New Insights into the Functions of Transcription Factors that Bind the RNA Polymerase Secondary Channel
Abstract
:1. Introduction
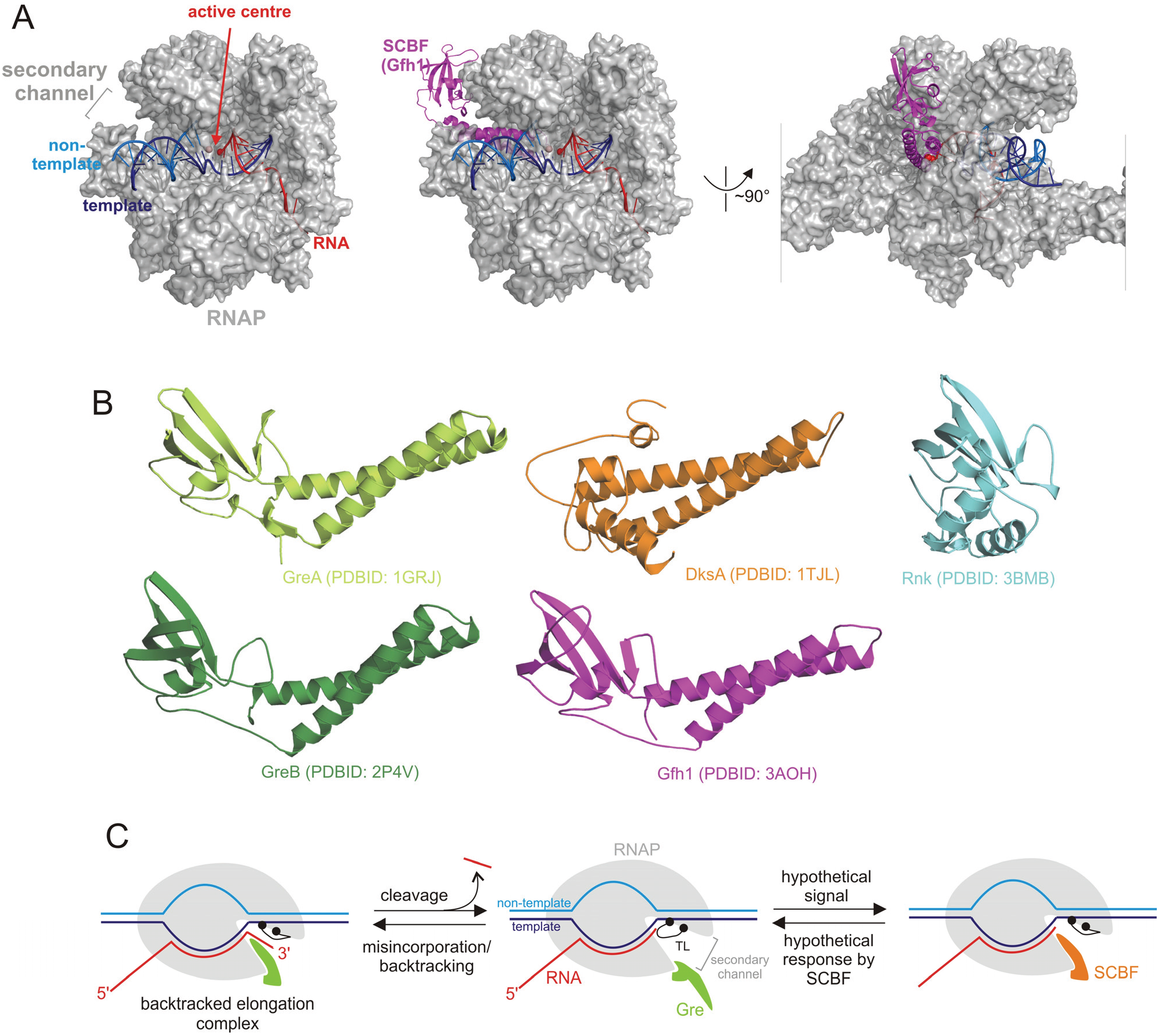
| Group | SCBF | Function |
|---|---|---|
| Cleavage factors | GreA/GreB | Resolve backtracked and misincorporated complexes via phosphodiester bond hydrolysis [11,30]. Increase clearance from some promoters [31] |
| DksA-like factors | DksA+ppGpp | Control isomerisation step of promoter open complexes formation on some promoters [16,32]. Increases fidelity of transcription elongation [29] |
| TraR | Encoded on the conjugative plasmid. Acts similarly to DksA on initiation of transcription with no requirement for ppGpp [17] | |
| Unknown function | Rnk | Binds RNAP but function is unknown [18] |
| Gfh1/Rv3788 | Inhibit catalysis by RNAP [33,34] |
2. Gre Factors
3. DksA/ppGpp and TraR
4. Factors with Unknown Functions
5. Regulation and Competition between SCBFs
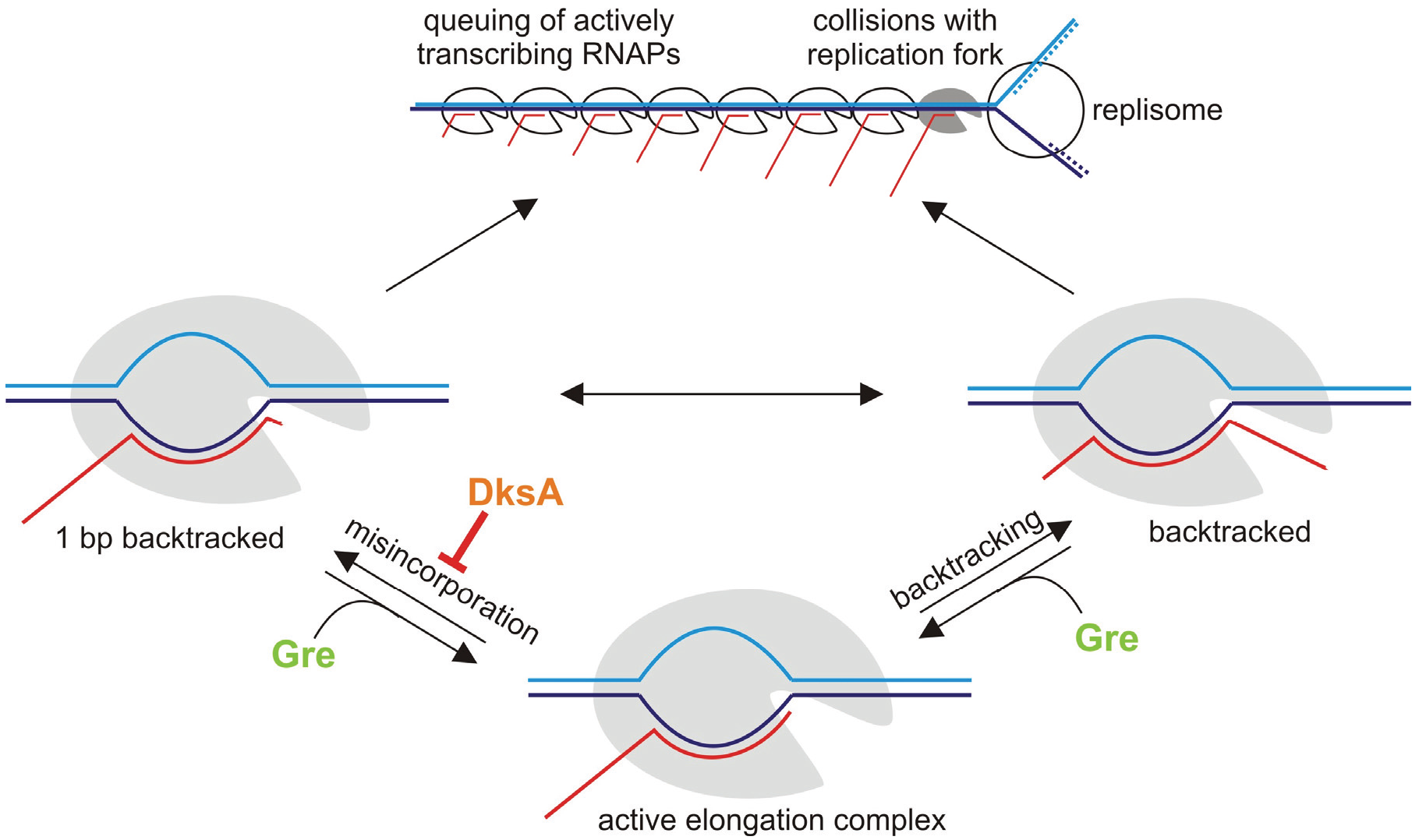
6. Misincorporation and Conflicts between Transcription and Replication
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Traviglia, S.L.; Datwyler, S.A.; Yan, D.; Ishihama, A.; Meares, C.F. Targeted protein footprinting: Where different transcription factors bind to RNA polymerase. Biochemistry 1999, 38, 15774–15778. [Google Scholar] [CrossRef] [PubMed]
- Belogurov, G.A.; Mooney, R.A.; Svetlov, V.; Landick, R.; Artsimovitch, I. Functional specialization of transcription elongation factors. EMBO J. 2009, 28, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Belogurov, G.A.; Vassylyeva, M.N.; Svetlov, V.; Klyuyev, S.; Grishin, N.V.; Vassylyev, D.G.; Artsimovitch, I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell 2007, 26, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Hirtreiter, A.; Damsma, G.E.; Cheung, A.C.; Klose, D.; Grohmann, D.; Vojnic, E.; Martin, A.C.; Cramer, P.; Werner, F. Spt4/5 stimulates transcription elongation through the rna polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010, 38, 4040–4051. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Marr, M.T.; Roberts, J.W.E. Coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 2002, 109, 757–767. [Google Scholar] [CrossRef]
- Roberts, J.; Park, J.S. Mfd, the bacterial transcription repair coupling factor: Translocation, repair and termination. Curr. Opin. Microbiol. 2004, 7, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Campbell, E.A.; Minakhin, L.; Richter, C.; Severinov, K.; Darst, S.A. Crystal structure of thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 1999, 98, 811–824. [Google Scholar] [CrossRef]
- Yuzenkova, J.; Delgado, M.; Nechaev, S.; Savalia, D.; Epshtein, V.; Artsimovitch, I.; Mooney, R.A.; Landick, R.; Farias, R.N.; Salomon, R.; et al. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 2002, 277, 50867–50875. [Google Scholar] [CrossRef] [PubMed]
- Korzheva, N.; Mustaev, A.; Kozlov, M.; Malhotra, A.; Nikiforov, V.; Goldfarb, A.; Darst, S.A. A structural model of transcription elongation. Science 2000, 289, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Adelman, K.; Yuzenkova, J.; la Porta, A.; Zenkin, N.; Lee, J.; Lis, J.T.; Borukhov, S.; Wang, M.D.; Severinov, K. Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol. Cell 2004, 14, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Borukhov, S.; Sagitov, V.; Goldfarb, A. Transcript cleavage factors from E. Coli. Cell 1993, 72, 459–466. [Google Scholar] [CrossRef]
- Borukhov, S.; Polyakov, A.; Nikiforov, V.; Goldfarb, A. Grea protein: A transcription elongation factor from Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 8899–8902. [Google Scholar] [CrossRef] [PubMed]
- Lange, U.; Hausner, W. Transcriptional fidelity and proofreading in Archaea and implications for the mechanism of TFS-induced RNA cleavage. Mol. Microbiol. 2004, 52, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Izban, M.G.; Luse, D.S. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J. Biol. Chem. 1993, 268, 12864–12873. [Google Scholar] [PubMed]
- Vinella, D.; Potrykus, K.; Murphy, H.; Cashel, M. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bacteriol. 2012, 194, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.J.; Barker, M.M.; Ross, W.; Schneider, D.A.; Webb, C.; Foster, J.W.; Gourse, R.L. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 2004, 118, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Blankschien, M.D.; Potrykus, K.; Grace, E.; Choudhary, A.; Vinella, D.; Cashel, M.; Herman, C. Trar, a homolog of a RNAp secondary channel interactor, modulates transcription. PLoS Genet. 2009, 5, e1000345. [Google Scholar] [CrossRef] [PubMed]
- Lamour, V.; Rutherford, S.T.; Kuznedelov, K.; Ramagopal, U.A.; Gourse, R.L.; Severinov, K.; Darst, S.A. Crystal structure of Escherichia coli Rnk, a new RNA polymerase-interacting protein. J. Mol. Biol. 2008, 383, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Kulish, D.; Lee, J.; Lomakin, I.; Nowicka, B.; Das, A.; Darst, S.; Normet, K.; Borukhov, S. The functional role of basic patch, a structural element of Escherichia coli transcript cleavage factors GreA and GreB. J. Biol. Chem. 2000, 275, 12789–12798. [Google Scholar] [CrossRef] [PubMed]
- Susa, M.; Kubori, T.; Shimamoto, N. A pathway branching in transcription initiation in Escherichia coli. Mol. Microbiol. 2006, 59, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Orlova, M.; Newlands, J.; Das, A.; Goldfarb, A.; Borukhov, S. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. USA 1995, 92, 4596–4600. [Google Scholar] [CrossRef] [PubMed]
- Glass, J.I.; Assad-Garcia, N.; Alperovich, N.; Yooseph, S.; Lewis, M.R.; Maruf, M.; Hutchison, C.A., 3rd; Smith, H.O.; Venter, J.C. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 2006, 103, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Peterson, S.N.; Gill, S.R.; Cline, R.T.; White, O.; Fraser, C.M.; Smith, H.O.; Venter, J.C. Global transposon mutagenesis and a minimal mycoplasma genome. Science 1999, 286, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- Yuzenkova, Y.; Gamba, P.; Herber, M.; Attaiech, L.; Shafeeq, S.; Kuipers, O.P.; Klumpp, S.; Zenkin, N.; Veening, J.W. Control of transcription elongation by GreA determines rate of gene expression in streptococcus pneumoniae. Nucleic Acids Res. 2014, 42, 10987–10999. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, E.V.; Shevelev, A.B.; Borukhov, S.I.; Severinov, K.V. Mechanisms of action of RNA polymerase-binding transcription factors that do not bind to DNA. Biofizika 2009, 54, 773–790. [Google Scholar] [CrossRef] [PubMed]
- Vassylyev, D.G.; Vassylyeva, M.N.; Perederina, A.; Tahirov, T.H.; Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 2007, 448, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Tagami, S.; Sekine, S.; Kumarevel, T.; Hino, N.; Murayama, Y.; Kamegamori, S.; Yamamoto, M.; Sakamoto, K.; Yokoyama, S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 2010, 468, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Yuzenkova, Y.; Roghanian, M.; Zenkin, N. Multiple active centers of multi-subunit RNA polymerases. Transcription 2012, 3, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Roghanian, M.; Zenkin, N.; Yuzenkova, Y. Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Komissarova, N.; Kashlev, M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3' end of the RNA intact and extruded. Proc. Natl. Acad. Sci. USA 1997, 94, 1755–1760. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, E.; Lee, J.; Ozerova, M.; Semenova, E.; Datsenko, K.; Wanner, B.L.; Severinov, K.; Borukhov, S. Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 2007, 189, 8772–8785. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.J.; Berkmen, M.B.; Gourse, R.L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA 2005, 102, 7823–7828. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Kim, S.S.; Lee, J.; Starodubtseva, M.; Cava, F.; Berenguer, J.; Kong, X.P.; Borukhov, S. PH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J. 2006, 25, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- China, A.; Mishra, S.; Tare, P.; Nagaraja, V. Inhibition of mycobacterium tuberculosis RNA polymerase by binding of a Gre factor homolog to the secondary channel. J. Bacteriol. 2012, 194, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Mustaev, A.; Lukhtanov, E.; Goldfarb, A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 1997, 89, 33–41. [Google Scholar] [CrossRef]
- Yuzenkova, Y.; Zenkin, N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc. Natl. Acad. Sci. USA 2010, 107, 10878–10883. [Google Scholar] [CrossRef] [PubMed]
- Roghanian, M.; Yuzenkova, Y.; Zenkin, N. Controlled interplay between trigger loop and Gre factor in the RNA polymerase active center. Nucleic Acids Res. 2011, 39, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Lee, J.; Lomakin, I.; Borukhov, S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003, 22, 6322–6334. [Google Scholar] [CrossRef] [PubMed]
- Sosunova, E.; Sosunov, V.; Kozlov, M.; Nikiforov, V.; Goldfarb, A.; Mustaev, A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc. Natl. Acad. Sci. USA 2003, 100, 15469–15474. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Landick, R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 2000, 97, 7090–7095. [Google Scholar] [CrossRef] [PubMed]
- Erie, D.A.; Hajiseyedjavadi, O.; Young, M.C.; von Hippel, P.H. Multiple RNA polymerase conformations and GreA: Control of the fidelity of transcription. Science 1993, 262, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Zenkin, N.; Yuzenkova, Y.; Severinov, K. Transcript-assisted transcriptional proofreading. Science 2006, 313, 518–520. [Google Scholar] [CrossRef] [PubMed]
- Epshtein, V.; Toulme, F.; Rahmouni, A.R.; Borukhov, S.; Nudler, E. Transcription through the roadblocks: The role of RNA polymerase cooperation. EMBO J. 2003, 22, 4719–4727. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA polymerase backtracking to genome instability in E. Coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, H.; Machon, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Stockum, A.; Nieduszynski, C.A.; Lloyd, R.G. Avoiding chromosome pathology when replication forks collide. Nature 2013, 500, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.K.; Lovell, M.A.; Hulme, S.D.; Zhang-Barber, L.; Barrow, P.A. Identification of salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immunity 1998, 66, 2099–2106. [Google Scholar]
- Kang, P.J.; Craig, E.A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a DNAk deletion mutation. J. Bacteriol. 1990, 172, 2055–2064. [Google Scholar] [PubMed]
- Brown, L.; Gentry, D.; Elliott, T.; Cashel, M. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 2002, 184, 4455–4465. [Google Scholar] [CrossRef] [PubMed]
- Mallik, P.; Paul, B.J.; Rutherford, S.T.; Gourse, R.L.; Osuna, R. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of FIS expression in Escherichia coli. J. Bacteriol. 2006, 188, 5775–5782. [Google Scholar] [CrossRef] [PubMed]
- Perron, K.; Comte, R.; van Delden, C. DksA represses ribosomal gene transcription in pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 2005, 56, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, L.U.; Gummesson, B.; Joksimovic, P.; Farewell, A.; Nystrom, T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 2007, 189, 5193–5202. [Google Scholar] [CrossRef] [PubMed]
- Aberg, A.; Shingler, V.; Balsalobre, C. Regulation of the fimB promoter: A case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 2008, 67, 1223–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mooney, R.A.; Grass, J.A.; Sivaramakrishnan, P.; Herman, C.; Landick, R.; Wang, J.D. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol. Cell 2014, 53, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Lamour, V.; Hogan, B.P.; Erie, D.A.; Darst, S.A. Crystal structure of thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J. Mol. Biol. 2006, 356, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, K.; Vinella, D.; Murphy, H.; Szalewska-Palasz, A.; D’Ari, R.; Cashel, M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 2006, 281, 15238–15248. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Lemke, J.J.; Vrentas, C.E.; Gaal, T.; Ross, W.; Gourse, R.L. Effects of DksA, GreA, and GreB on transcription initiation: Insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 2007, 366, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Rhodius, V.A.; Suh, W.C.; Nonaka, G.; West, J.; Gross, C.A. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006, 4, e2. [Google Scholar] [CrossRef] [PubMed]
- Len, A.C.; Harty, D.W.; Jacques, N.A. Stress-responsive proteins are upregulated in streptococcus mutans during acid tolerance. Microbiology 2004, 150, 1339–1351. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Campos, R.; BenAbdelkhalek, H.; Olivares, J.; Lluch, C.; Sanjuan, J. Rhizobium tropici genes involved in free-living salt tolerance are required for the establishment of efficient nitrogen-fixing symbiosis with phaseolus vulgaris. Mol. Plant Microbe Interact. 2002, 15, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, J.; Kurihara, T.; Kitagawa, M.; Kato, I.; Esaki, N. Proteomic studies of an antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins. Extrem. Life Extrem. Cond. 2007, 11, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Doherty, G.P.; Meredith, D.H.; Lewis, P.J. Subcellular partitioning of transcription factors in bacillus subtilis. J. Bacteriol. 2006, 188, 4101–4110. [Google Scholar] [CrossRef] [PubMed]
- Furman, R.; Tsodikov, O.V.; Wolf, Y.I.; Artsimovitch, I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J. Mol. Biol. 2013, 425, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.; Ortiz-Sanchez, J.M.; Da, L.; Wang, D.; McCammon, J.A. Molecular dynamics simulation study of conformational changes of transcription factor tfiis during RNA polymerase II transcriptional arrest and reactivation. PLoS ONE 2014, 9, e97975. [Google Scholar] [CrossRef] [PubMed]
- Perederina, A.; Svetlov, V.; Vassylyeva, M.N.; Tahirov, T.H.; Yokoyama, S.; Artsimovitch, I.; Vassylyev, D.G. Regulation through the secondary channel—Structural framework for ppGpp-DksA synergism during transcription. Cell 2004, 118, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Trautinger, B.W.; Jaktaji, R.P.; Rusakova, E.; Lloyd, R.G. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell 2005, 19, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Helmstetter, C.E. Chromosome replication and the division cycle of Escherichia coli B/R. J. Mol. Biol. 1968, 31, 519–540. [Google Scholar] [CrossRef]
- Srivatsan, A.; Tehranchi, A.; MacAlpine, D.M.; Wang, J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010, 6, e1000810. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, R.T.; O’Donnell, M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 2010, 327, 590–592. [Google Scholar] [CrossRef] [PubMed]
- French, S. Consequences of replication fork movement through transcription units in vivo. Science 1992, 258, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J. When polymerases collide: Replication and the transcriptional organization of the E. Coli chromosome. Cell 1988, 53, 679–686. [Google Scholar] [CrossRef]
- Hirose, S.; Hiraga, S.; Okazaki, T. Initiation site of deoxyribonucleotide polymerization at the replication origin of the Escherichia coli chromosome. Mol. Gen. Genet. 1983, 189, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, E.V.; Mirkin, S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Vilette, D.; Ehrlich, S.D.; Michel, B. Transcription-induced deletions in plasmid vectors: M13 DNA replication as a source of instability. Mol. Gen. Genet. 1996, 252, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Tehranchi, A.K.; Blankschien, M.D.; Zhang, Y.; Halliday, J.A.; Srivatsan, A.; Peng, J.; Herman, C.; Wang, J.D. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 2010, 141, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, E.V.; Roa, D.C.; Nudler, E.; Mirkin, S.M. Transcription regulatory elements are punctuation marks for DNA replication. Proc. Natl. Acad. Sci. USA 2006, 103, 7276–7281. [Google Scholar] [CrossRef] [PubMed]
- Furman, R.; Sevostyanova, A.; Artsimovitch, I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012, 40, 3392–3402. [Google Scholar] [CrossRef] [PubMed]
- Yuzenkova, Y.; Bochkareva, A.; Tadigotla, V.R.; Roghanian, M.; Zorov, S.; Severinov, K.; Zenkin, N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Mechold, U.; Potrykus, K.; Murphy, H.; Murakami, K.S.; Cashel, M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013, 41, 6175–6189. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Wang, Y.; Steitz, T.A. The mechanism of E. Coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 2013, 50, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Patlan, V.; Sekine, S.; Vassylyeva, M.N.; Hosaka, T.; Ochi, K.; Yokoyama, S.; Vassylyev, D.G. Structural basis for transcription regulation by alarmone ppGpp. Cell 2004, 117, 299–310. [Google Scholar] [CrossRef]
- Larson, M.H.; Mooney, R.A.; Peters, J.M.; Windgassen, T.; Nayak, D.; Gross, C.A.; Block, S.M.; Greenleaf, W.J.; Landick, R.; Weissman, J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 2014, 344, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, R.F.; Foskett, G. An estimate of the frequency of in vivo transcriptional errors at a nonsense codon in Escherichia coli. Mol. Gen. Genet. 1981, 183, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Libby, R.T.; Gallant, J.A. Phosphorolytic error correction during transcription. Mol. Microbiol. 1994, 12, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Herbert, K.M.; Zhou, J.; Mooney, R.A.; Porta, A.L.; Landick, R.; Block, S.M. E. Coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 2010, 399, 17–30. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zenkin, N.; Yuzenkova, Y. New Insights into the Functions of Transcription Factors that Bind the RNA Polymerase Secondary Channel. Biomolecules 2015, 5, 1195-1209. https://doi.org/10.3390/biom5031195
Zenkin N, Yuzenkova Y. New Insights into the Functions of Transcription Factors that Bind the RNA Polymerase Secondary Channel. Biomolecules. 2015; 5(3):1195-1209. https://doi.org/10.3390/biom5031195
Chicago/Turabian StyleZenkin, Nikolay, and Yulia Yuzenkova. 2015. "New Insights into the Functions of Transcription Factors that Bind the RNA Polymerase Secondary Channel" Biomolecules 5, no. 3: 1195-1209. https://doi.org/10.3390/biom5031195
APA StyleZenkin, N., & Yuzenkova, Y. (2015). New Insights into the Functions of Transcription Factors that Bind the RNA Polymerase Secondary Channel. Biomolecules, 5(3), 1195-1209. https://doi.org/10.3390/biom5031195





