2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation?
Abstract
1. Introduction
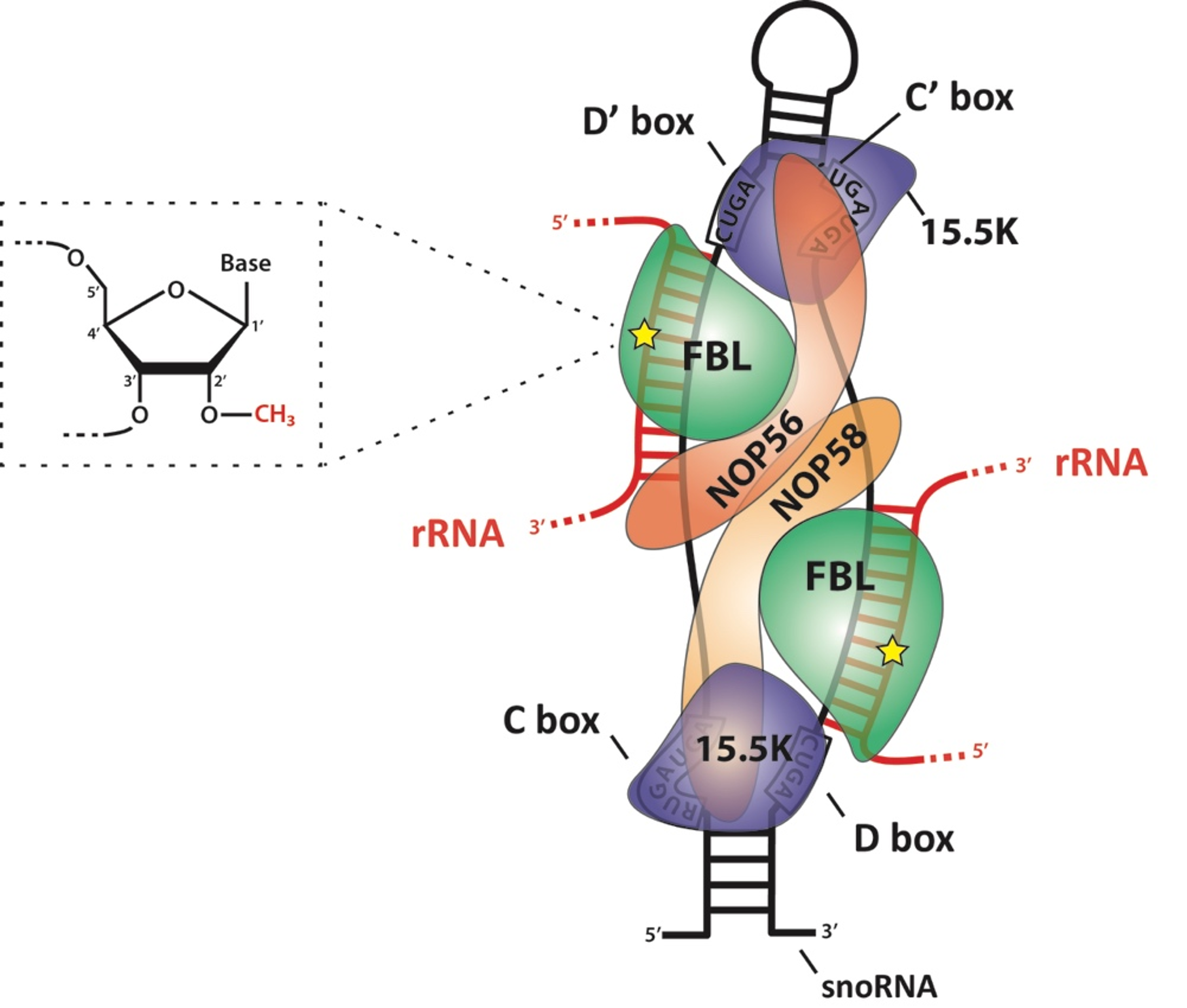
2. Adding 2′-O-Methylation to Eukaryotic rRNA: How, Where, and When?
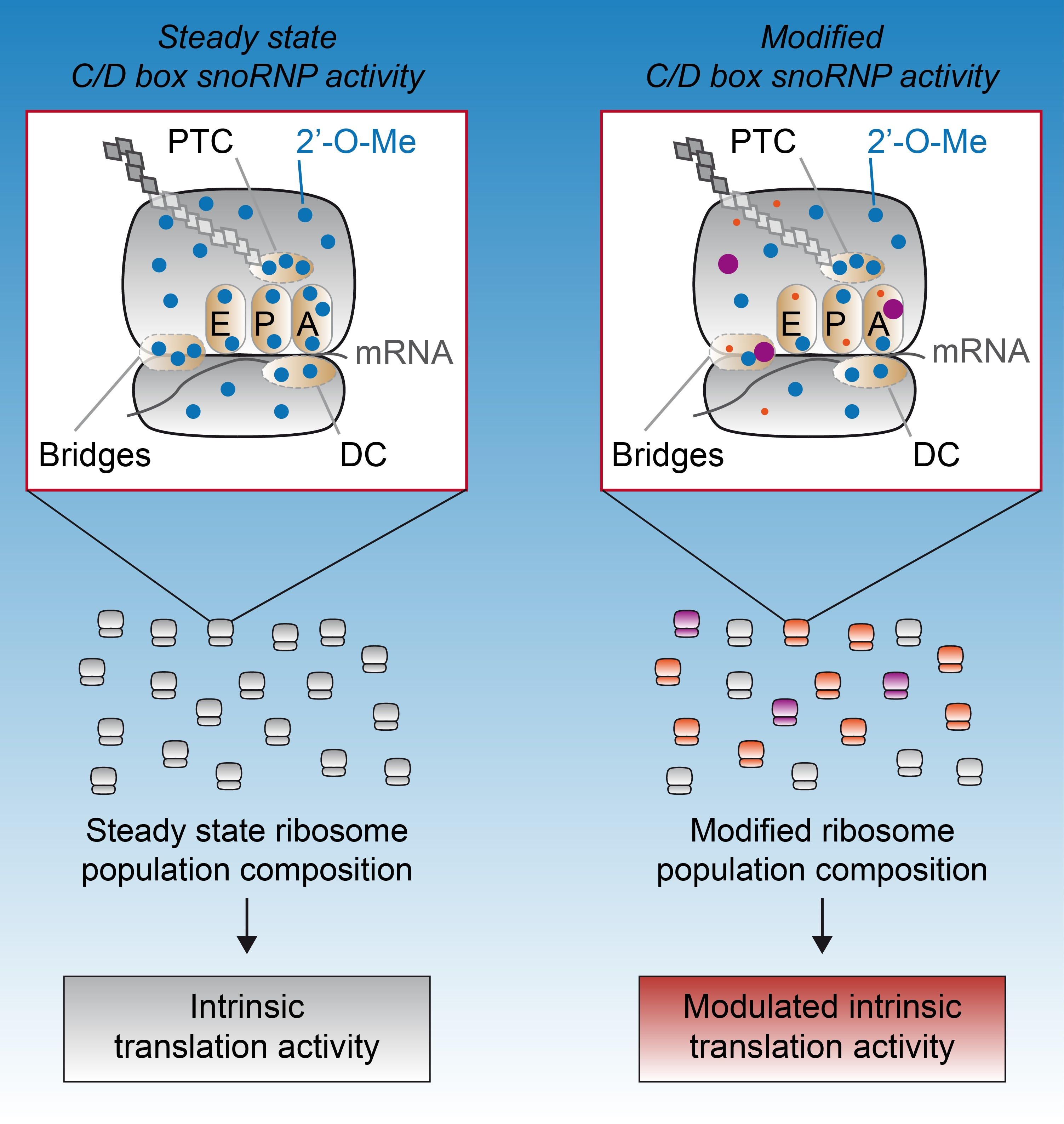
3. 2′-O-Methylation as a Source of Heterogeneity: All Ribosomes Are Not Created Equal
4. 2′-O-Methylation Contribution to Ribosome Structure and Function
5. Perspective: Towards an rRNA-Based Epitranscriptomic Control of mRNA Translation
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boccaletto, P.; MacHnicka, M.A.; Purta, E.; Pitkowski, P.; Baginski, B.; Wirecki, T.K.; De Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Decatur, W.A.; Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002, 27, 344–351. [Google Scholar] [CrossRef]
- Sharma, S.; Lafontaine, D.L. “View From A Bridge”: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Doudna Cate, J.H. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L. Turning uridines around: Role of rRNA pseudouridylation in ribosome biogenesis and ribosomal function. Biomolecules 2018, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Yang, J.; van Nues, R.; Watzinger, P.; Kötter, P.; Lafontaine, D.L.J.; Granneman, S.; Entian, K.D. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 2017, 13, e1006804. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Reichow, S.L.; Hamma, T.; Ferre-D’Amare, A.R.; Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007, 35, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Therizols, G.; Laforêts1, F.; Marcel, V.; Catez, F.; Bouvet, P.; Diaz, J.J.; Tollefsbol, T. Ribosomal RNA methylation and cancer. In Epigenetic Cancer Therapy; Gray, S., Ed.; Academic Press: London, UK, 2016. [Google Scholar]
- Bachellerie, J.P.; Cavaillé, J. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 1997, 22, 257–261. [Google Scholar] [CrossRef]
- Kiss-Laszlo, Z.; Henry, Y.; Bachellerie, J.P.; Caizergues-Ferrer, M.; Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef]
- Yang, Z.; Lin, J.; Ye, K. Box C/D guide RNAs recognize a maximum of 10 nt of substrates. Proc. Natl. Acad. Sci. USA 2016, 113, 10878–10883. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lai, S.; Jia, R.; Xu, A.; Zhang, L.; Lu, J.; Ye, K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 2011, 469, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Lapinaite, A.; Simon, B.; Skjaerven, L.; Rakwalska-Bange, M.; Gabel, F.; Carlomagno, T. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature 2013, 502, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Graziadei, A.; Masiewicz, P.; Lapinaite, A.; Carlomagno, T. Archaea box C/D enzymes methylate two distinct substrate rRNA sequences with different efficiency. RNA 2016, 22, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Quinternet, M.; Chagot, M.E.; Rothé, B.; Tiotiu, D.; Charpentier, B.; Manival, X. Structural Features of the Box C/D snoRNP Pre-assembly Process Are Conserved through Species. Structure 2016, 24, 1693–1706. [Google Scholar] [CrossRef] [PubMed]
- Bizarro, J.; Charron, C.; Boulon, S.; Westman, B.; Pradet-Balade, B.; Vandermoere, F.; Chagot, M.E.; Hallais, M.; Ahmad, Y.; Leonhardt, H.; et al. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 2014, 207, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Pogacic, V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002, 14, 319–327. [Google Scholar] [CrossRef]
- Salim, M.; Maden, B.E. Early and late methylations in HeLa cell ribosome maturation. Nature 1973, 244, 334–336. [Google Scholar] [CrossRef] [PubMed]
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Birkedal, U.; Christensen-Dalsgaard, M.; Krogh, N.; Sabarinathan, R.; Gorodkin, J.; Nielsen, H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. 2015, 54, 451–455. [Google Scholar] [CrossRef]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E.H. Mapping 2′-O-Methyl Groups in Ribosomal RNA. Methods 2001, 25, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Motorin, Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 2017, 14, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Kellner, S.; Burhenne, J.; Helm, M. Detection of RNA modifications. RNA Biol. 2010, 7, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Moshitch-Moshkovitz, S.; Han, D.; Kol, N.; Amariglio, N.; Rechavi, G.; Dominissini, D.; He, C. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 2017, 14, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Krogh, N.; Jansson, M.D.; Häfner, S.J.; Tehler, D.; Birkedal, U.; Christensen-Dalsgaard, M.; Lund, A.H.; Nielsen, H. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016, 44, 7884–7895. [Google Scholar] [CrossRef] [PubMed]
- Marchand, V.; Blanloeil-Oillo, F.; Helm, M.; Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Marchand, V.; Motorin, Y.; Lafontaine, D.L.J. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Pirnie, S.P.; Carmichael, G.G. High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA 2017, 23, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
- Incarnato, D.; Anselmi, F.; Morandi, E.; Neri, F.; Maldotti, M.; Rapelli, S.; Parlato, C.; Basile, G.; Oliviero, S. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gumienny, R.; Jedlinski, D.J.; Schmidt, A.; Gypas, F.; Martin, G.; Vina-Vilaseca, A.; Zavolan, M. High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq. Nucleic Acids Res. 2017, 45, 2341–2353. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E.H.; Salim, M. The methylated nucleotide sequences in HeLa cell ribosomal RNA and its precursors. J. Mol. Biol. 1974, 88. [Google Scholar] [CrossRef]
- Nazar, R.N.; Sitz, T.O.; Busch, H. Tissue specific differences in the 2′-O-methylation of eukaryotic 5.8S ribosomal RNA. FEBS Lett. 1975, 59, 83–87. [Google Scholar] [CrossRef]
- Nazar, R.N.; Lo, A.C.; Wildeman, A.G.; Sitz, T.O. Effect of 2′-O-methylation on the structure of mammalian 5.8S rRNAs and the 5.8S-28S rRNA junction. Nucleic Acids Res. 1983, 11, 5989–6001. [Google Scholar] [CrossRef] [PubMed]
- Buchhaupt, M.; Sharma, S.; Kellner, S.; Oswald, S.; Paetzold, M.; Peifer, C.; Watzinger, P.; Schrader, J.; Helm, M.; Entian, K.D. Partial methylation at Am100 in 18S rRNA of Baker’s yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Taoka, M.; Nobe, Y.; Yamaki, Y.; Yamauchi, Y.; Ishikawa, H.; Takahashi, N.; Nakayama, H.; Isobe, T. The complete chemical structure of Saccharomyces cerevisiae rRNA: Partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res. 2016, 44, 8951–8961. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Sharma, S.; Watzinger, P.; Hartmann, J.D.; Kötter, P.; Entian, K.D. Mapping of complete set of ribose and base modifications of yeast rRNA by RP-HPLC and mung bean nuclease assay. PLoS ONE 2016, 11, e0168873. [Google Scholar] [CrossRef] [PubMed]
- Belin, S.; Beghin, A.; Solano-Gonzalez, E.; Bezin, L.; Brunet-Manquat, S.; Textoris, J.; Prats, A.C.; Mertani, H.C.; Dumontet, C.; Diaz, J.J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 2009, 4, e7147. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.P.; Solano-Gonzàlez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. P53 Acts as a Safeguard of Translational Control by Regulating Fibrillarin and rRNA Methylation in Cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Kohn, M.; Misiak, D.; Baumer, N.; Cui, C.; Gollner, S.; et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Valleron, W.; Laprevotte, E.; Gautier, E.F.; Quelen, C.; Demur, C.; Delabesse, E.; Agirre, X.; Prosper, F.; Kiss, T.; Brousset, P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia 2012, 26, 2052–2060. [Google Scholar] [CrossRef] [PubMed]
- Valleron, W.; Ysebaert, L.; Berquet, L.; Fataccioli, V.; Quelen, C.; Martin, A.; Parrens, M.; Lamant, L.; de Leval, L.; Gisselbrecht, C.; et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood 2012, 120, 3997–4005. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, K.; Shen, J.; Liao, J.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol. Cancer 2014, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Corona, U.; Sobol, M.; Rodriguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin from Archaea to human. Biol. Cell 2015, 107, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Van Nues, R.W.; Granneman, S.; Kudla, G.; Sloan, K.E.; Chicken, M.; Tollervey, D.; Watkins, N.J. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J. 2011, 30, 2420–2430. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Das, P.; Chaudhuri, S.; Bevilacqua, E.; Andrews, J.; Barik, S.; Hatzoglou, M.; Komar, A.A.; Mazumder, B. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol. Cell. Biol. 2011, 31, 4482–4499. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Chowhan, R.K.; Singh, L.R. Macromolecular crowding: Macromolecules friend or foe. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1822–1831. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Nessling, M.; Lichter, P. Macromolecular crowding and its potential impact on nuclear function. Biochim. Biophys. Acta 2008, 1783, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V. Functional specialization of ribosomes? Trends Biochem. Sci. 2011, 36, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, P.; Yathindra, N.; Sundaralingam, M. Effect of ribose O(2′)-methylation on the conformation of nucleosides and nucleotides. BBA Sect. Nucleic Acids Protein Synth. 1974, 366, 115–123. [Google Scholar] [CrossRef]
- Adamiak, D.A.; Milecki, J.; Popenda, M.; Adamiak, R.W.; Dauter, Z.; Rypniewski, W.R. Crystal structure of 2′-O-Me(CGCGCG)2, an RNA duplex at 1.30 A resolution. Hydration pattern of 2′-O-methylated RNA. Nucleic Acids Res. 1997, 25, 4599–4607. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Biala, E.; Milecki, J.; Adamiak, R.W. Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2′-O-Me(CGCGCG)2 under low salt conditions. Nucleic Acids Res. 1997, 25, 4589–4598. [Google Scholar] [CrossRef] [PubMed]
- Polikanov, Y.S.; Melnikov, S.V.; Soll, D.; Steitz, T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015, 22, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Weixlbaumer, A.; Loakes, D.; Kelley, A.C.; Ramakrishnan, V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009, 16, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Natchiar, S.K.; Myasnikov, A.G.; Kratzat, H.; Hazemann, I.; Klaholz, B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017, 551, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Liu, Q.; Fournier, M.J. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 2007, 28, 965–977. [Google Scholar] [CrossRef] [PubMed]
- King, T.H.; Liu, B.; McCully, R.R.; Fournier, M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 2003, 11, 425–435. [Google Scholar] [CrossRef]
- Liang, X.H.; Vickers, T.A.; Guo, S.; Crooke, S.T. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2011, 39, e13. [Google Scholar] [CrossRef] [PubMed]
- Baudin-Baillieu, A.; Fabret, C.; Liang, X.H.; Piekna-Przybylska, D.; Fournier, M.J.; Rousset, J.P. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009, 37, 7665–7677. [Google Scholar] [CrossRef] [PubMed]
- Esguerra, J.; Warringer, J.; Blomberg, A. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA 2008, 14, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Baxter-Roshek, J.L.; Petrov, A.N.; Dinman, J.D. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Higa-Nakamine, S.; Suzuki, T.; Uechi, T.; Chakraborty, A.; Nakajima, Y.; Nakamura, M.; Hirano, N.; Suzuki, T.; Kenmochi, N. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012, 40, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.; Thompson, I.; Yuan, Z.M. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014, 33, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, L.; Pacilli, A.; Sethi, R.; Penzo, M.; Schneider, R.J.; Treré, D.; Brigotti, M.; Montanaro, L. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res. 2013, 41, 8308–8318. [Google Scholar] [CrossRef] [PubMed]
- Panthu, B.; Decimo, D.; Balvay, L.; Ohlmann, T. In vitro translation in a hybrid cell free lysate with exogenous cellular ribosomes. Biochem. J. 2015, 467, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Rocchi, L.; Brugiere, S.; Carnicelli, D.; Onofrillo, C.; Coute, Y.; Brigotti, M.; Montanaro, L. Human ribosomes from cells with reduced dyskerin levels are intrinsically altered in translation. FASEB J. 2015, 29, 3472–3482. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Carnicelli, D.; Montanaro, L.; Brigotti, M. A reconstituted cell-free assay for the evaluation of the intrinsic activity of purified human ribosomes. Nat. Protoc. 2016, 11, 1309–1325. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Recher, G.; Jouralet, J.; Brombin, A.; Heuze, A.; Mugniery, E.; Hermel, J.M.; Desnoulez, S.; Savy, T.; Herbomel, P.; Bourrat, F.; et al. Zebrafish midbrain slow-amplifying progenitors exhibit high levels of transcripts for nucleotide and ribosome biogenesis. Development 2013, 140, 4860–4869. [Google Scholar] [CrossRef] [PubMed]
- Bouffard, S.; Dambroise, E.; Brombin, A.; Lempereur, S.; Hatin, I.; Simion, M.; Corre, R.; Bourrat, F.; Joly, J.-S.; Jamen, F. Fibrillarin is essential for S-phase progression and neuronal differentiation in zebrafish dorsal midbrain and retina. Dev. Biol. 2018, 437, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Susaki, K.; Takada, H.; Enomoto, K.; Miwata, K.; Ishimine, H.; Intoh, A.; Ohtaka, M.; Nakanishi, M.; Sugino, H.; Asashima, M.; et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 2014, 32, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Catez, F.; Diaz, J.-J. Ribosome heterogeneity in tumorigenesis: The rRNA point of view. Mol. Cell. Oncol. 2015, 2, e983755. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Welden, J.R.; Duncan, M.J.; Stamm, S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. BioEssays 2017, 39, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sornjai, W.; Lithanatudom, P.; Erales, J.; Joly, P.; Francina, A.; Hacot, S.; Fucharoen, S.; Svasti, S.; Diaz, J.J.; Mertani, H.C.; et al. Hypermethylation of 28S ribosomal RNA in beta-thalassemia trait carriers. Int. J. Biol. Macromol. 2017, 94, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 2017, 67, 71–83. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monaco, P.L.; Marcel, V.; Diaz, J.-J.; Catez, F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. https://doi.org/10.3390/biom8040106
Monaco PL, Marcel V, Diaz J-J, Catez F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules. 2018; 8(4):106. https://doi.org/10.3390/biom8040106
Chicago/Turabian StyleMonaco, Piero Lo, Virginie Marcel, Jean-Jacques Diaz, and Frédéric Catez. 2018. "2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation?" Biomolecules 8, no. 4: 106. https://doi.org/10.3390/biom8040106
APA StyleMonaco, P. L., Marcel, V., Diaz, J.-J., & Catez, F. (2018). 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules, 8(4), 106. https://doi.org/10.3390/biom8040106







