-molecule three-color FRET. Biophys. J. 2004, 87, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Koehler, C.; Gambin, Y.; Deniz, A.A.; Lemke, E.A. Intramolecular three-colour single pair FRET of intrinsically disordered proteins with increased dynamic range. Mol. Biosyst. 2012, 8, 2531. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B.; Müller-Späth, S.; Soranno, A.; Nettels, D. Application of confocal single-molecule FRET to intrinsically disordered proteins. Methods Mol. Biol. 2012, 896, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Deniz, A.A.; Laurence, T.A.; Beligere, G.S.; Dahan, M.; Martin, A.B.; Chemla, D.S.; Dawson, P.E.; Schultz, P.G.; Weiss, S. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 5179–5184. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; Weninger, K.R.; Bowen, M.E. Immobilization of Proteins for Single-Molecule Fluorescence Resonance Energy Transfer Measurements of Conformation and Dynamics. In Intrinsically Disordered Protein Analysis; Springer: New York, NY, USA, 2012; Volume 896, pp. 3–20. [Google Scholar]
- Hoffmann, A.; Nettels, D.; Clark, J.; Borgia, A.; Radford, S.E.; Clarke, J.; Schuler, B. Quantifying heterogeneity and conformational dynamics from single molecule FRET of diffusing molecules: Recurrence analysis of single particles (RASP). Phys. Chem. Chem. Phys. 2011, 13, 1857. [Google Scholar] [CrossRef] [PubMed]
- McCann, J.J.; Choi, U.B.; Zheng, L.Q.; Weninger, K.; Bowen, M.E. Optimizing methods to recover absolute FRET efficiency from immobilized single molecules. Biophys. J. 2010, 99, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Brunger, A.T.; Strop, P.; Vrljic, M.; Chu, S.; Weninger, K.R. Three-dimensional molecular modeling with single molecule FRET. J. Struct. Biol. 2011, 173, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Hellenkamp, B.; Schmid, S.; Doroshenko, O.; Opanasyuk, O.; Kühnemuth, R.; Rezaei Adariani, S.; Ambrose, B.; Aznauryan, M.; Barth, A.; Birkedal, V.; et al. Precision and accuracy of single-molecule FRET measurements—A multi-laboratory benchmark study. Nat. Methods 2018, 15, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Kalinin, S.; Peulen, T.; Sindbert, S.; Rothwell, P.J.; Berger, S.; Restle, T.; Goody, R.S.; Gohlke, H.; Seidel, C.A.M. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods 2012, 9, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Makarov, D.E.; Plaxco, K.W. Measuring distances within unfolded biopolymers using fluorescence resonance energy transfer: The effect of polymer chain dynamics on the observed fluorescence resonance energy transfer efficiency. J. Chem. Phys. 2009, 131, 085105. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Borgia, A.; Buholzer, K.; Grishaev, A.; Schuler, B.; Best, R.B. Probing the action of chemical denaturant on an intrinsically disordered protein by simulation and experiment. J. Am. Chem. Soc. 2016, 138, 11702–11713. [Google Scholar] [CrossRef] [PubMed]
- Zerze, G.H.; Best, R.B.; Mittal, J. Modest influence of FRET chromophores on the properties of unfolded proteins. Biophys. J. 2014, 107, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.P.; Morrison, G.; Brooks, B.R.; Thirumalai, D. How accurate are polymer models in the analysis of Förster resonance energy transfer experiments on proteins? J. Chem. Phys. 2009, 130, 124903. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Soranno, A.; Borgia, A.; Gast, K.; Nettels, D.; Schuler, B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 16155–16160. [Google Scholar] [CrossRef] [PubMed]
- Soranno, A.; Buchli, B.; Nettels, D.; Cheng, R.R.; Müller-Späth, S.; Pfeil, S.H.; Hoffmann, A.; Lipman, E.A.; Makarov, D.E.; Schuler, B. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 17800–17806. [Google Scholar] [CrossRef] [PubMed]
- Soranno, A.; Holla, A.; Dingfelder, F.; Nettels, D.; Makarov, D.E.; Schuler, B. Integrated view of internal friction in unfolded proteins from single-molecule FRET, contact quenching, theory, and simulations. Proc. Natl. Acad. Sci. USA 2017, 114, E1833–E1839. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zerze, G.H.; Borgia, A.; Mittal, J.; Schuler, B.; Best, R.B. Inferring properties of disordered chains from FRET transfer efficiencies. J. Chem. Phys. 2018, 148, 123329. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Zheng, W.; Buholzer, K.; Borgia, M.B.; Schüler, A.; Hofmann, H.; Soranno, A.; Nettels, D.; Gast, K.; Grishaev, A.; et al. Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J. Am. Chem. Soc. 2016, 138, 11714–11726. [Google Scholar] [CrossRef] [PubMed]
- Yoo, T.Y.; Meisburger, S.P.; Hinshaw, J.; Pollack, L.; Haran, G.; Sosnick, T.R.; Plaxco, K. Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J. Mol. Biol. 2012, 418, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Kohn, J.E.; Millett, I.S.; Jacob, J.; Zagrovic, B.; Dillon, T.M.; Cingel, N.; Dothager, R.S.; Seifert, S.; Thiyagarajan, P.; Sosnick, T.R.; et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 12491–12496. [Google Scholar] [CrossRef] [PubMed]
- McCarney, E.R.; Werner, J.H.; Bernstein, S.L.; Ruczinski, I.; Makarov, D.E.; Goodwin, P.M.; Plaxco, K.W. Site-specific dimensions across a highly denatured protein; A single molecule study. J. Mol. Biol. 2005, 352, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Merchant, K.A.; Best, R.B.; Louis, J.M.; Gopich, I.V.; Eaton, W.A. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. USA 2007, 104, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Watkins, H.M.; Simon, A.J.; Sosnick, T.R.; Lipman, E.A.; Hjelm, R.P.; Plaxco, K.W. Random coil negative control reproduces the discrepancy between scattering and FRET measurements of denatured protein dimensions. Proc. Natl. Acad. Sci. USA 2015, 112, 6631–6636. [Google Scholar] [CrossRef] [PubMed]
- Knowles, D.B.; Shkel, I.A.; Phan, N.M.; Sternke, M.; Lingeman, E.; Cheng, X.; Cheng, L.; O’Connor, K.; Record, M.T. Chemical interactions of polyethylene glycols (PEGs) and glycerol with protein functional groups: Applications to effects of PEG and glycerol on protein processes. Biochemistry 2015, 54, 3528–3542. [Google Scholar] [CrossRef] [PubMed]
- Hammes, G.G.; Schimmel, P.R. An Investigation of water-urea and water-urea-polyethylene glycol interactions. J. Am. Chem. Soc. 1967, 89, 442–446. [Google Scholar] [CrossRef]
- Schuler, B.; Lipman, E.A.; Eaton, W.A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 2002, 419, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, G.; Banterle, N.; Ruff, K.M.; Chowdhury, A.; Mercadante, D.; Koehler, C.; Kachala, M.; Estrada Girona, G.; Milles, S.; Mishra, A.; et al. Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. USA 2017, 114, E6342–E6351. [Google Scholar] [CrossRef] [PubMed]
- Aznauryan, M.; Delgado, L.; Soranno, A.; Nettels, D.; Huang, J.-R.; Labhardt, A.M.; Grzesiek, S.; Schuler, B. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA 2016, 113, E5389–E5398. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, T.; Jin, F.; Yu, D.; Liu, Z. Dimension conversion and scaling of disordered protein chains. Mol. Biosyst. 2016, 12, 2932–2940. [Google Scholar] [CrossRef] [PubMed]
- Riback, J.A.; Bowman, M.A.; Zmyslowski, A.M.; Knoverek, C.R.; Jumper, J.M.; Hinshaw, J.R.; Kaye, E.B.; Freed, K.F.; Clark, P.L.; Sosnick, T.R. Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water. Science 2017, 358, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Gomes, G.-N.; Shi, T.; Gradinaru, C.C.; Chan, H.S. Conformational heterogeneity and FRET data interpretation for dimensions of unfolded proteins. Biophys. J. 2017, 113, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Gomes, G.-N.; Gradinaru, C.C.; Chan, H.S. An Adequate account of excluded volume is necessary to infer compactness and asphericity of disordered proteins by Förster resonance energy transfer. J. Phys. Chem. B 2015, 119, 15191–15202. [Google Scholar] [CrossRef] [PubMed]
- Riback, J.A.; Bowman, M.A.; Zmyslowski, A.; Knoverek, C.R.; Jumper, J.; Kaye, E.B.; Freed, K.F.; Clark, P.L.; Sosnick, T.R. Response to comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”. Science 2018, 361, eaar7949. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zheng, W.; Borgia, A.; Buholzer, K.; Borgia, M.B.; Hofmann, H.; Soranno, A.; Nettels, D.; Gast, K.; Grishaev, A.; et al. Comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”. Science 2018, 361, eaar7101. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, G.; Banterle, N.; Ruff, K.M.; Chowdhury, A.; Pappu, R.V.; Svergun, D.I.; Lemke, E.A. Comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”. Science 2018, 361, eaau8230. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Jolly, M.K.; Jia, D.; Mooney, S.M.; Bhargava, A.; Kagohara, L.T.; Chen, Y.; Hao, P.; He, Y.; Veltri, R.W.; et al. Phosphorylation-induced conformational dynamics in an intrinsically disordered protein and potential role in phenotypic heterogeneity. Proc. Natl. Acad. Sci. USA 2017, 114, E2644–E2653. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Krishnan, R.; Lemke, E.A.; Lindquist, S.; Deniz, A.A. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. USA 2007, 104, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Moosa, M.M.; Ferreon, A.C.M.; Deniz, A.A. Forced folding of a disordered protein accesses an alternative folding landscape. ChemPhysChem 2015, 16, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Lamboy, J.A.; Kim, H.; Lee, K.S.; Ha, T.; Komives, E.A. Visualization of the nanospring dynamics of the IkappaBalpha ankyrin repeat domain in real time. Proc. Natl. Acad. Sci. USA 2011, 108, 10178–10183. [Google Scholar] [CrossRef] [PubMed]
- Lamboy, J.A.; Kim, H.; Dembinski, H.; Ha, T.; Komives, E.A. Single-molecule FRET reveals the native-state dynamics of the IκBα ankyrin repeat domain. J. Mol. Biol. 2013, 425, 2578–2590. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; McCann, J.J.; Weninger, K.R.; Bowen, M.E. Beyond the random coil: Stochastic conformational switching in intrinsically disordered proteins. Structure 2011, 19, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chia, D.; Csizmok, V.; Farber, P.; Forman-Kay, J.D.; Gradinaru, C.C. The effect of intrachain electrostatic repulsion on conformational disorder and dynamics of the Sic1 protein. J. Phys. Chem. B 2014, 118, 4088–4097. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, D.; Liu, J.; Feng, Y.; Weng, J.; Li, Y.; Gao, X.; Liu, J.; Wang, W. The dynamic multisite interactions between two intrinsically disordered proteins. Angew. Chem. Int. Ed. Engl. 2017, 56, 7515–7519. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Uversky, V.N. Intrinsic disorder and posttranslational modifications: The darker side of the biological dark matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Monahan, Z.; Ryan, V.H.; Janke, A.M.; Burke, K.A.; Rhoads, S.N.; Zerze, G.H.; O’Meally, R.; Dignon, G.L.; Conicella, A.E.; Zheng, W.; et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017, 36, 2951–2967. [Google Scholar] [CrossRef] [PubMed]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; Xiao, S.; Wollmuth, L.P.; Bowen, M.E. Effect of Src kinase phosphorylation on disordered C-terminal domain of N-methyl-d-aspartic acid (NMDA) receptor subunit GluN2B protein. J. Biol. Chem. 2011, 286, 29904–29912. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.B.; Kazi, R.; Stenzoski, N.; Wollmuth, L.P.; Uversky, V.N.; Bowen, M.E. Modulating the intrinsic disorder in the cytoplasmic domain alters the biological activity of the N-methyl-d-aspartate-sensitive glutamate receptor. J. Biol. Chem. 2013, 288, 22506–22515. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; He, Y.; Yang, F.; Mooney, S.M.; Getzenberg, R.H.; Orban, J.; Kulkarni, P. The cancer/testis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J. Biol. Chem. 2011, 286, 13985–13994. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.; Qiu, R.; Mooney, S.M.; Rao, S.; Shiraishi, T.; Sacho, E.; Huang, H.; Shapiro, E.; Weninger, K.R.; Kulkarni, P. The stress-response protein prostate-associated gene 4, interacts with c-Jun and potentiates its transactivation. Biochim. Biophys. Acta 2014, 1842, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Dunker, A.K.; Weninger, K.; Orban, J. Prostate-associated gene 4 (PAGE4), an intrinsically disordered cancer/testis antigen, is a novel therapeutic target for prostate cancer. Asian J. Androl. 2016, 18, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Jolly, M.K.; Dorff, T.; Lau, C.; Weninger, K.; Orban, J.; Kulkarni, P. Prostate-associated gene 4 (PAGE4): Leveraging the conformational dynamics of a dancing protein cloud as a therapeutic target. J. Clin. Med. 2018, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Kulkarni, P.; Weninger, K.; Orban, J.; Levine, H. Phenotypic plasticity, bet-hedging, and androgen independence in prostate cancer: Role of non-genetic heterogeneity. Front. Oncol. 2018, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Mooney, S.M.; Qiu, R.; Kim, J.J.; Sacho, E.J.; Rajagopalan, K.; Johng, D.; Shiraishi, T.; Kulkarni, P.; Weninger, K.R. Cancer/testis antigen PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1, a component of the stress-response pathway. Biochemistry 2014, 53, 1670–1679. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, Y.; Mooney, S.M.; Rajagopalan, K.; Bhargava, A.; Sacho, E.; Weninger, K.; Bryan, P.N.; Kulkarni, P.; Orban, J. Phosphorylation-induced conformational ensemble switching in an intrinsically disordered cancer/testis antigen. J. Biol. Chem. 2015, 290, 25090–25102. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Roy, S.; Jolly, M.K.; Bocci, F.; Schafer, N.P.; Tsai, M.-Y.; Chen, Y.; He, Y.; Grishaev, A.; Weninger, K.; et al. PAGE4 and conformational switching: Insights from molecular dynamics simulations and implications for prostate cancer. J. Mol. Biol. 2018, 430, 2422–2438. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [PubMed]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.-H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Intrinsically disordered proteins in crowded milieu: When chaos prevails within the cellular gumbo. Cell. Mol. Life Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.-T.; Elbaum-Garfinkle, S.; Holehouse, A.S.; Chen, C.C.-H.; Feric, M.; Arnold, C.B.; Priestley, R.D.; Pappu, R.V.; Brangwynne, C.P. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017, 9, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Gapsys, V.; Gajda, M.J.; Zweckstetter, M.; de Groot, B.L.; Grubmüller, H. Structural ensembles of intrinsically disordered proteins depend strongly on force field: A comparison to experiment. J. Chem. Theory Comput. 2015, 11, 5513–5524. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Trbovic, N.; Maragakis, P.; Piana, S.; Shaw, D.E. Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J. Am. Chem. Soc. 2012, 134, 3787–3791. [Google Scholar] [CrossRef] [PubMed]
- Robustelli, P.; Piana, S.; Shaw, D.E. Developing a molecular dynamics force field for both folded and disordered protein states. Proc. Natl. Acad. Sci. USA 2018, 115, E4758–E4766. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B. Computational and theoretical advances in studies of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2017, 42, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Ferrie, J.J.; Haney, C.M.; Yoon, J.; Pan, B.; Lin, Y.-C.; Fakhraai, Z.; Rhoades, E.; Nath, A.; Petersson, E.J. Using a FRET library with multiple probe pairs to drive monte carlo simulations of α-synuclein. Biophys. J. 2018, 114, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G. Ensemble models of proteins and protein domains based on distance distribution restraints. Proteins Struct. Funct. Bioinform. 2016, 84, 544–560. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Sammalkorpi, M.; DeWitt, D.C.; Trexler, A.J.; Elbaum-Garfinkle, S.; O’Hern, C.S.; Rhoades, E. The conformational ensembles of α-synuclein and tau: Combining single-molecule FRET and simulations. Biophys. J. 2012, 103, 1940–1949. [Google Scholar] [CrossRef] [PubMed]
- Jao, C.C.; Hegde, B.G.; Chen, J.; Haworth, I.S.; Langen, R. Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. USA 2008, 105, 19666–19671. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Tsafou, K.; Tiwari, P.B.; Forman-Kay, J.D.; Metallo, S.J.; Toretsky, J.A. Targeting Intrinsically Disordered Transcription Factors: Changing the Paradigm. J. Mol. Biol. 2018, 430, 2321–2341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Boskovic, Z.; Hussain, M.M.; Hu, W.; Inouye, C.; Kim, H.-J.; Abole, A.K.; Doud, M.K.; Lewis, T.A.; Koehler, A.N.; et al. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. eLife 2015, 4, e07777. [Google Scholar] [CrossRef] [PubMed]
- Sakon, J.J.; Weninger, K.R. Detecting the conformation of individual proteins in live cells. Nat. Methods 2010, 7, 203–205. [Google Scholar] [CrossRef] [PubMed]
- König, I.; Zarrine-Afsar, A.; Aznauryan, M.; Soranno, A.; Wunderlich, B.; Dingfelder, F.; Stüber, J.C.; Plückthun, A.; Nettels, D.; Schuler, B. Single-molecule spectroscopy of protein conformational dynamics in live eukaryotic cells. Nat. Methods 2015, 12, 773–779. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
LeBlanc, S.J.; Kulkarni, P.; Weninger, K.R. Single Molecule FRET: A Powerful Tool to Study Intrinsically Disordered Proteins. Biomolecules 2018, 8, 140. https://doi.org/10.3390/biom8040140
LeBlanc SJ, Kulkarni P, Weninger KR. Single Molecule FRET: A Powerful Tool to Study Intrinsically Disordered Proteins. Biomolecules. 2018; 8(4):140. https://doi.org/10.3390/biom8040140
Chicago/Turabian StyleLeBlanc, Sharonda J., Prakash Kulkarni, and Keith R. Weninger. 2018. "Single Molecule FRET: A Powerful Tool to Study Intrinsically Disordered Proteins" Biomolecules 8, no. 4: 140. https://doi.org/10.3390/biom8040140
APA StyleLeBlanc, S. J., Kulkarni, P., & Weninger, K. R. (2018). Single Molecule FRET: A Powerful Tool to Study Intrinsically Disordered Proteins. Biomolecules, 8(4), 140. https://doi.org/10.3390/biom8040140





