Protein Phosphatase Sit4 Affects Lipid Droplet Synthesis and Soraphen A Resistance Independent of Its Role in Regulating Elongator Dependent tRNA Modification
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Reagents
2.3. Growth Conditions and Lipid Droplets Quantification
2.4. Drug Assays
3. Results
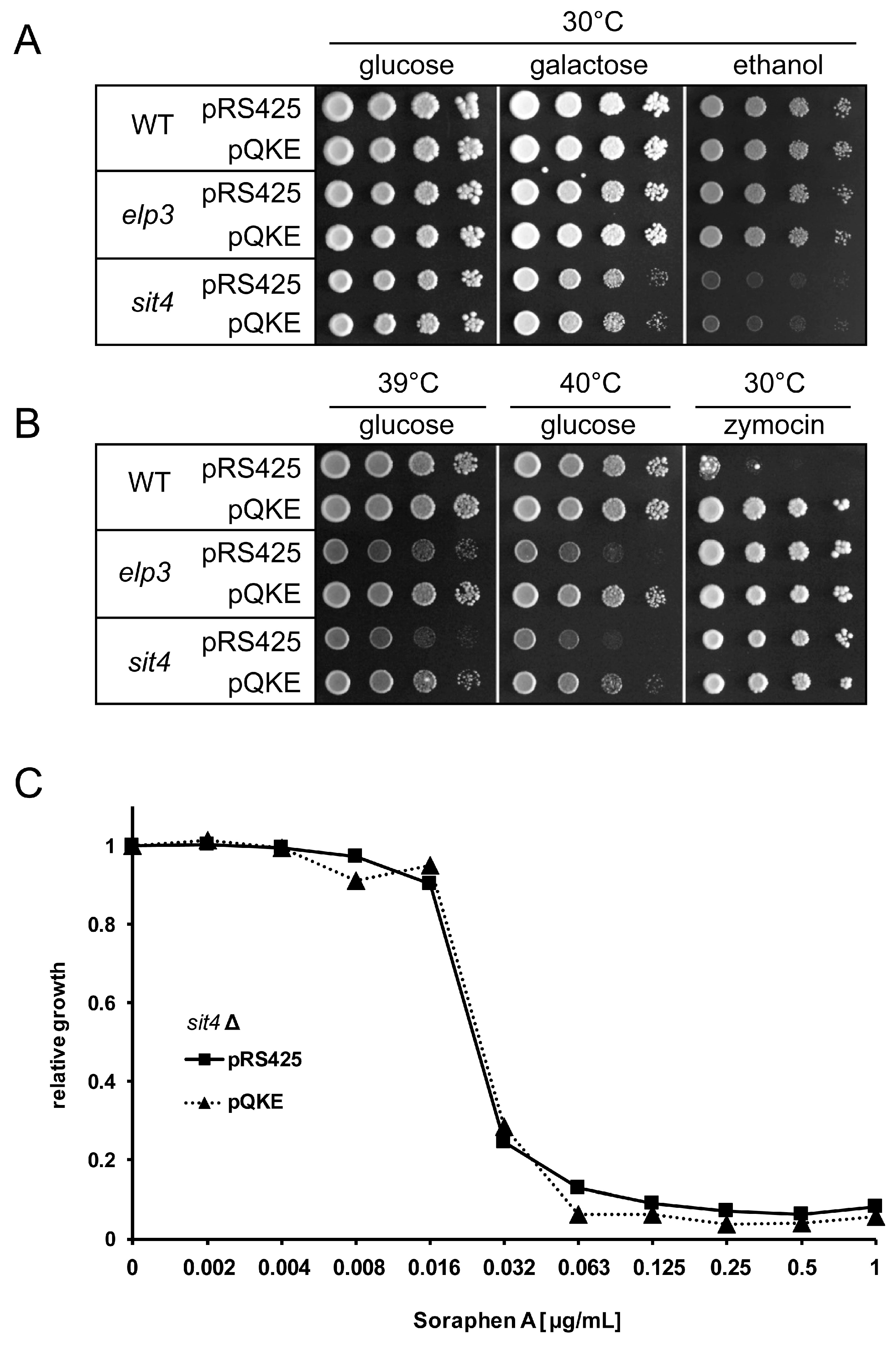
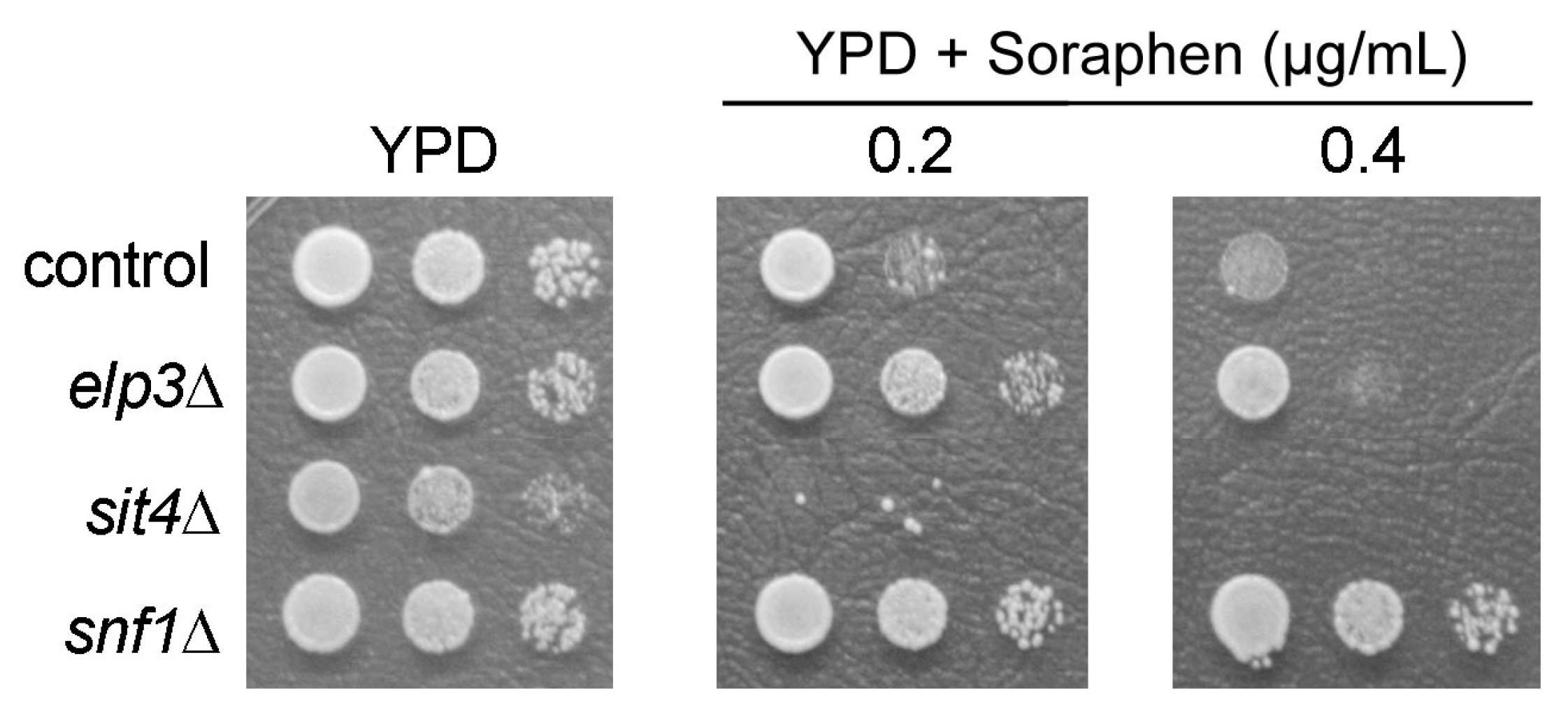
3.1. Analysis of Shared and Non-Shared Phenotypes of sit4 and elp3 Mutants
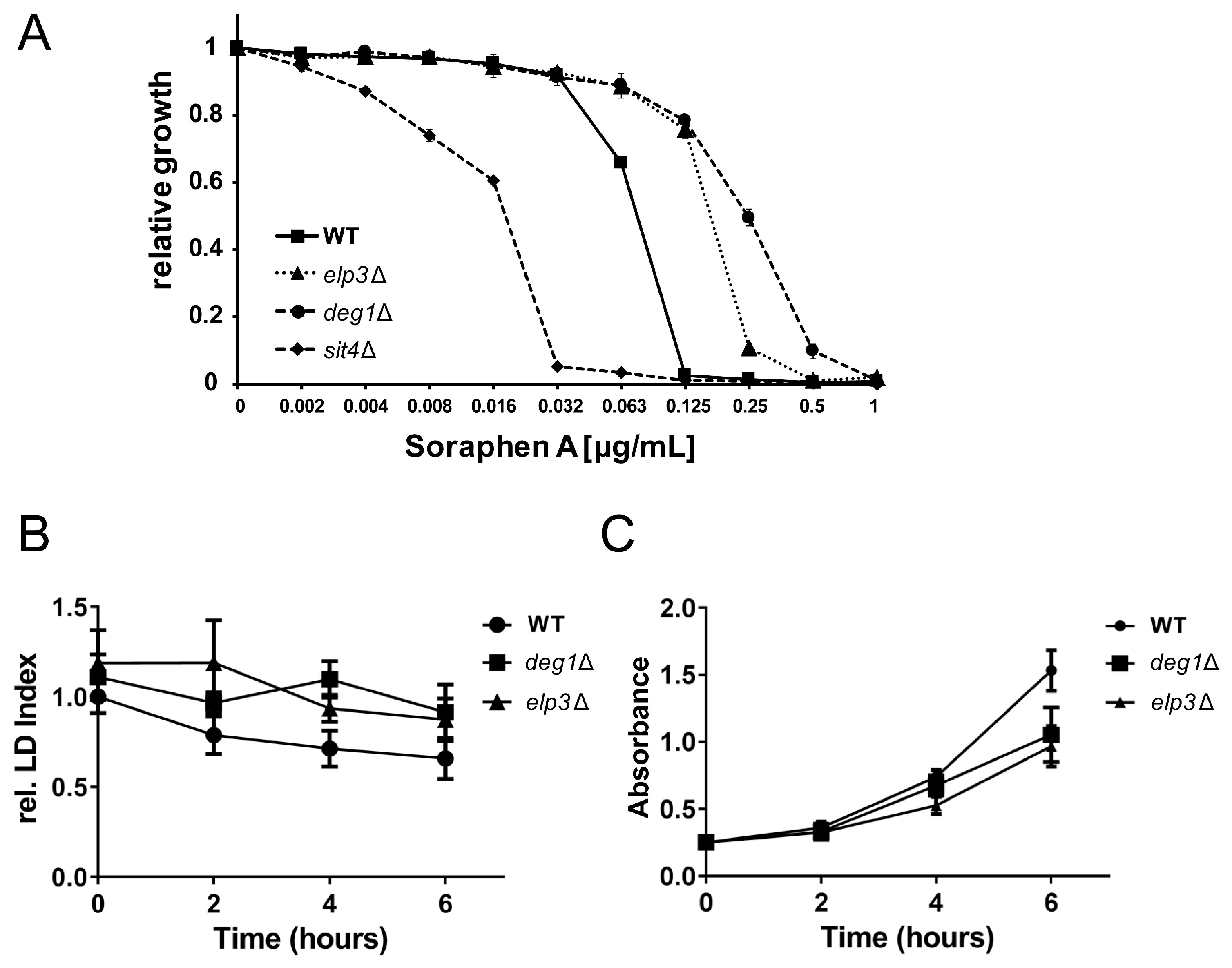
3.2. Role of 5-Methoxycarbonylmethyluridine Modification in Soraphen A Resistance and Lipid Droplet Dynamics
3.3. Screening Other tRNA Modification Mutants for Soraphen A Resistance
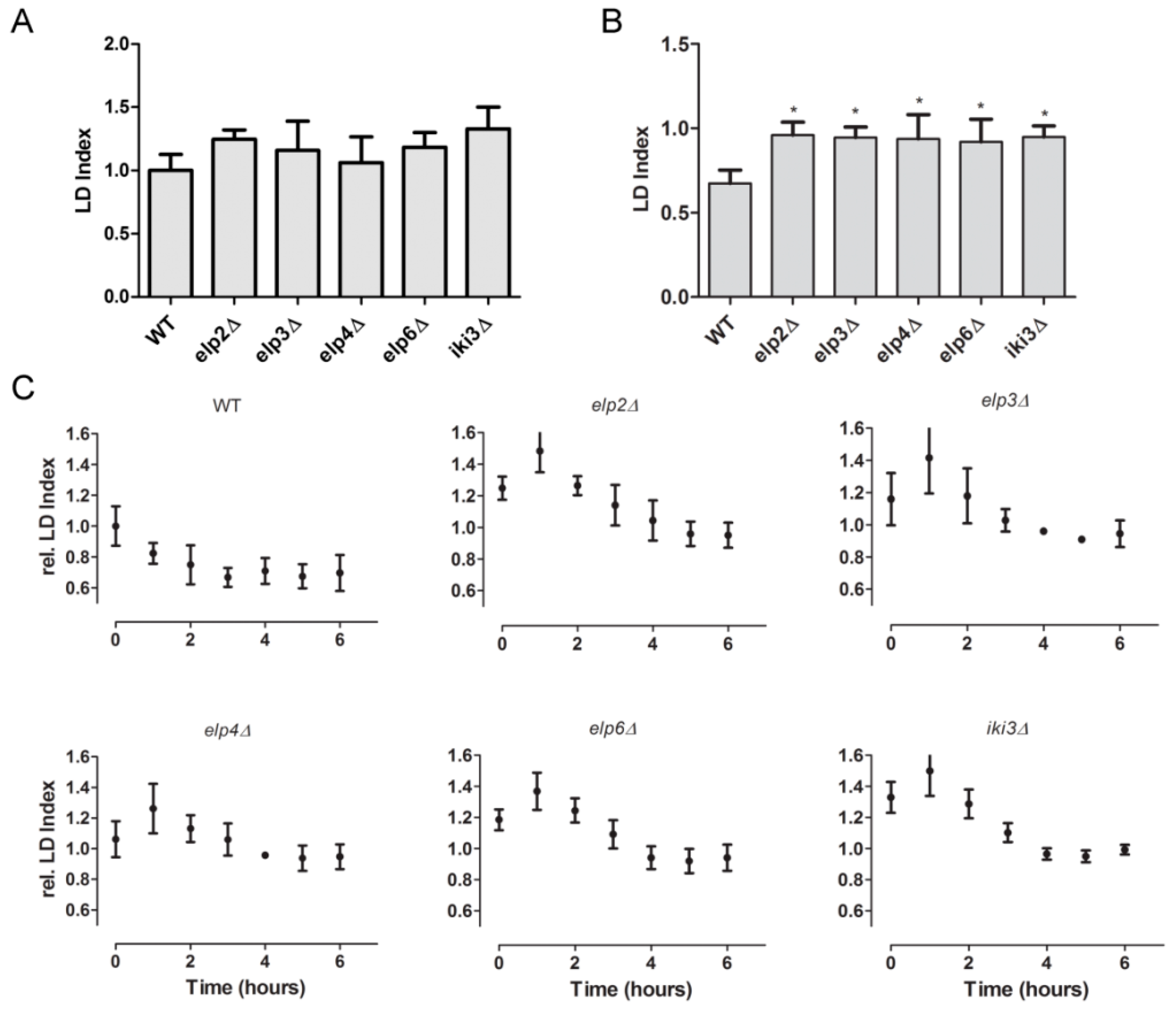
3.4. Changes in Lipid Droplet Dynamics Upon Loss of Ψ38/39
3.5. Rescue of Lipolysis Defect of deg1 Mutants by tRNA Overexpression
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hasslacher, M.; Ivessa, A.S.; Paltauf, F.; Kohlwein, S.D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 1993, 268, 10946–10952. [Google Scholar] [PubMed]
- Bozaquel-Morais, B.L.; Madeira, J.B.; Maya-Monteiro, C.M.; Masuda, C.A.; Montero-Lomeli, M. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS ONE 2010, 5, e13692. [Google Scholar] [CrossRef] [PubMed]
- Woods, A.; Munday, M.R.; Scott, J.; Yang, X.; Carlson, M.; Carling, D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 1994, 269, 19509–19515. [Google Scholar] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed]
- Madeira, J.B.; Masuda, C.A.; Maya-Monteiro, C.M.; Matos, G.S.; Montero-Lomelí, M.; Bozaquel-Morais, B.L. TORC1 inhibition induces lipid droplet replenishment in yeast. Mol. Cell. Biol. 2015, 35, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Fichtner, L.; Stark, M.J.R.; Schaffrath, R. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell 2004, 15, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Täubert, J.-E.; Bär, C.; Stark, M.J.R.; Schaffrath, R. Distinct subsets of Sit4 holophosphatases are required for inhibition of Saccharomyces cerevisiae growth by rapamycin and zymocin. Eukaryot. Cell 2009, 8, 1637–1647. [Google Scholar] [CrossRef] [PubMed]
- Mehlgarten, C.; Jablonowski, D.; Breunig, K.D.; Stark, M.J.R.; Schaffrath, R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 2009, 73, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Schaffrath, R.; Leidel, S.A. Wobble uridine modifications–a reason to live, a reason to die?! RNA Biol. 2017, 14, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Dauden, M.I.; Kosinski, J.; Kolaj-Robin, O.; Desfosses, A.; Ori, A.; Faux, C.; Hoffmann, N.A.; Onuma, O.F.; Breunig, K.D.; Beck, M.; et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017, 18, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Setiaputra, D.T.; Cheng, D.T.; Lu, S.; Hansen, J.M.; Dalwadi, U.; Lam, C.H.; To, J.L.; Dong, M.-Q.; Yip, C.K. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017, 18, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Esberg, A.; Huang, B.; Johansson, M.J.O.; Byström, A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 2006, 24, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Johansson, M.J.O.; Byström, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Frohloff, F.; Fichtner, L.; Jablonowski, D.; Breunig, K.D.; Schaffrath, R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001, 20, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Pedrioli, P.G.A.; Bucher, T.; Brost, R.; Costanzo, M.; Schmidt, A.; Aebersold, R.; Boone, C.; Hofmann, K.; Peter, M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009, 458, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Scheidt, V.; Jüdes, A.; Bär, C.; Klassen, R.; Schaffrath, R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb. Cell 2014, 1, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Grunewald, P.; Thüring, K.L.; Eichler, C.; Helm, M.; Schaffrath, R. Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0119261. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Ciftci, A.; Funk, J.; Bruch, A.; Butter, F.; Schaffrath, R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016, 44, 10946–10959. [Google Scholar] [CrossRef] [PubMed]
- Zinshteyn, B.; Gilbert, W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013, 9, e1003675. [Google Scholar] [CrossRef] [PubMed]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, N.; Rodnina, M.V. Thio-Modification of tRNA at the Wobble Position as Regulator of the Kinetics of Decoding and Translocation on the Ribosome. J. Am. Chem. Soc. 2017, 139, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Lu, J.; Byström, A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008, 14, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Lecointe, F.; Simos, G.; Sauer, A.; Hurt, E.C.; Motorin, Y.; Grosjean, H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of Ψ38 and Ψ39 in tRNA anticodon loop. J. Biol. Chem. 1998, 273, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Schaffrath, R. Role of Pseudouridine Formation by Deg1 for Functionality of Two Glutamine Isoacceptor tRNAs. Biomolecules 2017, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Huang, B.; Esberg, A.; Johansson, M.J.O.; Byström, A.S. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 2005, 11, 1648–1654. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Krampe, S.; Meinhardt, F. Homologous recombination and the yKu70/80 complex exert opposite roles in resistance against the killer toxin from Pichia acaciae. DNA Repair 2007, 6, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Wemhoff, S.; Krause, J.; Meinhardt, F. DNA repair defects sensitize cells to anticodon nuclease yeast killer toxins. Mol. Genet. Genom. MGG 2011, 285, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Romero-Aguilar, L.; Montero-Lomeli, M.; Pardo, J.P.; Guerra-Sánchez, G. Lipid Index Determination by Liquid Fluorescence Recovery in the Fungal Pathogen Ustilago Maydis. J. Vis. Exp. JoVE 2018, 134. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.R.; White, J.H.; Folawiyo, Y.; Edlin, A.; Gardiner, D.; Stark, M.J. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 1994, 14, 6306–6316. [Google Scholar] [CrossRef] [PubMed]
- Jablonowski, D.; Zink, S.; Mehlgarten, C.; Daum, G.; Schaffrath, R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator’s key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006, 59, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Kurat, C.F.; Natter, K.; Petschnigg, J.; Wolinski, H.; Scheuringer, K.; Scholz, H.; Zimmermann, R.; Leber, R.; Zechner, R.; Kohlwein, S.D. Obese yeast: Triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 2006, 281, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Bozaquel-Morais, B.L.; Madeira, J.B.; Venâncio, T.M.; Pacheco-Rosa, T.; Masuda, C.A.; Montero-Lomeli, M. A Chemogenomic Screen Reveals Novel Snf1p/AMPK Independent Regulators of Acetyl-CoA Carboxylase. PLoS ONE 2017, 12, e0169682. [Google Scholar] [CrossRef] [PubMed]
- Jüdes, A.; Bruch, A.; Klassen, R.; Helm, M.; Schaffrath, R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microb. Cell 2016, 3, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Kon, Y.; Phizicky, E.M. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 2015, 21, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Rajakumari, S.; Daum, G. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 15769–15776. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Legendre, R.; Rojas-Benítez, D.; Baudin-Baillieu, A.; Hatin, I.; Chalancon, G.; Glavic, A.; Namy, O.; de Crécy-Lagard, V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 2016, 3, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-J.; Donnard, E.; Gustafsson, H.T.; Garber, M.; Rando, O.J. Transcriptome-wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol. Cell 2017, 68, 978–992.e4. [Google Scholar] [CrossRef] [PubMed]
- Sokołowski, M.; Klassen, R.; Bruch, A.; Schaffrath, R.; Glatt, S. Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta 2018, 4, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Karlsborn, T.; Mahmud, A.K.M.F.; Tükenmez, H.; Byström, A.S. Loss of ncm5 and mcm5 wobble uridine side chains results in an altered metabolic profile. Metabol. Off. J. Metabol. Soc. 2016, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Mülleder, M.; Calvani, E.; Alam, M.T.; Wang, R.K.; Eckerstorfer, F.; Zelezniak, A.; Ralser, M. Functional Metabolomics Describes the Yeast Biosynthetic Regulome. Cell 2016, 167, 553–565.e12. [Google Scholar] [CrossRef] [PubMed]
- Uršič, K.; Ogrizović, M.; Kordiš, D.; Natter, K.; Petrovič, U. Tum1 is involved in the metabolism of sterol esters in Saccharomyces cerevisiae. BMC Microbiol. 2017, 17, 181. [Google Scholar] [CrossRef] [PubMed]

| Gene | Nomenclature | Description | Sensitivity Score | Response |
|---|---|---|---|---|
| DEG1 | YFL001W | depressed growth rate | +6 | RESISTANT |
| TUM1 | YOR251C | thiouridine modification | +2 | RESISTANT |
| SIT4 | YDL047W | suppressor initiation of transcription | −5 | SENSITIVE |
| SAP185 | YKR028W | Sit4 associated protein | −4 | SENSITIVE |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bozaquel-Morais, B.L.; Vogt, L.; D’Angelo, V.; Schaffrath, R.; Klassen, R.; Montero-Lomelí, M. Protein Phosphatase Sit4 Affects Lipid Droplet Synthesis and Soraphen A Resistance Independent of Its Role in Regulating Elongator Dependent tRNA Modification. Biomolecules 2018, 8, 49. https://doi.org/10.3390/biom8030049
Bozaquel-Morais BL, Vogt L, D’Angelo V, Schaffrath R, Klassen R, Montero-Lomelí M. Protein Phosphatase Sit4 Affects Lipid Droplet Synthesis and Soraphen A Resistance Independent of Its Role in Regulating Elongator Dependent tRNA Modification. Biomolecules. 2018; 8(3):49. https://doi.org/10.3390/biom8030049
Chicago/Turabian StyleBozaquel-Morais, Bruno Leonardo, Leonie Vogt, Valentina D’Angelo, Raffael Schaffrath, Roland Klassen, and Mónica Montero-Lomelí. 2018. "Protein Phosphatase Sit4 Affects Lipid Droplet Synthesis and Soraphen A Resistance Independent of Its Role in Regulating Elongator Dependent tRNA Modification" Biomolecules 8, no. 3: 49. https://doi.org/10.3390/biom8030049
APA StyleBozaquel-Morais, B. L., Vogt, L., D’Angelo, V., Schaffrath, R., Klassen, R., & Montero-Lomelí, M. (2018). Protein Phosphatase Sit4 Affects Lipid Droplet Synthesis and Soraphen A Resistance Independent of Its Role in Regulating Elongator Dependent tRNA Modification. Biomolecules, 8(3), 49. https://doi.org/10.3390/biom8030049





