A Novel Cell Penetrating Peptide for the Differentiation of Human Neural Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
2.2. Peptide Purification and Isolation
2.3. Mass Spetrometry
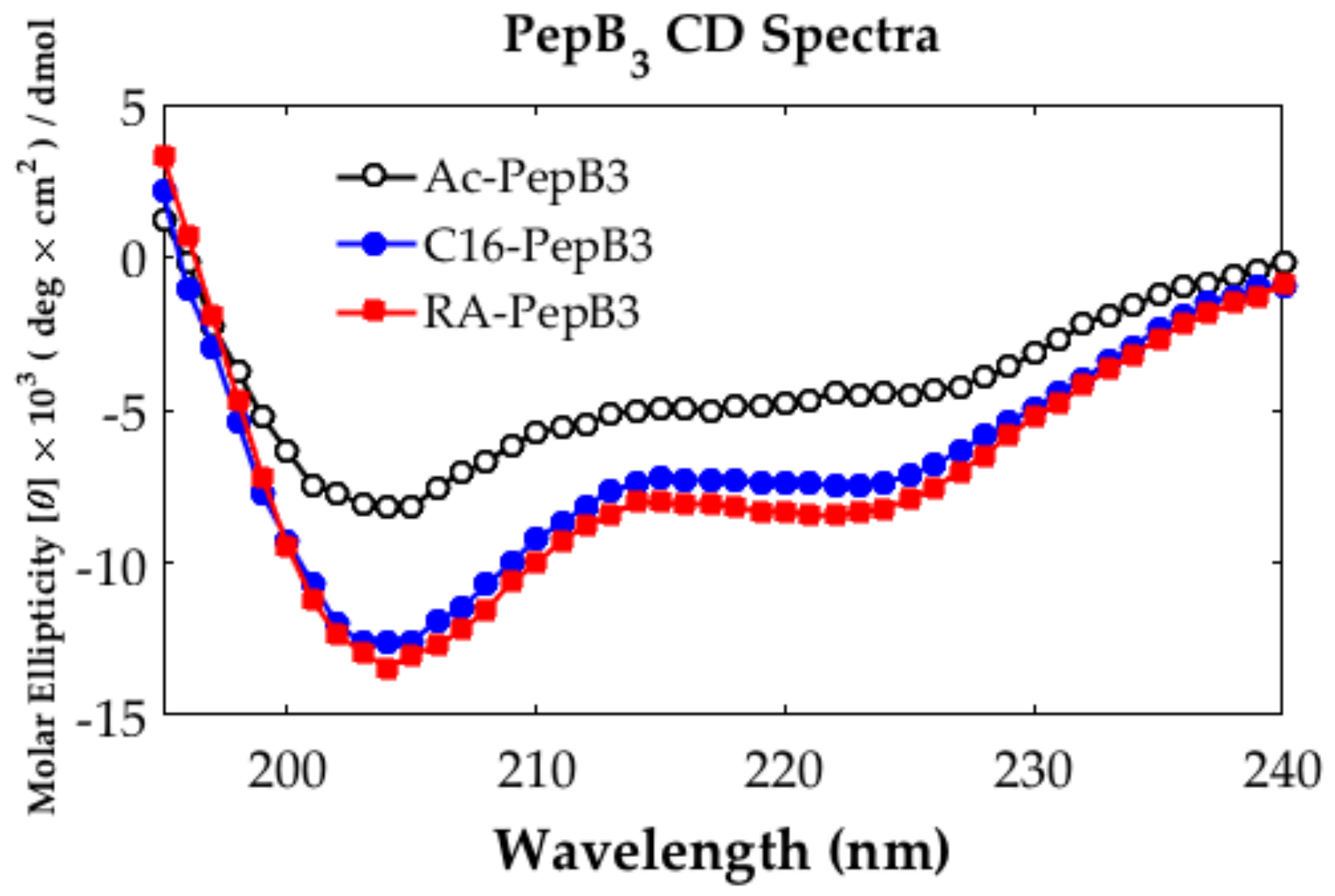
2.4. Circular Dichroism
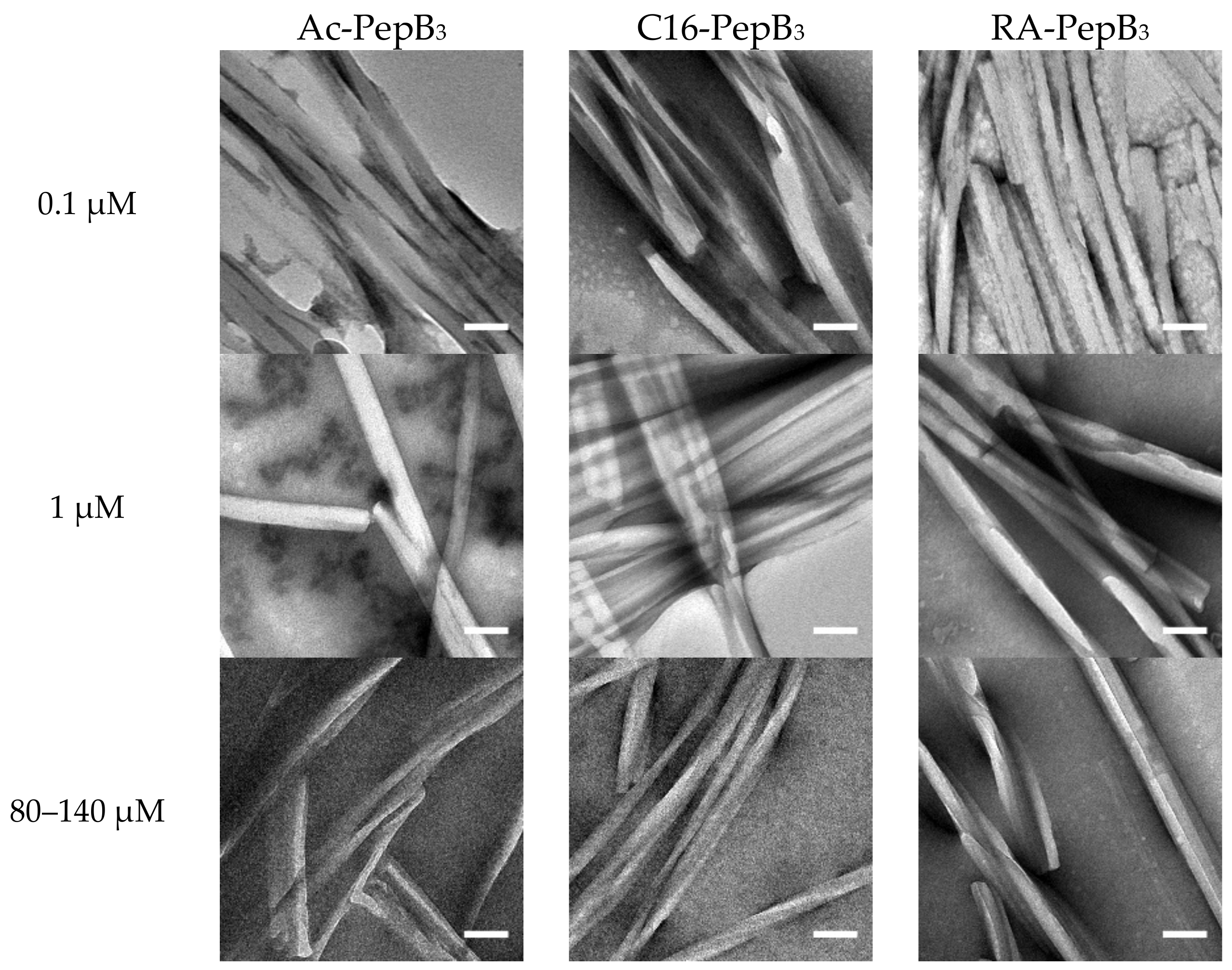
2.5. Transmission Electron Microscopy
2.6. Cell Culture and Differentiation
2.7. Immunocytochemistry
2.8. Peptide Uptake and Flow Cytometry
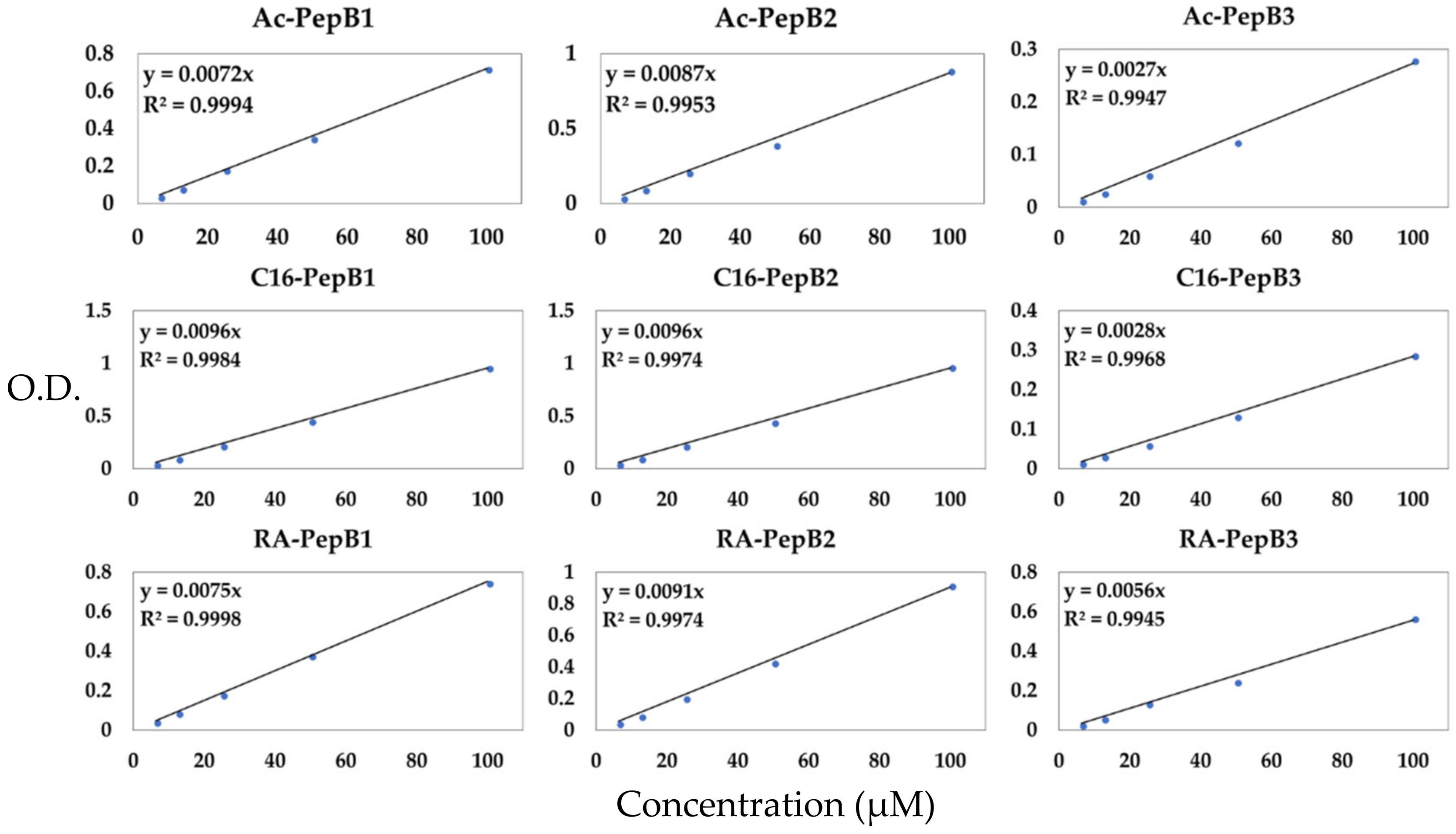
2.9. Quantification of Peptide Stock Solutions
2.10. Cytotoxicity
2.11. Statistical Analysis
3. Results
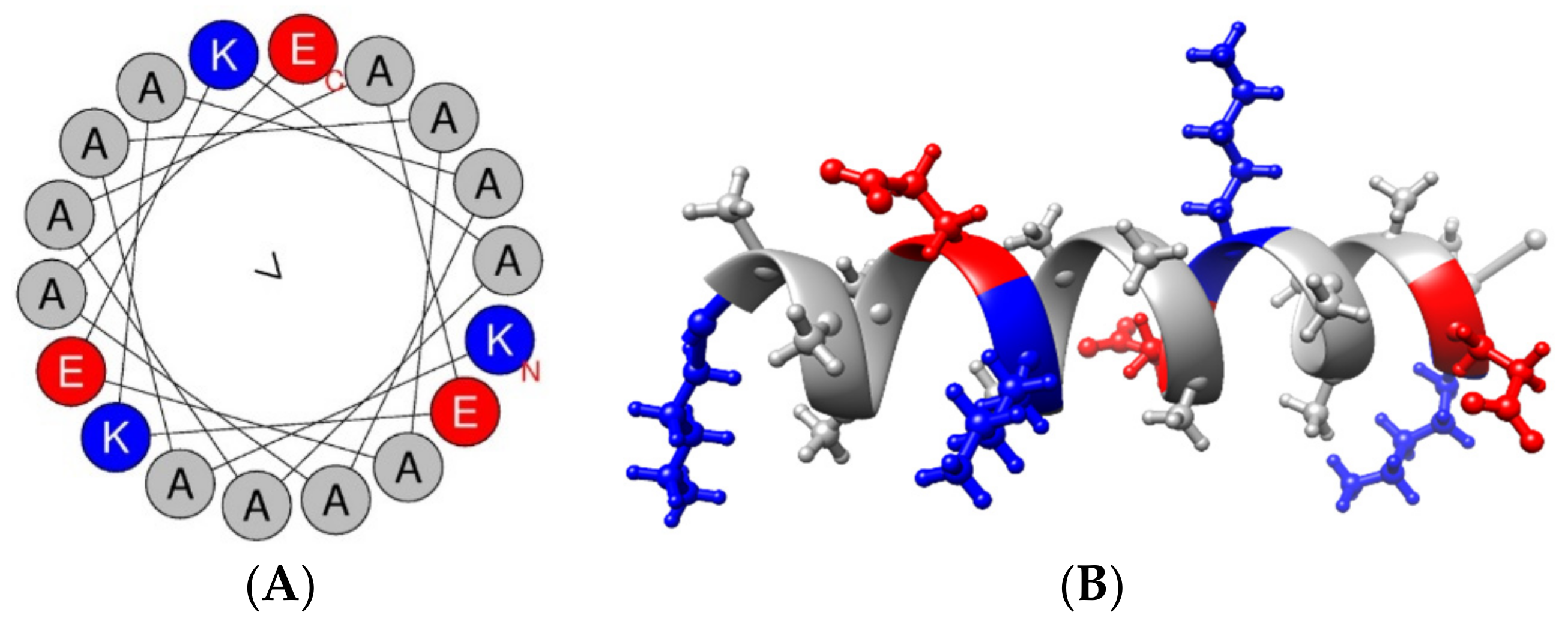
3.1. Peptide Characterization
3.2. Peptide Concentration Quantification
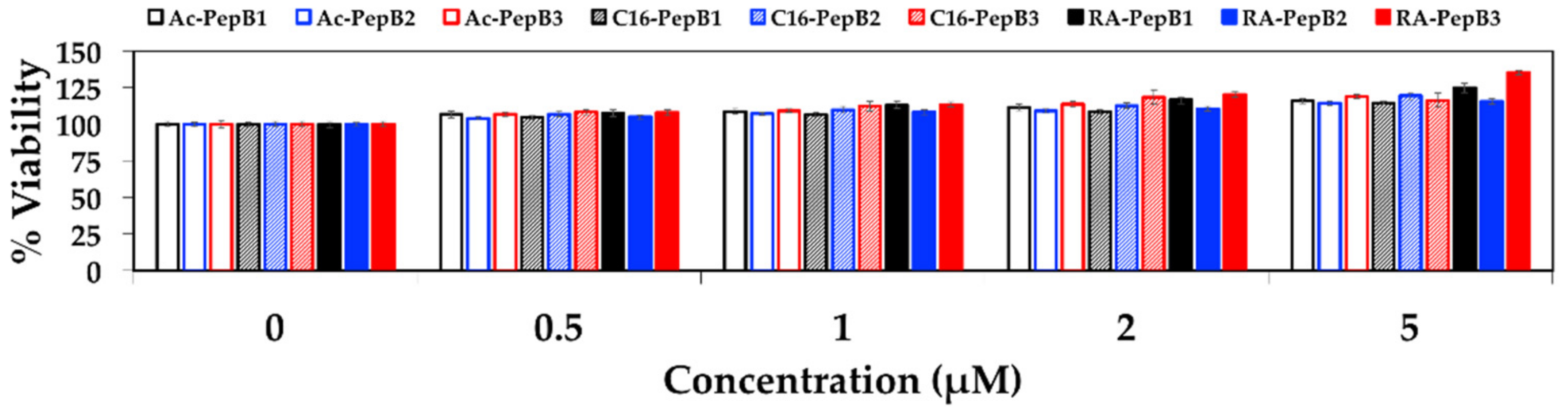
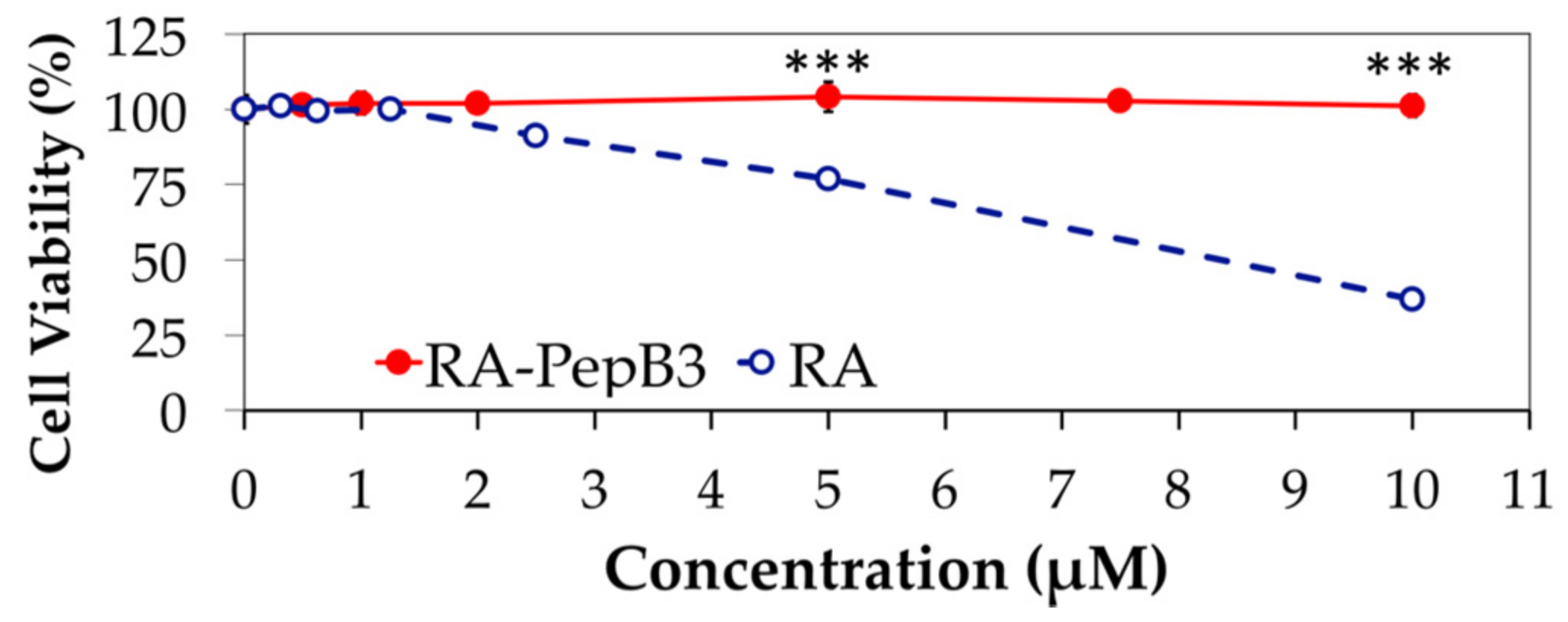
3.3. Cytotoxicity
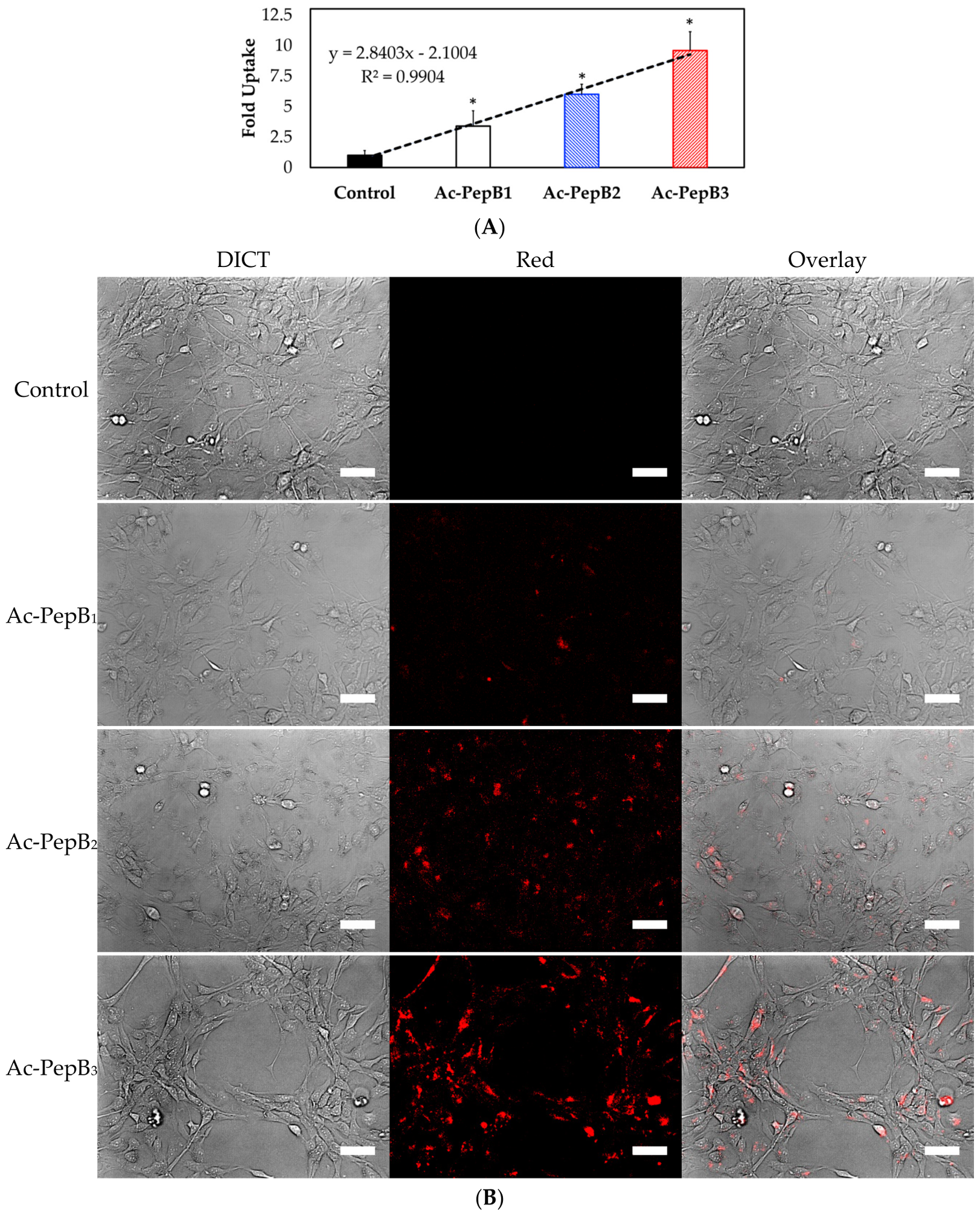
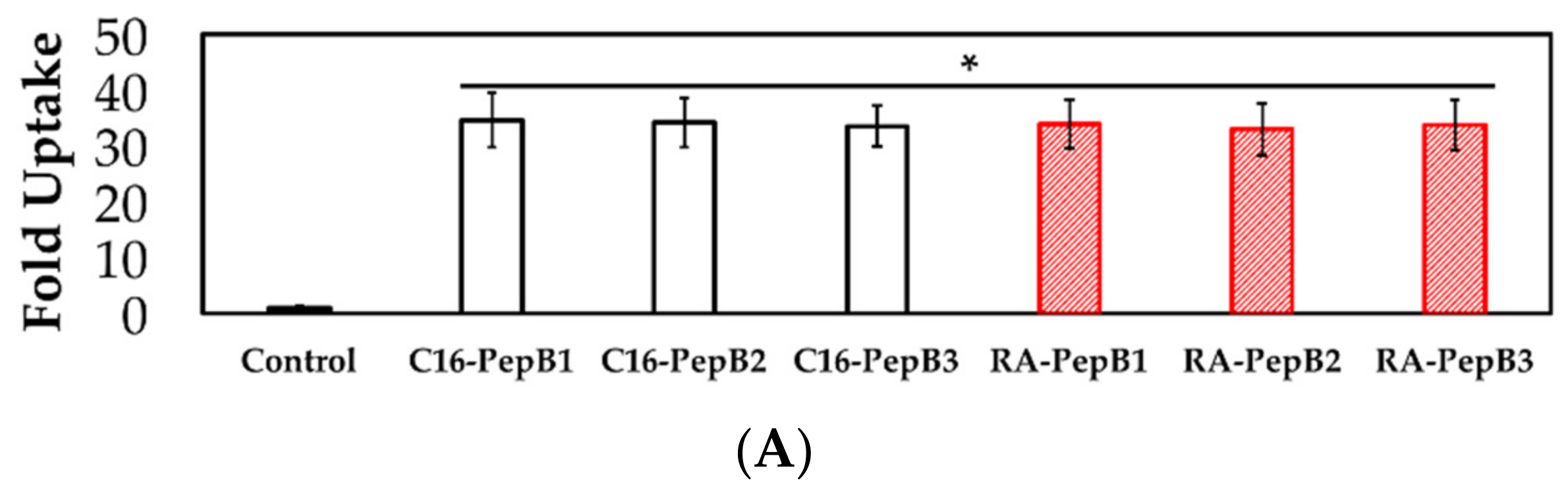
3.4. Cell Uptake and Flow Cytometry
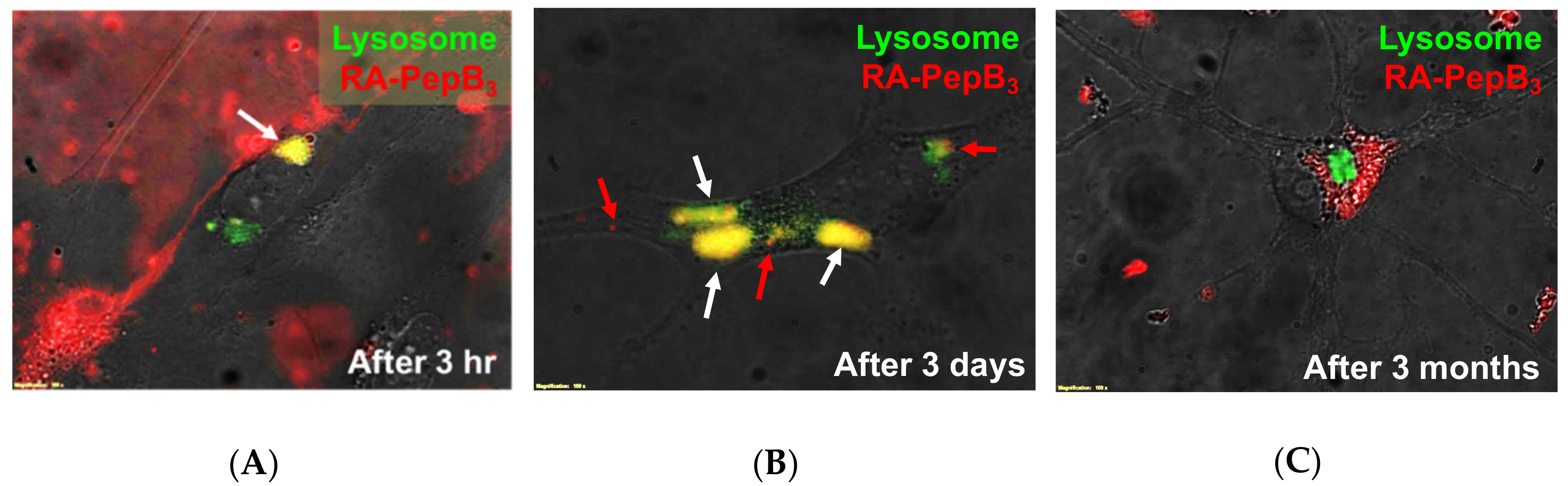
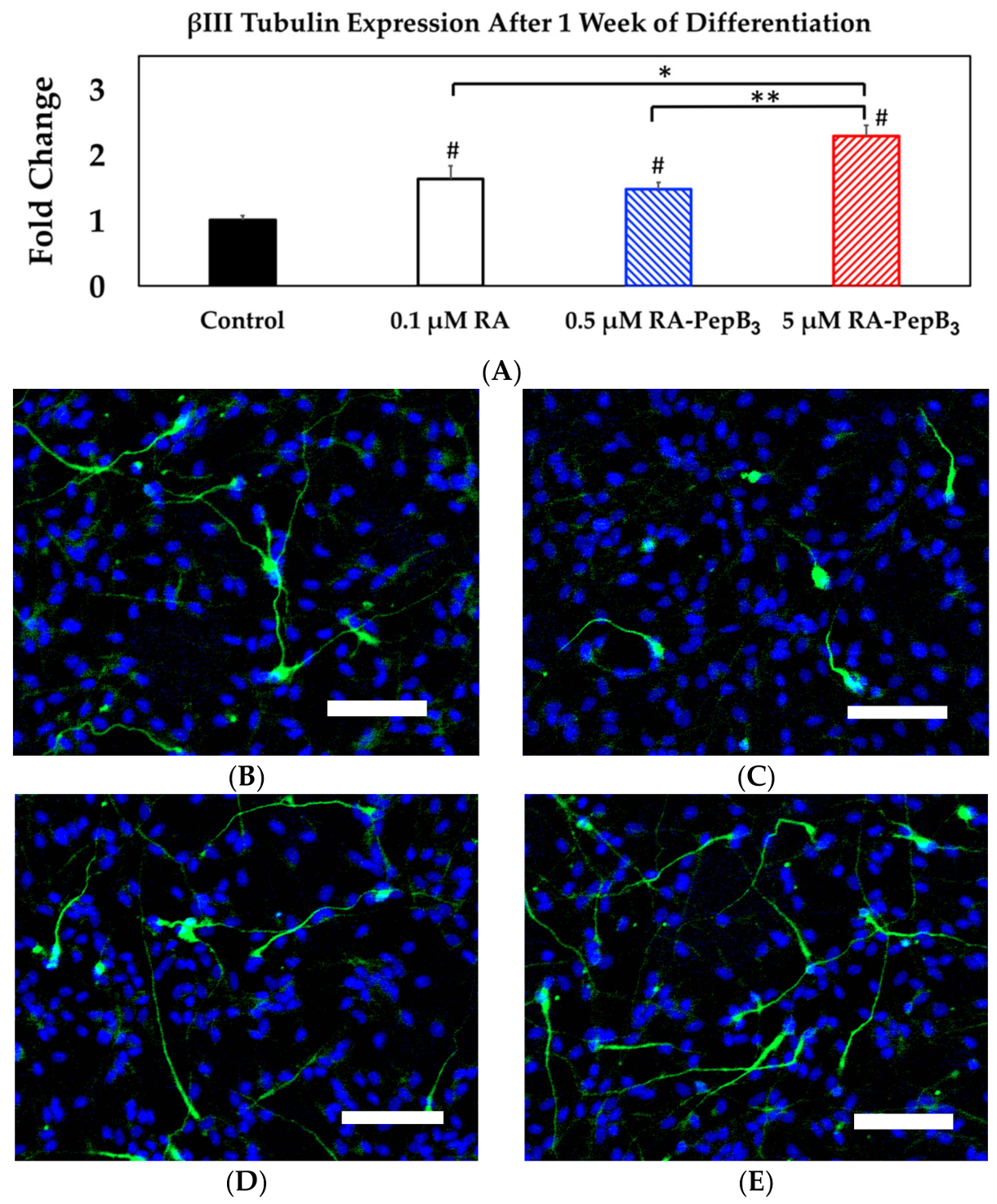
3.5. Long-Term and Short-Term Differentiation Results
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gorman, A.M. Neuronal cell death in neurodegenerative diseases: Recurring themes around protein handling. J. Cell. Mol. Med. 2008, 12, 2263–2280. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.U.; Lee, H.J.; Kim, Y.B. Neural stem cell-based treatment for neurodegenerative diseases. Neuropathology 2013, 33, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.M.; Yeon, B.K.; Cho, S.J.; Suh, Y.H. Stem cell therapy for Alzheimer’s disease: A review of recent clinical trials. J. Alzheimers Dis. 2016, 54, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.T.; Wang, L.; Li, S.; Long, Z.Y.; Wu, Y.M.; Liu, Y. Retinoic acid induced the differentiation of neural stem cells from embryonic spinal cord into functional neurons in vitro. Int. J. Clin. Exp. Pathol. 2015, 8, 8129–8135. [Google Scholar] [PubMed]
- Janesick, A.; Wu, S.C.; Blumberg, B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 2015, 72, 1559–1576. [Google Scholar] [CrossRef] [PubMed]
- Engberg, N.; Kahn, M.; Petersen, D.R.; Hansson, M.; Serup, P. Retinoic acid synthesis promotes development of neural progenitors from mouse embryonic stem cells by suppressing endogenous, Wnt-dependent nodal signaling. Stem Cells 2010, 28, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhao, J.; Fu, B.; Yin, S.; Song, C.; Zhang, J.; Zhao, S.; Zhang, Y. Retinoic acid-induced upregulation of mir-219 promotes the differentiation of embryonic stem cells into neural cells. Cell Death Dis. 2017, 8, e2953. [Google Scholar] [CrossRef] [PubMed]
- Tonge, P.D.; Andrews, P.W. Retinoic acid directs neuronal differentiation of human pluripotent stem cell lines in a non-cell-autonomous manner. Differentiation 2010, 80, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Rochette-Egly, C. Retinoic acid signaling and mouse embryonic stem cell differentiation: Cross talk between genomic and non-genomic effects of RA. Biochim. Biophys. Acta 2015, 1851, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Levi, L.; Siegel, R.; Noy, N. Retinoic acid induces neurogenesis by activating both retinoic acid receptors (RARs) and peroxisome proliferator-activated receptor β/δ (PPARβ/δ). J. Biol. Chem. 2012, 287, 42195–42205. [Google Scholar] [CrossRef] [PubMed]
- Szuts, E.Z.; Harosi, F.I. Solubility of retinoids in water. Arch. Biochem. Biophys. 1991, 287, 297–304. [Google Scholar] [CrossRef]
- Errico, C.; Gazzarri, M.; Chiellini, F. A novel method for the preparation of retinoic acid-loaded nanoparticles. Int. J. Mol. Sci. 2009, 10, 2336–2347. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, S.; Kim, S.Y.; Jeon, N.L.; Song, J.M.; Won, C.; Min, D.H. Highly efficient and rapid neural differentiation of mouse embryonic stem cells based on retinoic acid encapsulated porous nanoparticle. ACS Appl. Mater. Interfaces 2017, 9, 34634–34640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, Y.; Hu, B.; Lu, Z.; Zhang, J.; Zhang, X. Traceable nanoparticle delivery of small interfering RNA and retinoic acid with temporally release ability to control neural stem cell differentiation for Alzheimer’s disease therapy. Adv. Mater. 2016, 28, 6345–6352. [Google Scholar] [CrossRef] [PubMed]
- Carpenedo, R.L.; Bratt-Leal, A.M.; Marklein, R.A.; Seaman, S.A.; Bowen, N.J.; McDonald, J.F.; McDevitt, T.C. Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules. Biomaterials 2009, 30, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.; Edgar, J.; Agbay, A.; Bibault, E.; Montgomery, A.; Mohtaram, N.; Willerth, S. Incorporation of retinoic acid releasing microspheres into pluripotent stem cell aggregates for inducing neuronal differentiation. Cell. Mol. Bioeng. 2015, 8, 307–319. [Google Scholar] [CrossRef]
- Castleberry, S.A.; Quadir, M.A.; Sharkh, M.A.; Shopsowitz, K.E.; Hammond, P.T. Polymer conjugated retinoids for controlled transdermal delivery. J. Control Release 2017, 262, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Kim, K.; Kang, H.; Park, T.; Han, S.; Chang, I. Chemical immobilization of retinoic acid within poly(ε-caprolactone) nanoparticles based on drug-polymer bioconjugates. J. Appl. Polym. Sci. 2003, 89, 1631–1637. [Google Scholar]
- Louisse, J.; Gönen, S.; Rietjens, I.M.; Verwei, M. Relative developmental toxicity potencies of retinoids in the embryonic stem cell test compared with their relative potencies in in vivo and two other in vitro assays for developmental toxicity. Toxicol. Lett. 2011, 203, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.A.; Smith, A.M.; Egnash, L.A.; Colwell, M.R.; Donley, E.L.R.; Kirchner, F.R.; Burrier, R.E. A human induced pluripotent stem cell-based in vitro assay predicts developmental toxicity through a retinoic acid receptor-mediated pathway for a series of related retinoid analogues. Reprod. Toxicol. 2017, 73, 350–361. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.T.; Parrotta, E.I.; Santamaria, G.; Cuda, G. Short-term retinoic acid treatment sustains pluripotency and suppresses differentiation of human induced pluripotent stem cells. Cell Death Dis. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Suh, W.H. Engineered cell penetrating peptides. In Control of Amphiphile Self-Assembling at the Molecular Level: Supra-Molecular Assemblies with Tuned Physicochemical Properties for Delivery Applications; American Chemical Society: Washington, DC, USA, 2017; Volume 1271, pp. 297–319. [Google Scholar]
- Perret, P.; Ahmadi, M.; Riou, L.; Bacot, S.; Pecher, J.; Poillot, C.; Broisat, A.; Ghezzi, C.; De Waard, M. Biodistribution, stability, and blood distribution of the cell penetrating peptide maurocalcine in mice. Int. J. Mol. Sci. 2015, 16, 27730–27740. [Google Scholar] [CrossRef] [PubMed]
- Palm, C.; Jayamanne, M.; Kjellander, M.; Hällbrink, M. Peptide degradation is a critical determinant for cell-penetrating peptide uptake. Biochim. Biophys. Acta 2007, 1768, 1769–1776. [Google Scholar] [CrossRef] [PubMed]
- Kirschberg, T.; VanDeusen, C.; Rothbard, J.; Yang, M.; Wender, P. Arginine-based molecular transporters: The synthesis and chemical evaluation of releasable taxol-transporter conjugates. Organ. Lett. 2003, 5, 3459–3462. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Console, S.; Marty, C.; García-Echeverría, C.; Schwendener, R.; Ballmer-Hofer, K. Antennapedia and HIV transactivator of transcription (TAT) “protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J. Biol. Chem. 2003, 278, 35109–35114. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.; Galdiero, M.; Galdiero, S. Membranotropic cell penetrating peptides: The outstanding journey. Int. J. Mol. Sci. 2015, 16, 25323–25337. [Google Scholar] [CrossRef] [PubMed]
- Mo, R.H.; Zaro, J.L.; Shen, W.C. Comparison of cationic and amphipathic cell penetrating peptides for siRNA delivery and efficacy. Mol. Pharm. 2012, 9, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, A.; Cheong, D.W.; Accardo, A.; Di Fabrizio, E.; Riekel, C.; Hauser, C.A.E. Aliphatic peptides show similar self-assembly to amyloid core sequences, challenging the importance of aromatic interactions in amyloidosis. Proc. Natl. Acad. Sci. USA 2013, 110, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Colletier, J.P.; Laganowsky, A.; Landau, M.; Zhao, M.L.; Soriaga, A.B.; Goldschmidt, L.; Flot, D.; Cascio, D.; Sawaya, M.R.; Eisenberg, D. Molecular basis for amyloid-beta polymorphism. Proc. Natl. Acad. Sci. USA 2011, 108, 16938–16943. [Google Scholar] [CrossRef] [PubMed]
- Paravastu, A.K.; Leapman, R.D.; Yau, W.M.; Tycko, R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 18349–18354. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Miljan, E.A.; Hines, S.J.; Aouabdi, S.; Pollock, K.; Patel, S.; Edwards, F.A.; Sinden, J.D. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Suh, W.H.; Kang, J.K.; Suh, Y.H.; Tirrell, M.; Suslick, K.S.; Stucky, G.D. Porous carbon produced in air: Physicochemical properties and stem cell engineering. Adv. Mater. 2011, 23, 2332–2338. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.-W.; Ma, W.; Suh, W.H. Three dimensional cell culture of human neural stem cells using polysaccharide-based hydrogels and subsequent bioanalyses. In Microrna Technologies; Kye, M.J., Ed.; Springer: New York, NY, USA, 2017; pp. 183–204. [Google Scholar]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, H.C.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014, 515, 274. [Google Scholar] [CrossRef] [PubMed]
- Suh, W.H.; Tirrell, M. Surface engineering using peptide amphiphiles. In Comprehensive Biomaterials; Elsevier: Oxford, UK, 2011; Volume 4, pp. 219–245. [Google Scholar]
- Rexeisen, E.L.; Fan, W.; Pangburn, T.O.; Taribagil, R.R.; Bates, F.S.; Lodge, T.P.; Tsapatsis, M.; Kokkoli, E. Self-assembly of fibronectin mimetic peptide-amphiphile nanofibers. Langmuir 2010, 26, 1953–1959. [Google Scholar] [CrossRef] [PubMed]
- Yasa, I.C.; Gunduz, N.; Kilinc, M.; Guler, M.O.; Tekinay, A.B. Basal lamina mimetic nanofibrous peptide networks for skeletal myogenesis. Sci. Rep. 2015, 5, 16460. [Google Scholar] [CrossRef] [PubMed]
- Morrisett, J.D.; David, J.S.K.; Pownall, H.J.; Gotto, A.M. Interaction of an apolipoprotein (apolp-alanine) with phosphatidylcholine. Biochemistry 1973, 12, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Rohl, C.A.; Baldwin, R.L. Comparison of NH exchange and circular dichroism as techniques for measuring the parameters of the helix-coil transition in peptides. Biochemistry 1997, 36, 8435–8442. [Google Scholar] [CrossRef] [PubMed]
- Marullo, R.; Kastantin, M.; Drews, L.B.; Tirrell, M. Peptide contour length determines equilibrium secondary structure in protein-analogous micelles. Biopolymers 2013, 99, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, H.; Ishiyama, M.; Ohseto, F.; Sasamoto, K.; Hamamoto, T.; Suzuki, K.; Watanabe, M. A water-soluble tetrazolium salt useful for colorimetric cell viability assay. Anal. Commun. 1999, 36, 47–50. [Google Scholar] [CrossRef]
- Nierode, G.J.; Perea, B.C.; McFarland, S.K.; Pascoal, J.F.; Clark, D.S.; Schaffer, D.V.; Dordick, J.S. High-throughput toxicity and phenotypic screening of 3D human neural progenitor cell cultures on a microarray chip platform. Stem Cell Rep. 2016, 7, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Meli, L.; Barbosa, H.S.C.; Hickey, A.M.; Gasimli, L.; Nierode, G.; Diogo, M.M.; Linhardt, R.J.; Cabral, J.M.S.; Dordick, J.S. Three dimensional cellular microarray platform for human neural stem cell differentiation and toxicology. Stem Cell Res. 2014, 13, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Oehlke, J.; Scheller, A.; Wiesner, B.; Krause, E.; Beyermann, M.; Klauschenz, E.; Melzig, M.; Bienert, M. Cellular uptake of an alpha-helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior non-endocytically. Biochim. Biophys. Acta 1998, 1414, 127–139. [Google Scholar] [CrossRef]
- Oehlke, J.; Krause, E.; Wiesner, B.; Beyermann, M.; Bienert, M. Extensive cellular uptake into endothelial cells of an amphipathic beta-sheet forming peptide. FEBS Lett. 1997, 415, 196–199. [Google Scholar] [CrossRef]
- De Coupade, C.; Fittipaldi, A.; Chagnas, V.; Michel, M.; Carlier, S.; Tasciotti, E.; Darmon, A.; Ravel, D.; Kearsey, J.; Giacca, M.; et al. Novel human-derived cell-penetrating peptides for specific subcellular delivery of therapeutic biomolecules. Biochem. J. 2005, 390, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zeng, X.Z.; Li, J.X.; Jiang, Y.H.; Zhao, H.; Wang, D.Y.; Huang, X.H.; Li, Z.G. Achieving enhanced cell penetration of short conformationally constrained peptides through amphiphilicity tuning. Chem. Sci. 2017, 8, 7576–7581. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Kato, T.; Oba, M.; Misawa, T.; Hattori, T.; Ohoka, N.; Tanaka, M.; Naito, M.; Kurihara, M.; Demizu, Y. Development of a cell-penetrating peptide that exhibits responsive changes in its secondary structure in the cellular environment. Sci. Rep. 2016, 6, 33003. [Google Scholar] [CrossRef] [PubMed]
- Eiríksdóttir, E.; Konate, K.; Langel, U.; Divita, G.; Deshayes, S. Secondary structure of cell-penetrating peptides controls membrane interaction and insertion. Biochim. Biophys. Acta 2010, 1798, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Manning, M.C.; Illangasekare, M.; Woody, R.W. Circular dichroism studies of distorted alpha-helices, twisted beta-sheets, and beta turns. Biophys. Chem. 1988, 31, 77–86. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. Pep-fold3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Marqusee, S.; Robbins, V.H.; Baldwin, R.L. Unusually stable helix formation in short alanine-based peptides. Proc. Natl. Acad. Sci. USA 1989, 86, 5286–5290. [Google Scholar] [CrossRef] [PubMed]
- Serrano, L.; Neira, J.L.; Sancho, J.; Fersht, A.R. Effect of alanine versus glycine in α-helices on protein stability. Nature 1992, 356, 453–455. [Google Scholar] [CrossRef] [PubMed]
- López-Llano, J.; Campos, L.A.; Sancho, J. α-helix stabilization by alanine relative to glycine: Roles of polar and apolar solvent exposures and of backbone entropy. Proteins 2006, 64, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Do, T.D.; Chamas, A.; Zheng, X.; Barnes, A.; Chang, D.; Veldstra, T.; Takhar, H.; Dressler, N.; Trapp, B.; Miller, K.; et al. Elucidation of the aggregation pathways of helix-turn-helix peptides: Stabilization at the turn region is critical for fibril formation. Biochemistry 2015, 54, 4050–4062. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Lee, S.; Bates, F.S.; Hotta, A.; Tirrell, M. Wormlike micelle formation in peptide-lipid conjugates driven by secondary structure transformation of the headgroups. J. Phys. Chem. B 2009, 113, 13711–13714. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Sakamoto, N.; Motokawa, R.; Koizumi, S.; Tirrell, M. Self-assembly process of peptide amphiphile worm-like micelles. J. Phys. Chem. B 2012, 116, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Feng, Y. Thermo-switchable surfactant gel. Chem. Commun. 2011, 47, 7191–7193. [Google Scholar] [CrossRef] [PubMed]
- Casey, P.J. Protein lipidation in cell signaling. Science 1995, 268, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Draper, J.M.; Xia, Z.; Smith, C.D. Cellular palmitoylation and trafficking of lipidated peptides. J. Lipid Res. 2007, 48, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Covic, L.; Misra, M.; Badar, J.; Singh, C.; Kuliopulos, A. Pepducin-based intervention of thrombin-receptor signaling and systemic platelet activation. Nat. Med. 2002, 8, 1161–1165. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.; Kent, S. Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 2000, 69, 923–960. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.; White, P.D. Fmoc Solid Phase Peptide Synthesis: A Practical Approach; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Lepage, P.; Heckel, C.; Humbert, S.; Stahl, S.; Rautmann, G. Recombinant technology as an alternative to chemical peptide synthesis: Expression and characterization of HIV-1 rev recombinant peptides. Anal. Biochem. 1993, 213, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tan, L.; Li, P.; Gao, H.; Fang, B.; Ye, S.; Geng, Z.; Zheng, P.; Song, H. All-trans retinoic acid promotes neural lineage entry by pluripotent embryonic stem cells via multiple pathways. BMC Cell Biol. 2009, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Bieberich, E. It’s a lipid’s world: Bioactive lipid metabolism and signaling in neural stem cell differentiation. Neurochem. Res. 2012, 37, 1208–1229. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.; Zintl, F. Effects of retinoic acid on proliferation, apoptosis, cytotoxicity, migration, and invasion of neuroblastoma cells. Med. Pediatr. Oncol. 2003, 40, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Bidad, K.; Salehi, E.; Oraei, M.; Saboor-Yaraghi, A.A.; Nicknam, M.H. Effect of all-trans retinoic acid (ATRA) on viability, proliferation, activation and lineage-specific transcription factors of CD4+ T cells. Iran. J. Allergy Asthma Immunol. 2011, 10, 243–249. [Google Scholar] [PubMed]
- Lotan, R. Different susceptibilities of human melanoma and breast carcinoma cell lines to retinoic acid-induced growth inhibition. Cancer Res. 1979, 39, 1014–1019. [Google Scholar] [PubMed]
- Hsu, S.L.; Wu, W.S.; Tyan, Y.S.; Chou, C.K. Retinoic acid-induced apoptosis is prevented by serum albumin and enhanced by Lipiodol in human hepatoma Hep3B cells. Cancer Lett. 1998, 129, 205–214. [Google Scholar] [CrossRef]
- Seewaldt, V.L.; Johnson, B.S.; Parker, M.B.; Collins, S.J.; Swisshelm, K. Expression of retinoic acid receptor beta mediates retinoic acid-induced growth arrest and apoptosis in breast cancer cells. Cell Growth Differ. 1995, 6, 1077–1088. [Google Scholar] [PubMed]
- Atencia, R.; García-Sanz, M.; Unda, F.; Aréchaga, J. Apoptosis during retinoic acid-induced differentiation of F9 embryonal carcinoma cells. Exp. Cell Res. 1994, 214, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.C.; Skarin, T.; Sumitran, S.; Toftgård, R. Retinoids induce apoptosis in cultured keratinocytes. Br. J. Dermatol. 2000, 143, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Wolf, G. Retinoic acid as cause of cell proliferation or cell growth inhibition depending on activation of one of two different nuclear receptors. Nutr. Rev. 2008, 66, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Miyatake, J.I.; Maeda, Y. Inhibition of proliferation and CD25 down-regulation by retinoic acid in human adult t cell leukemia cells. Leukemia 1997, 11, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Sidell, N. Retinoic acid-induced growth inhibition and morphologic differentiation of human neuroblastoma cells in vitro. J. Natl. Cancer Inst. 1982, 68, 589–596. [Google Scholar] [PubMed]
- Zhang, H.; Satyamoorthy, K.; Herlyn, M.; Rosdahl, I. All-trans retinoic acid (ATRA) differentially induces apoptosis in matched primary and metastatic melanoma cells—A speculation on damage effect of atra via mitochondrial dysfunction and cell cycle redistribution. Carcinogenesis 2003, 24, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Chen, Y.; Lu, H.; Cao, X. Coupling mitochondrial respiratory chain to cell death: An essential role of mitochondrial complex i in the interferon-beta and retinoic acid-induced cancer cell death. Cell Death Differ. 2007, 14, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.S.; Guo, D.D.; Song, H.H.; Kim, I.Y.; Jiang, H.L.; Jin, H.L.; Kim, Y.K.; Chung, C.S.; Choi, Y.J.; Lee, H.K.; et al. Regulation of adipocyte differentiation by pegylated all-trans retinoic acid: Reduced cytotoxicity and attenuated lipid accumulation. J. Nutr. Biochem. 2007, 18, 322–331. [Google Scholar] [CrossRef] [PubMed]
- El-Andaloussi, S.; Järver, P.; Johansson, H.J.; Langel, U. Cargo-dependent cytotoxicity and delivery efficacy of cell-penetrating peptides: A comparative study. Biochem. J. 2007, 407, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chai, C.; Jiang, X.; Teoh, S.; Leong, K. Fibronectin immobilized by covalent conjugation or physical adsorption shows different bioactivity on aminated-PET. Mater. Sci. Eng. C-Biomim. Supramol. Syst. 2007, 27, 213–219. [Google Scholar] [CrossRef]
- LeCher, J.C.; Nowak, S.J.; McMurry, J.L. Breaking in and busting out: Cell-penetrating peptides and the endosomal escape problem. Biomol. Concepts 2017, 8, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Lu, Y.; Wang, W.; Zhu, H.; Wang, Z.; Cao, Z. Simple protein modification using zwitterionic polymer to mitigate the bioactivity loss of conjugated insulin. Adv. Healthc. Mater. 2017, 6, 11. [Google Scholar] [CrossRef] [PubMed]
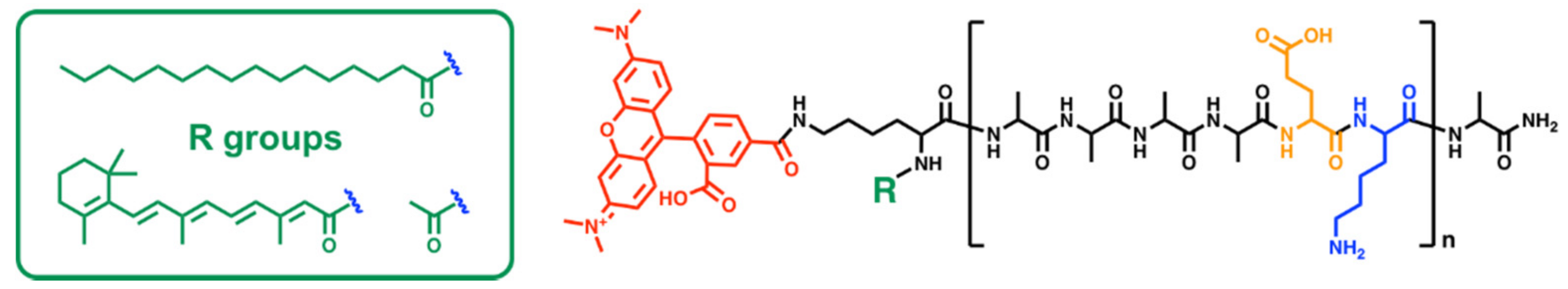



| PepB Repeat Structures (n) | NH2-Terminus | Predicted Mass (Da) | Abbreviated Name |
|---|---|---|---|
 | Acetyl | 1212.60 | Ac-PepB1 |
| Palmitoyl | 1409.76 | C16-PepB1 | |
| Retinoyl | 1452.79 | RA-PepB1 | |
 | Acetyl | 1753.89 | Ac-PepB2 |
| Palmitoyl | 1950.11 | C16-PepB2 | |
| Retinoyl | 1994.08 | RA-PepB2 | |
 | Acetyl | 2295.18 | Ac-PepB3 |
| Palmitoyl | 2491.40 | C16-PepB3 | |
| Retinoyl | 2535.36 | RA-PepB3* |
| Abbreviated Name | PepB Repeats (n) | NH2-Terminus Tail | Predicted Mass (Da) | ESI-ToF Measured Mass (m/z) | MALDI-ToF Measured Mass |
|---|---|---|---|---|---|
| Ac-PepB1 | 1 | Ac | 1212.60 | 606.81 | 1212.66 |
| C16-PepB1 | 1 | C16 | 1409.76 | 704.92 | 1409.73 |
| RA-PepB1 | 1 | RA | 1452.79 | 728.92 | 1457.01 |
| Ac-PepB2 | 2 | Ac | 1753.89 | 585.64 | 1754.53 |
| C16-PepB2 | 2 | C16 | 1950.11 | 651.04 | 1950.52 |
| RA-PepB2 | 2 | RA | 1994.08 | 667.04 | 1998.38 |
| Ac-PepB3 | 3 | Ac | 2295.18 | 574.80 | 2295.28 |
| C16-PepB3 | 3 | C16 | 2491.40 | 623.85, 831.47 | 2491.94 |
| RA-PepB3* | 3 | RA | 2535.36 | 635.60 | 2539.78 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, W.; Jin, G.-w.; Gehret, P.M.; Chada, N.C.; Suh, W.H. A Novel Cell Penetrating Peptide for the Differentiation of Human Neural Stem Cells. Biomolecules 2018, 8, 48. https://doi.org/10.3390/biom8030048
Ma W, Jin G-w, Gehret PM, Chada NC, Suh WH. A Novel Cell Penetrating Peptide for the Differentiation of Human Neural Stem Cells. Biomolecules. 2018; 8(3):48. https://doi.org/10.3390/biom8030048
Chicago/Turabian StyleMa, Weili, Geun-woo Jin, Paul M. Gehret, Neil C. Chada, and Won Hyuk Suh. 2018. "A Novel Cell Penetrating Peptide for the Differentiation of Human Neural Stem Cells" Biomolecules 8, no. 3: 48. https://doi.org/10.3390/biom8030048
APA StyleMa, W., Jin, G.-w., Gehret, P. M., Chada, N. C., & Suh, W. H. (2018). A Novel Cell Penetrating Peptide for the Differentiation of Human Neural Stem Cells. Biomolecules, 8(3), 48. https://doi.org/10.3390/biom8030048





