Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story
Abstract
:1. Introduction
2. History of JMJD6
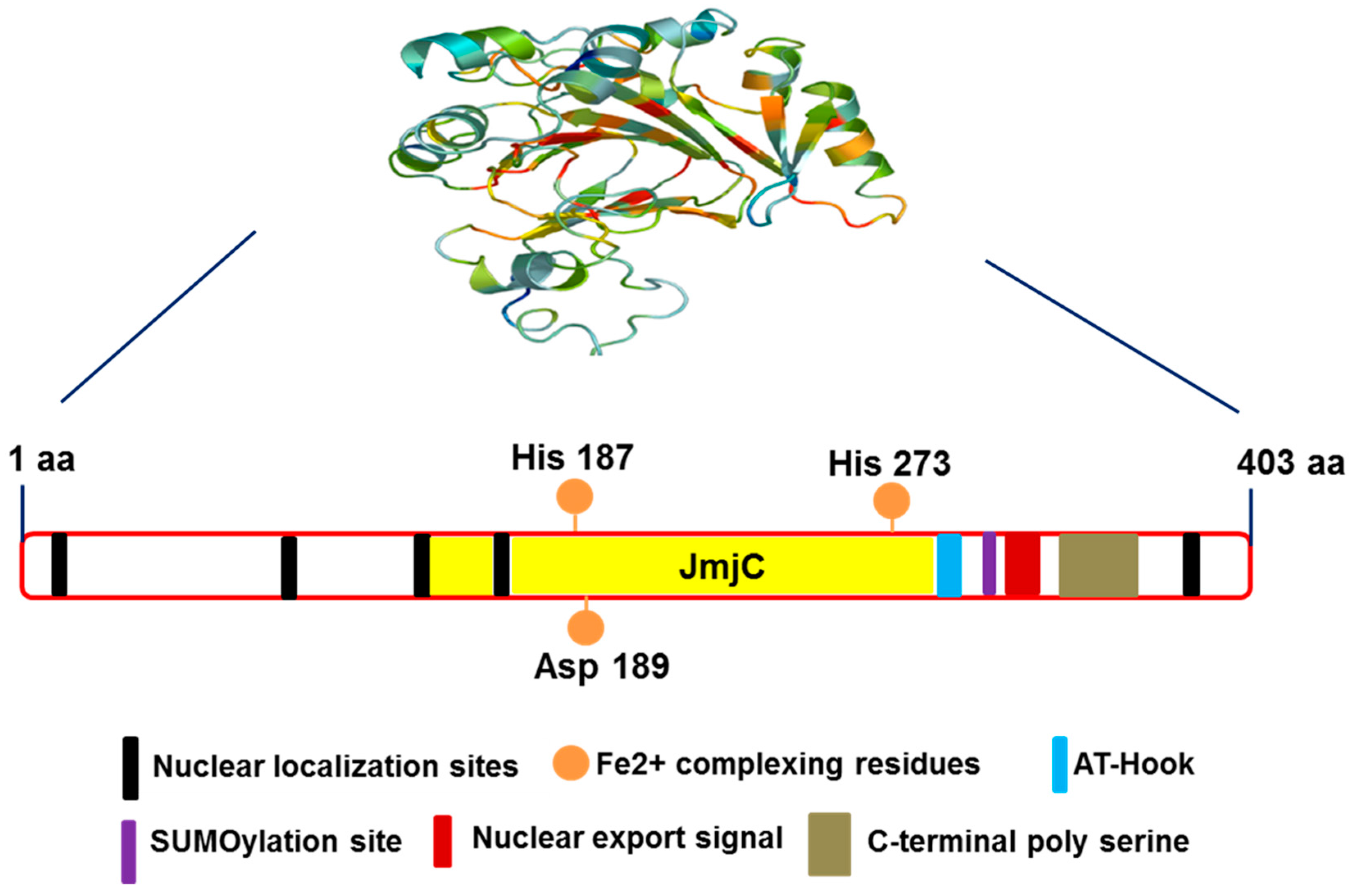
3. Structure of JMJD6
4. Enzymatic Activity and Interactions of JMJD6
5. Cancer and JMJD6
6. Oral Cancer and JMJD6
7. Colon Cancer and JMJD6
8. Lung Cancer and JMJD6
9. Breast Cancer and JMJD6
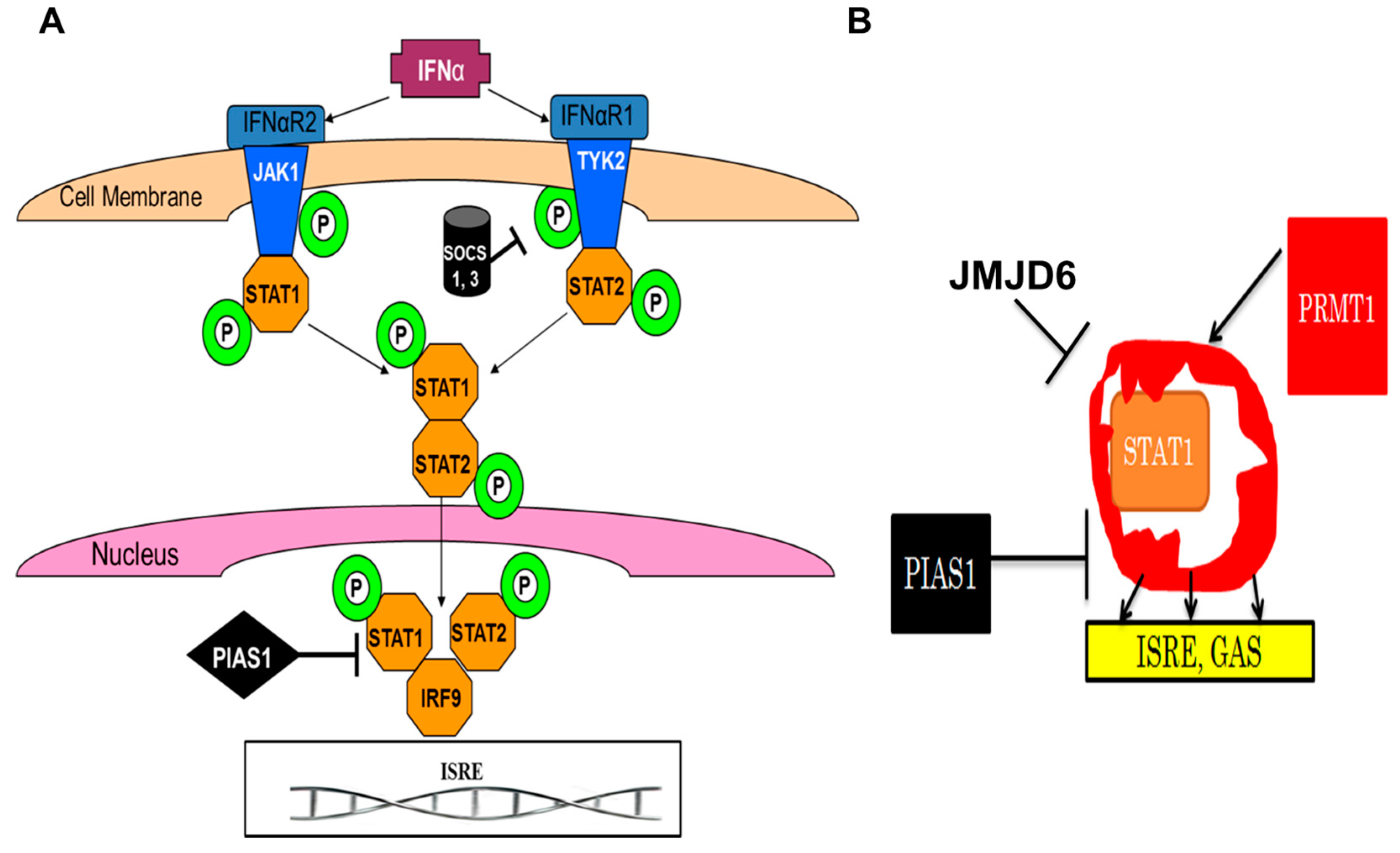
10. JMJD6 and Viruses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ambler, R.P.; Rees, M.W. Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature 1959, 184, 56–57. [Google Scholar] [CrossRef] [PubMed]
- Murray, K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry 1964, 3, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, G.S.; Carnegie, P.R. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science 1971, 171, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Brostoff, S.; Eylar, E.H. Localization of methylated arginine in the A1 protein from myelin. Proc. Natl. Acad. Sci. USA 1971, 68, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, Y.; Akazawa, S. Isolation and identification of N-G, n-G-and N-G,n’-G-dimethyl-arginine, N-epsilon-mono-, di-, and trimethyllysine, and glucosylgalactosyl-and galactosyl-delta-hydroxylysine from human urine. J. Biol. Chem. 1970, 245, 5751–5758. [Google Scholar] [PubMed]
- Lee, D.Y.; Teyssier, C.; Strahl, B.D.; Stallcup, M.R. Role of protein methylation in regulation of transcription. Endocr. Rev. 2005, 26, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Aletta, J.M.; Cimato, T.R.; Ettinger, M.J. Protein methylation: A signal event in post-translational modification. Trends Biochem. Sci. 1998, 23, 89–91. [Google Scholar] [CrossRef]
- McBride, A.E.; Silver, P.A. State of the arg: Protein methylation at arginine comes of age. Cell 2001, 106, 5–8. [Google Scholar] [CrossRef]
- Mowen, K.A.; Schurter, B.T.; Fathman, J.W.; David, M.; Glimcher, L.H. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 2004, 15, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Blanchet, F.; Cardona, A.; Letimier, F.A.; Hershfield, M.S.; Acuto, O. Cd28 costimulatory signal induces protein arginine methylation in t cells. J. Exp. Med. 2005, 202, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.A.; Schurter, B.T.; Wong-Staal, F.; David, M. Arginine methylation of RNA helicase A determines its subcellular localization. J. Biol. Chem. 2004, 279, 22795–22798. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S. Protein methylation. Curr. Opin. Cell. Biol. 1993, 5, 977–983. [Google Scholar] [CrossRef]
- Cheng, X.; Roberts, R.J. Adomet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001, 29, 3784–3795. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.K.; Gordon, R.K.; Tal, J.; Zeng, G.C.; Doctor, B.P.; Pardhasaradhi, K.; McCann, P.P. S-adenosylmethionine and methylation. FASEB J. 1996, 10, 471–480. [Google Scholar] [PubMed]
- Katz, J.E.; Dlakic, M.; Clarke, S. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteom. 2003, 2, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Shilatifard, A. Posttranslational modifications of histones by methylation. Adv. Protein Chem. 2004, 67, 201–222. [Google Scholar] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [PubMed]
- Poulard, C.; Corbo, L.; Le Romancer, M. Protein arginine methylation/demethylation and cancer. Oncotarget 2016, 7, 67532–67550. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wysocka, J.; Sayegh, J.; Lee, Y.H.; Perlin, J.R.; Leonelli, L.; Sonbuchner, L.S.; McDonald, C.H.; Cook, R.G.; Dou, Y.; et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004, 306, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone deimination antagonizes arginine methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Chen, Y.; Zhao, Y.; Bruick, R.K. JMJD6 is a histone arginine demethylase. Science 2007, 318, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Walport, L.J.; Hopkinson, R.J.; Chowdhury, R.; Schiller, R.; Ge, W.; Kawamura, A.; Schofield, C.J. Arginine demethylation is catalysed by a subset of jmjc histone lysine demethylases. Nat. Commun. 2016, 7, 11974. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Bratton, D.L.; Rose, D.M.; Pearson, A.; Ezekewitz, R.A.; Henson, P.M. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 2000, 405, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cikala, M.; Alexandrova, O.; David, C.N.; Proschel, M.; Stiening, B.; Cramer, P.; Bottger, A. The phosphatidylserine receptor from Hydra is a nuclear protein with potential Fe(II) dependent oxygenase activity. BMC Cell Biol. 2004, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Gruber, A.D.; Helming, L.; Schiebe, S.; Wegener, I.; Hafner, M.; Beales, M.; Kontgen, F.; Lengeling, A. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Kunisaki, Y.; Masuko, S.; Noda, M.; Inayoshi, A.; Sanui, T.; Harada, M.; Sasazuki, T.; Fukui, Y. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood 2004, 103, 3362–3364. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.R.; Lin, G.H.; Lin, C.J.; Wang, W.P.; Lee, C.C.; Lin, T.L.; Wu, J.L. Phosphatidylserine receptor is required for the engulfment of dead apoptotic cells and for normal embryonic development in zebrafish. Development 2004, 131, 5417–5427. [Google Scholar] [CrossRef] [PubMed]
- Webby, C.J.; Wolf, A.; Gromak, N.; Dreger, M.; Kramer, H.; Kessler, B.; Nielsen, M.L.; Schmitz, C.; Butler, D.S.; Yates, J.R.; et al. JMJD6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 2009, 325, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Bottger, A.; Islam, M.S.; Chowdhury, R.; Schofield, C.J.; Wolf, A. The oxygenase JMJD6-a case study in conflicting assignments. Biochem. J. 2015, 468, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ma, Q.; Wong, K.; Li, W.; Ohgi, K.; Zhang, J.; Aggarwal, A.K.; Rosenfeld, M.G. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 2013, 155, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Rambaud, J.; Hussein, N.; Corbo, L.; Le Romancer, M. JMJD6 regulates ERα methylation on arginine. PLoS ONE 2014, 9, e87982. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.; Conderino, J.S.; Rieder, E. Redistribution of demethylated RNA helicase a during foot-and-mouth disease virus infection: Role of jumonji C-domain containing protein 6 in RHA demethylation. Virology 2014, 452–453, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wesche, J.; Kuhn, S.; Kessler, B.M.; Salton, M.; Wolf, A. Protein arginine methylation: A prominent modification and its demethylation. Cell. Mol. Life Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hahn, P.; Bose, J.; Edler, S.; Lengeling, A. Genomic structure and expression of JMJD6 and evolutionary analysis in the context of related JmjC domain containing proteins. BMC Genom. 2008, 9, 293. [Google Scholar] [CrossRef] [PubMed]
- Kwok, J.; O’Shea, M.; Hume, D.A.; Lengeling, A. JMJD6, a JmjC dioxygenase with many interaction partners and pleiotropic functions. Front. Genet. 2017, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Mantri, M.; Krojer, T.; Bagg, E.A.; Webby, C.A.; Butler, D.S.; Kochan, G.; Kavanagh, K.L.; Oppermann, U.; McDonough, M.A.; Schofield, C.J. Crystal structure of the 2-oxoglutarate-and Fe(II)-dependent lysyl hydroxylase JMJD6. J. Mol. Biol. 2010, 401, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Zang, J.; White, J.; Wang, C.; Pan, C.H.; Zhao, R.; Murphy, R.C.; Dai, S.; Henson, P.; Kappler, J.W.; et al. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 14568–14572. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Li, J.; Wang, Y.; Li, X.; Mao, H.; Liu, Y.; Chen, C.D. The hydroxylation activity of JMJD6 is required for its homo-oligomerization. J. Cell. Biochem. 2012, 113, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Mantri, M.; Heim, A.; Muller, U.; Fichter, E.; Mackeen, M.M.; Schermelleh, L.; Dadie, G.; Leonhardt, H.; Venien-Bryan, C.; et al. The polyserine domain of the lysyl-5 hydroxylase JMJD6 mediates subnuclear localization. Biochem. J. 2013, 453, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.R.; McDonough, M.A.; King, O.N.; Kawamura, A.; Schofield, C.J. Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev. 2011, 40, 4364–4397. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson, R.J.; Walport, L.J.; Munzel, M.; Rose, N.R.; Smart, T.J.; Kawamura, A.; Claridge, T.D.; Schofield, C.J. Is JmjC oxygenase catalysis limited to demethylation? Angew. Chem. Int. Ed. Engl. 2013, 52, 7709–7713. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Hewitson, K.S.; Granatino, N.; Welford, R.W.; McDonough, M.A.; Schofield, C.J. Oxidation by 2-oxoglutarate oxygenases: Non-haem iron systems in catalysis and signalling. Philos. Trans. A Math. Phys. Eng. Sci. 2005, 363, 807–828. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bagan, J.; Sarrion, G.; Jimenez, Y. Oral cancer: Clinical features. Oral Oncol. 2010, 46, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Chiou, S.H.; Yu, C.C.; Huang, C.Y.; Lin, S.C.; Liu, C.J.; Tsai, T.H.; Chou, S.H.; Chien, C.S.; Ku, H.H.; Lo, J.F. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin. Cancer Res. 2008, 14, 4085–4095. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, S.H.; Rigas, N.K.; Kim, R.H.; Kang, M.K.; Park, N.H.; Shin, K.H. Elevated expression of JMJD6 is associated with oral carcinogenesis and maintains cancer stemness properties. Carcinogenesis 2016, 37, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Haggar, F.A.; Boushey, R.P. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Zhou, J.; Chen, Z.R.; Chng, W.J. P53 mutations in colorectal cancer-molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol. 2015, 21, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Zilfou, J.T.; Lowe, S.W. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 2009, 1, a001883. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; He, L.; Huangyang, P.; Liang, J.; Si, W.; Yan, R.; Han, X.; Liu, S.; Gui, B.; Li, W.; et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 2014, 12, e1001819. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.F.; Miller, L.D.; Chan, X.B.; Black, M.A.; Pang, B.; Ong, C.W.; Salto-Tellez, M.; Liu, E.T.; Desai, K.V. JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 2012, 14, R85. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.F.; Miller, L.D.; Chan, X.B.; Black, M.A.; Pang, B.; Ong, C.W.; Salto-Tellez, M.; Liu, E.T.; Desai, K.V. Erratum to: JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 2017, 19, 42. [Google Scholar] [CrossRef] [PubMed]
- Davidson, M.R.; Gazdar, A.F.; Clarke, B.E. The pivotal role of pathology in the management of lung cancer. J. Thorac. Dis. 2013, 5, 463–478. [Google Scholar]
- Travis, W.D.; Brambilla, E.; Riely, G.J. New pathologic classification of lung cancer: Relevance for clinical practice and clinical trials. J. Clin. Oncol. 2013, 31, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, J.; Govindan, R. Lung cancer in never smokers: A review. J. Clin. Oncol. 2007, 25, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ni, S.S.; Zhao, W.L.; Dong, X.C.; Wang, J.L. High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol. 2013, 34, 2397–2401. [Google Scholar] [CrossRef] [PubMed]
- Lappin, T.R.; Grier, D.G.; Thompson, A.; Halliday, H.L. HOX genes: Seductive science, mysterious mechanisms. Ulster Med. J. 2006, 75, 23–31. [Google Scholar] [PubMed]
- Zhan, J.; Wang, P.; Niu, M.; Wang, Y.; Zhu, X.; Guo, Y.; Zhang, H. High expression of transcriptional factor HoxB9 predicts poor prognosis in patients with lung adenocarcinoma. Histopathology 2015, 66, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Xu, W.; Zhan, J.; Ma, J.; Li, X.; Xie, Y.; Wang, J.; Zhu, W.G.; Luo, J.; Zhang, H. PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6. Nucleic Acids Res. 2016, 44, 10662–10675. [Google Scholar] [CrossRef] [PubMed]
- Poulard, C.; Rambaud, J.; Lavergne, E.; Jacquemetton, J.; Renoir, J.M.; Tredan, O.; Chabaud, S.; Treilleux, I.; Corbo, L.; Le Romancer, M. Role of JMJD6 in breast tumourigenesis. PLoS ONE 2015, 10, e0126181. [Google Scholar] [CrossRef] [PubMed]
- Aprelikova, O.; Chen, K.; El Touny, L.H.; Brignatz-Guittard, C.; Han, J.; Qiu, T.; Yang, H.H.; Lee, M.P.; Zhu, M.; Green, J.E. The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenet. 2016, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Feng, X.; Liao, H.Y.; Jin, W.J.; Zhang, J.; Wang, Y.; Gong, L.L.; Liu, J.J.; Yuan, X.H.; Zhao, B.B.; et al. Regulation of T cell proliferation by JMJD6 and PDGF-BB during chronic hepatitis B infection. Sci. Rep. 2014, 4, 6359. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.; Pacheco, J.; Stenfeldt, C.; Arzt, J.; Rai, D.K.; Rieder, E. Pathogenesis and micro-anatomic characterization of a cell-adapted mutant foot-and-mouth disease virus in cattle: Impact of the Jumonji C-domain containing protein 6 (JMJD6) and route of inoculation. Virology 2016, 492, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.; Rai, D.; Conderino, J.S.; Uddowla, S.; Rieder, E. Role of Jumonji C-domain containing protein 6 (JMJD6) in infectivity of foot-and-mouth disease virus. Virology 2016, 492, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S. Evasion of innate and adaptive immunity by flaviviruses. Immunol. Cell. Biol. 2003, 81, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Xagorari, A.; Chlichlia, K. Toll-like receptors and viruses: Induction of innate antiviral immune responses. Open Microbiol. J. 2008, 2, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Tikhanovich, I.; Kuravi, S.; Artigues, A.; Villar, M.T.; Dorko, K.; Nawabi, A.; Roberts, B.; Weinman, S.A. Dynamic arginine methylation of tumor necrosis factor (TNF) receptor-associated factor 6 regulates Toll-like receptor signaling. J. Biol. Chem. 2015, 290, 22236–22249. [Google Scholar] [CrossRef] [PubMed]
- Stanton, G.J.; Weigent, D.A.; Fleischmann, W.R.; Dianzani, F.; Baron, S. Interferon review. Investig. Radiol. 1987, 22, 259–273. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Mowen, K.A.; David, M. Unconventional post-translational modifications in immunological signaling. Nat. Immunol. 2014, 15, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, M.; Zhang, J.; Bronich, T.; Poluektova, L.I.; Donohue, T.M.; Tuma, D.J.; Kharbanda, K.K.; Osna, N.A. Acetaldehyde accelerates HCV-induced impairment of innate immunity by suppressing methylation reactions in liver cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, M.; Natarajan, S.K.; Zhang, J.; Mott, J.L.; Poluektova, L.I.; McVicker, B.L.; Kharbanda, K.K.; Tuma, D.J.; Osna, N.A. Role of apoptotic hepatocytes in HCV dissemination: Regulation by acetaldehyde. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Osna, N.A. Demethylase JMJD6 as a regulator of innate immunity in HCV-associated liver injury. Int. J. Hepatobiliary Pancreat Dis. 2017, 7, 15–17. [Google Scholar]
- Osna, N.A.; Ganesan, M. University of Nabraska Medical Center: Omaha, NE, USA, Unpublished work. 2017.
- Hahn, P.; Wegener, I.; Burrells, A.; Bose, J.; Wolf, A.; Erck, C.; Butler, D.; Schofield, C.J.; Bottger, A.; Lengeling, A. Analysis of JMJD6 cellular localization and testing for its involvement in histone demethylation. PLoS ONE 2010, 5, e13769. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vangimalla, S.S.; Ganesan, M.; Kharbanda, K.K.; Osna, N.A. Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story. Biomolecules 2017, 7, 41. https://doi.org/10.3390/biom7020041
Vangimalla SS, Ganesan M, Kharbanda KK, Osna NA. Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story. Biomolecules. 2017; 7(2):41. https://doi.org/10.3390/biom7020041
Chicago/Turabian StyleVangimalla, Shiva Shankar, Murali Ganesan, Kusum K. Kharbanda, and Natalia A. Osna. 2017. "Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story" Biomolecules 7, no. 2: 41. https://doi.org/10.3390/biom7020041
APA StyleVangimalla, S. S., Ganesan, M., Kharbanda, K. K., & Osna, N. A. (2017). Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story. Biomolecules, 7(2), 41. https://doi.org/10.3390/biom7020041







