Artificial Affinity Proteins as Ligands of Immunoglobulins
Abstract
:1. Introduction
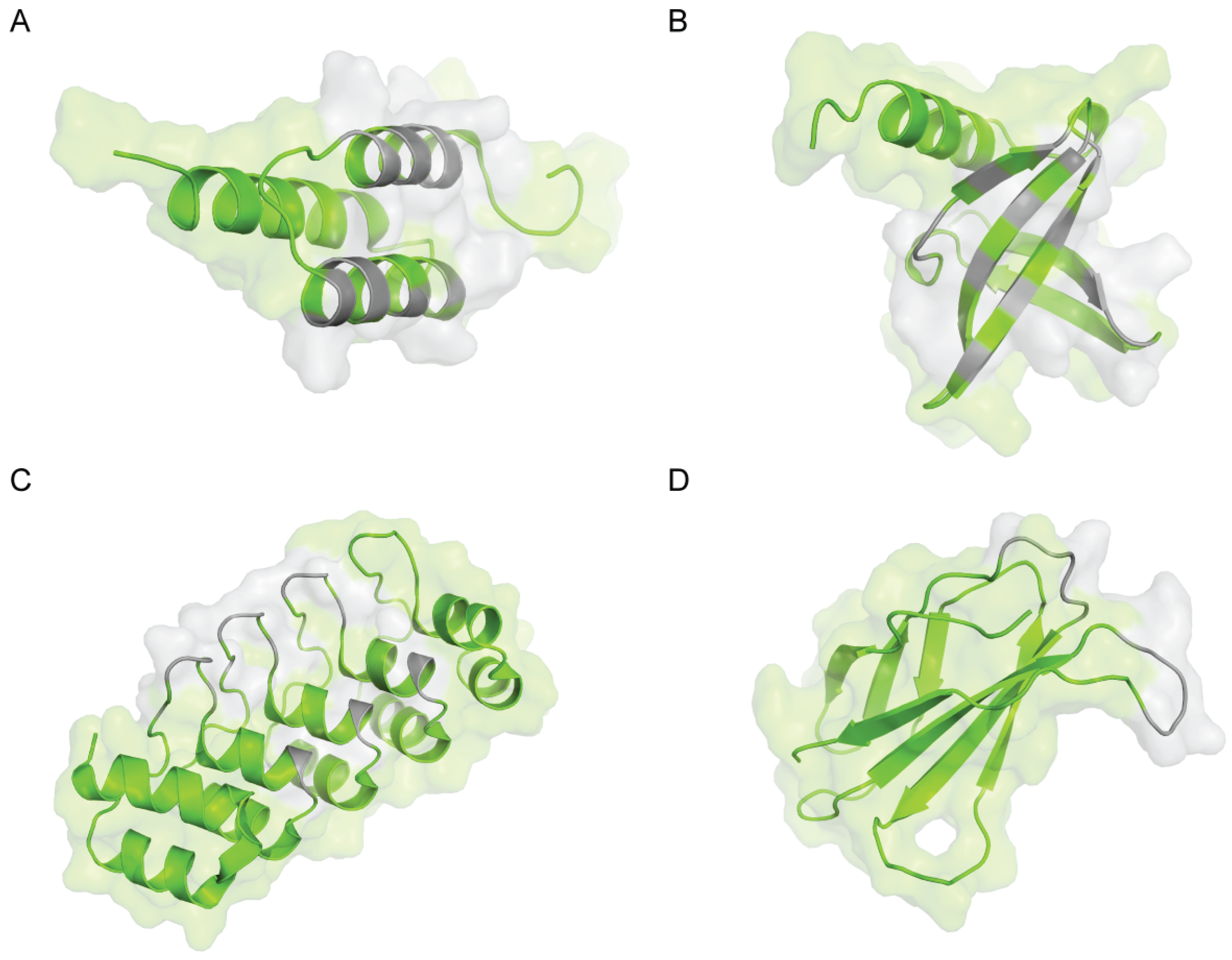
2. Z-domain of Staphylococcal Protein A (Affibody)
| Scaffold Name | Acronym | Size (aa) | Origin | Mutated Region | Disulfide Bridge | Ig specificity | KD (nM) | References |
|---|---|---|---|---|---|---|---|---|
| Z-domain of staphylococcal Protein A | Affibody | 58 | Bacteria (Staphylococcus aureus) | 13 aa on two α-helices | No | Human IgA | 500–3000 | [24,32,33] |
| Human IgE | 400 | |||||||
| Mouse IgG1 | 1.8–1300 | |||||||
| Archeal “7 kDa DNA binder” protein family | Affitin | ~65 | Archaea (Sulfolobus acidocaldarius, Sulfolobus solfataricus) | 10-14 aa on a β-sheet surface, and on two loops | No | Human IgG1,2,4 | 34–74 | [34,35,36,37] |
| Human IgG1–4 | 400–5280 | |||||||
| Mouse IgG | 26 | |||||||
| Chicken IgY | 30 | |||||||
| Rabbit IgG | 2638 | |||||||
| Carbohydrate-binding module | CBM | 168 | Bacteria (Rhodothermus marinus) | 12 aa at the binding site | No | Human IgG4 | N.D. | [38] |
| Designed ankyrin repeat protein | DARPin | 67 + (n × 33) | Artificial sequence from a consensus analysis | 7 aa on a β-turn and on an α-helix of repeats | No | Human IgG1 | 2.1–137 | [39,40,41] |
| Human IgE | 0.9–6.8 | |||||||
| Cystine-knot miniprotein | knottin (Min-23) | 23 | Plant (Ecballium elaterium) | 10 random aa inserted | Yes | Several monoclonal antibodies | 15.5 | [42] |
| Fibronectin type III domain | monobody | 94 | Human | Various number of aa randomized in three loops | No | Goat IgG | 1.2–35 | [43,44] |
| Bovin IgG | N.D. | |||||||
| Rabbit IgG | 0.051–1.08 | |||||||
| Mouse IgG | 4.1 |
3. Archeal “7 kDa DNA Binder” Protein Family (Affitin, Commercial Name: Nanofitin)
4. Carbohydrate-Binding Module (CBM)
5. Designed Ankyrin Repeat Protein (DARPin)
6. Cystine-Knot Miniprotein (Knottin)
7. Fibronectin Type III Domain (Monobody, Commercial Name: Adnectin)
8. Computationally-Designed IgG-Binding Protein
9. Other Alternative Scaffolds
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Roque, A.C.; Silva, C.S.; Taipa, M.A. Affinity-based methodologies and ligands for antibody purification: Advances and perspectives. J. Chromatogr. A 2007, 1160, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Bjorck, L.; Kronvall, G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol. 1984, 133, 969–974. [Google Scholar] [PubMed]
- Bjorck, L. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J. Immunol. 1988, 140, 1194–1197. [Google Scholar] [PubMed]
- Forsgren, A.; Sjoquist, J. “Protein A” from S. aureus. I. Pseudo-immune reaction with human γ-globulin. J. Immunol. 1966, 97, 822–827. [Google Scholar] [PubMed]
- Gulich, S.; Linhult, M.; Nygren, P.; Uhlen, M.; Hober, S. Stability towards alkaline conditions can be engineered into a protein ligand. J. Biotechnol. 2000, 80, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Gulich, S.; Linhult, M.; Stahl, S.; Hober, S. Engineering streptococcal protein G for increased alkaline stability. Protein Eng. 2002, 15, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Linhult, M.; Gulich, S.; Graslund, T.; Simon, A.; Karlsson, M.; Sjoberg, A.; Nord, K.; Hober, S. Improving the tolerance of a protein a analogue to repeated alkaline exposures using a bypass mutagenesis approach. Proteins 2004, 55, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.; Angus, K.; Taylor, L.; Warwicker, J.; Derrick, J.P. Design of stability at extreme alkaline pH in streptococcal protein G. J. Biotechnol. 2008, 134, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Carter-Franklin, J.N.; Victa, C.; McDonald, P.; Fahrner, R. Fragments of protein A eluted during protein A affinity chromatography. J. Chromatogr. A 2007, 1163, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Hanes, J.; Pluckthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 1997, 94, 4937–4942. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Aramini, J.M.; Montelione, G.T. Validation of helical tilt angles in the solution NMR structure of the Z domain of Staphylococcal protein A by combined analysis of residual dipolar coupling and NOE data. Protein Sci. 2004, 13, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Robinson, H.; Gao, Y.G.; McCrary, B.S.; Edmondson, S.P.; Shriver, J.W.; Wang, A.H. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 1998, 392, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Kohl, A.; Binz, H.K.; Forrer, P.; Stumpp, M.T.; Pluckthun, A.; Grutter, M.G. Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. USA 2003, 100, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Leahy, D.J.; Aukhil, I.; Erickson, H.P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 1996, 84, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Skerra, A. Alternative non-antibody scaffolds for molecular recognition. Curr. Opin. Biotechnol. 2007, 18, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, M.; Skerra, A. Engineered protein scaffolds as next-generation antibody therapeutics. Curr. Opin. Chem. Biol. 2009, 13, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Amstutz, P.; Pluckthun, A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 2005, 23, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Pluckthun, A. Engineered proteins as specific binding reagents. Curr. Opin. Biotechnol. 2005, 16, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Jost, C.; Pluckthun, A. Engineered proteins with desired specificity: DARPins, other alternative scaffolds and bispecific IgGs. Curr. Opin. Struct. Biol. 2014, 27, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Guss, B.; Nilsson, B.; Gatenbeck, S.; Philipson, L.; Lindberg, M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J. Biol. Chem. 1984, 259, 1695–1702. [Google Scholar] [PubMed]
- Nilsson, B.; Moks, T.; Jansson, B.; Abrahmsen, L.; Elmblad, A.; Holmgren, E.; Henrichson, C.; Jones, T.A.; Uhlen, M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987, 1, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Nord, K.; Nilsson, J.; Nilsson, B.; Uhlen, M.; Nygren, P.A. A combinatorial library of an α-helical bacterial receptor domain. Protein Eng. 1995, 8, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Grimm, S.; Yu, F.; Nygren, P.A. Ribosome display selection of a murine IgG1 Fab binding affibody molecule allowing species selective recovery of monoclonal antibodies. Mol. Biotechnol. 2011, 48, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Wikman, M.; Steffen, A.C.; Gunneriusson, E.; Tolmachev, V.; Adams, G.P.; Carlsson, J.; Stahl, S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 2004, 17, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Orlova, A.; Magnusson, M.; Eriksson, T.L.; Nilsson, M.; Larsson, B.; Hoiden-Guthenberg, I.; Widstrom, C.; Carlsson, J.; Tolmachev, V.; Stahl, S.; et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006, 66, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Lendel, C.; Dincbas-Renqvist, V.; Flores, A.; Wahlberg, E.; Dogan, J.; Nygren, P.A.; Hard, T. Biophysical characterization of Z(SPA-1)—A phage-display selected binder to protein A. Protein Sci. 2004, 13, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Hogbom, M.; Eklund, M.; Nygren, P.A.; Nordlund, P. Structural basis for recognition by an in vitro evolved affibody. Proc. Natl. Acad. Sci. USA 2003, 100, 3191–3196. [Google Scholar] [CrossRef] [PubMed]
- Feldwisch, J.; Tolmachev, V.; Lendel, C.; Herne, N.; Sjoberg, A.; Larsson, B.; Rosik, D.; Lindqvist, E.; Fant, G.; Hoiden-Guthenberg, I.; et al. Design of an optimized scaffold for affibody molecules. J. Mol. Biol. 2010, 398, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Löfblom, J.; Feldwisch, J.; Tolmachev, V.; Carlsson, J.; Ståhl, S.; Frejd, F.Y. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010, 584, 2670–2680. [Google Scholar] [CrossRef] [PubMed]
- Seijsing, J.; Lindborg, M.; Hoiden-Guthenberg, I.; Bonisch, H.; Guneriusson, E.; Frejd, F.Y.; Abrahmsen, L.; Ekblad, C.; Lofblom, J.; Uhlen, M.; et al. An engineered affibody molecule with pH-dependent binding to FcRn mediates extended circulatory half-life of a fusion protein. Proc. Natl. Acad. Sci. USA 2014, 111, 17110–17115. [Google Scholar] [CrossRef] [PubMed]
- Ronnmark, J.; Gronlund, H.; Uhlen, M.; Nygren, P.A. Human immunoglobulin A (IgA)-specific ligands from combinatorial engineering of protein A. Eur. J. Biochem. 2002, 269, 2647–2655. [Google Scholar] [CrossRef] [PubMed]
- Gunneriusson, E.; Samuelson, P.; Ringdahl, J.; Gronlund, H.; Nygren, P.A.; Stahl, S. Staphylococcal surface display of immunoglobulin A (IgA)- and IgE-specific in vitro-selected binding proteins (affibodies) based on Staphylococcus aureus protein A. Appl. Environ. Microbiol. 1999, 65, 4134–4140. [Google Scholar] [PubMed]
- Gera, N.; Hussain, M.; Wright, R.C.; Rao, B.M. Highly stable binding proteins derived from the hyperthermophilic Sso7d scaffold. J. Mol. Biol. 2011, 409, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Gera, N.; Hill, A.B.; White, D.P.; Carbonell, R.G.; Rao, B.M. Design of pH Sensitive Binding Proteins from the Hyperthermophilic Sso7d Scaffold. PLOS ONE 2012, 7, e48928. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Gera, N.; Hill, A.B.; Rao, B.M. Scaffold diversification enhances effectiveness of a superlibrary of hyperthermophilic proteins. ACS Synth. Biol. 2013, 2, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Béhar, G.; Bellinzoni, M.; Maillasson, M.; Paillard-Laurance, L.; Alzari, P.M.; He, X.; Mouratou, B.; Pecorari, F. Tolerance of the archaeal Sac7d scaffold protein to alternative library designs: Characterization of anti-immunoglobulin G Affitins. Protein Eng. Des. Sel. 2013, 26, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Cicortas Gunnarsson, L.; Nordberg Karlsson, E.; Albrekt, A.S.; Andersson, M.; Holst, O.; Ohlin, M. A carbohydrate binding module as a diversity-carrying scaffold. Protein Eng. Des. Sel. 2004, 17, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Steiner, D.; Forrer, P.; Pluckthun, A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J. Mol. Biol. 2008, 382, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Baumann, M.J.; Eggel, A.; Amstutz, P.; Stadler, B.M.; Vogel, M. DARPins against a functional IgE epitope. Immunol. Lett. 2010, 133, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Eggel, A.; Buschor, P.; Baumann, M.J.; Amstutz, P.; Stadler, B.M.; Vogel, M. Inhibition of ongoing allergic reactions using a novel anti-IgE DARPin-Fc fusion protein. Allergy 2011, 66, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Souriau, C.; Chiche, L.; Irving, R.; Hudson, P. New binding specificities derived from Min-23, a small cystine-stabilized peptidic scaffold. Biochemistry 2005, 44, 7143–7155. [Google Scholar] [CrossRef] [PubMed]
- Hackel, B.J.; Ackerman, M.E.; Howland, S.W.; Wittrup, K.D. Stability and CDR composition biases enrich binder functionality landscapes. J. Mol. Biol. 2010, 401, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Hackel, B.J.; Wittrup, K.D. The full amino acid repertoire is superior to serine/tyrosine for selection of high affinity immunoglobulin G binders from the fibronectin scaffold. Protein Eng. Des. Sel. 2010, 23, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Mouratou, B.; Schaeffer, F.; Guilvout, I.; Tello-Manigne, D.; Pugsley, A.P.; Alzari, P.M.; Pecorari, F. Remodeling a DNA-binding protein as a specific in vivo inhibitor of bacterial secretin PulD. Proc. Natl. Acad. Sci. USA 2007, 104, 17983–17988. [Google Scholar] [CrossRef] [PubMed]
- Pecorari, F.; Alzari, P.M. OB-Fold Used as Scaffold for Engineering New Specific Binders. Patent Publication Nos. PCT/IB2007/004388, 12 June 2008. [Google Scholar]
- Krehenbrink, M.; Chami, M.; Guilvout, I.; Alzari, P.M.; Pecorari, F.; Pugsley, A.P. Artificial binding proteins (Affitins) as probes for conformational changes in secretin PulD. J. Mol. Biol. 2008, 383, 1058–1068. [Google Scholar] [CrossRef] [PubMed]
- Béhar, G.; Pacheco, S.; Maillasson, M.; Mouratou, B.; Pecorari, F. Switching an anti-IgG binding site between archaeal extremophilic proteins results in Affitins with enhanced pH stability. J. Biotechnol. 2014, 192A, 123–129. [Google Scholar] [CrossRef]
- McCrary, B.S.; Edmondson, S.P.; Shriver, J.W. Hyperthermophile protein folding thermodynamics: Differential scanning calorimetry and chemical denaturation of Sac7d. J. Mol. Biol. 1996, 264, 784–805. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, S.P.; Shriver, J.W. DNA binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 2001, 334, 129–145. [Google Scholar] [PubMed]
- Correa, A.; Pacheco, S.; Mechaly, Ariel E.; Obal, G.; Béhar, G.; Mouratou, B.; Oppezzo, P.; Alzari, P.M.; Pecorari, F. Potent and specific inhibition of glycosidases by small artificial binding proteins (Affitins). PLOS ONE 2014, 9, e97438. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Lockney, D.; Wang, R.; Gera, N.; Rao, B.M. Avidity-mediated virus separation using a hyperthermophilic affinity ligand. Biotechnol. Prog. 2013, 29, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Buddelmeijer, N.; Krehenbrink, M.; Pecorari, F.; Pugsley, A.P. Type II secretion system secretin PulD localizes in clusters in the Escherichia coli outer membrane. J. Bacteriol. 2009, 191, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Cinier, M.; Petit, M.; Williams, M.N.; Fabre, R.M.; Pecorari, F.; Talham, D.R.; Bujoli, B.; Tellier, C. Bisphosphonate adaptors for specific protein binding on zirconium phosphonate-based microarrays. Bioconjug. Chem. 2009, 20, 2270–2277. [Google Scholar] [CrossRef] [PubMed]
- Miranda, F.F.; Brient-Litzler, E.; Zidane, N.; Pecorari, F.; Bedouelle, H. Reagentless fluorescent biosensors from artificial families of antigen binding proteins. Biosens. Bioelectron. 2011, 26, 4184–4190. [Google Scholar] [CrossRef] [PubMed]
- Abou-Hachem, M.; Karlsson, E.N.; Simpson, P.J.; Linse, S.; Sellers, P.; Williamson, M.P.; Jamieson, S.J.; Gilbert, H.J.; Bolam, D.N.; Holst, O. Calcium binding and thermostability of carbohydrate binding module CBM4–2 of Xyn10A from Rhodothermus marinus. Biochemistry 2002, 41, 5720–5729. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, L.C.; Dexlin, L.; Karlsson, E.N.; Holst, O.; Ohlin, M. Evolution of a carbohydrate binding module into a protein-specific binder. Biomol. Eng. 2006, 23, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mahajan, A.; Tsai, M.D. Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry 2006, 45, 15168–15178. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Stumpp, M.T.; Forrer, P.; Amstutz, P.; Pluckthun, A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003, 332, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Schilling, J.; Schöppe, J.; Plückthun, A. From DARPins to LoopDARPins: Novel LoopDARPin design allows the selection of low picomolar binders in a single round of ribosome display. J. Mol. Biol. 2014, 426, 691–721. [Google Scholar] [CrossRef] [PubMed]
- Zahnd, C.; Wyler, E.; Schwenk, J.M.; Steiner, D.; Lawrence, M.C.; McKern, N.M.; Pecorari, F.; Ward, C.W.; Joos, T.O.; Pluckthun, A. A designed ankyrin repeat protein evolved to picomolar affinity to HER2. J. Mol. Biol. 2007, 369, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Amstutz, P.; Kohl, A.; Stumpp, M.T.; Briand, C.; Forrer, P.; Grutter, M.G.; Pluckthun, A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 2004, 22, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Kohl, A.; Amstutz, P.; Parizek, P.; Binz, H.K.; Briand, C.; Capitani, G.; Forrer, P.; Pluckthun, A.; Grutter, M.G. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure 2005, 13, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, P.; Binz, H.K.; Parizek, P.; Stumpp, M.T.; Kohl, A.; Grutter, M.G.; Forrer, P.; Pluckthun, A. Intracellular kinase inhibitors selected from combinatorial libraries of designed ankyrin repeat proteins. J. Biol. Chem. 2005, 280, 24715–24722. [Google Scholar] [CrossRef] [PubMed]
- Zahnd, C.; Pecorari, F.; Straumann, N.; Wyler, E.; Pluckthun, A. Selection and characterisation of HER2-binding designed ankyrin repeat proteins. J. Biol. Chem. 2006, 281, 9. [Google Scholar] [CrossRef] [PubMed]
- Zahnd, C.; Kawe, M.; Stumpp, M.T.; de Pasquale, C.; Tamaskovic, R.; Nagy-Davidescu, G.; Dreier, B.; Schibli, R.; Binz, H.K.; Waibel, R.; et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: Effects of affinity and molecular size. Cancer Res. 2010, 70, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, R.; Sosabowski, J.; Livanos, M.; Leyton, J.; Vigor, K.; Bhavsar, G.; Nagy-Davidescu, G.; Rashid, M.; Miranda, E.; Yeung, J.; et al. Development of the designed ankyrin repeat protein (DARPin) G3 for HER2 molecular imaging. Eur. J. Nucl. Med. Mol. Imaging 2014. [Google Scholar] [CrossRef]
- Martin-Killias, P.; Stefan, N.; Rothschild, S.; Pluckthun, A.; Zangemeister-Wittke, U. A novel fusion toxin derived from an EpCAM-specific designed ankyrin repeat protein has potent antitumor activity. Clin. Cancer Res. 2011, 17, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Dreier, B.; Mikheeva, G.; Belousova, N.; Parizek, P.; Boczek, E.; Jelesarov, I.; Forrer, P.; Pluckthun, A.; Krasnykh, V. Her2-specific multivalent adapters confer designed tropism to adenovirus for gene targeting. J. Mol. Biol. 2011, 405, 410–426. [Google Scholar] [CrossRef] [PubMed]
- Colgrave, M.L.; Craik, D.J. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 2004, 43, 5965–5975. [Google Scholar] [CrossRef] [PubMed]
- Pallaghy, P.K.; Nielsen, K.J.; Craik, D.J.; Norton, R.S. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef] [PubMed]
- Heitz, A.; le-Nguyen, D.; Chiche, L. Min-21 and min-23, the smallest peptides that fold like a cystine-stabilized β-sheet motif: Design, solution structure, and thermal stability. Biochemistry 1999, 38, 10615–10625. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Bailey, C.W.; Huang, X.; Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998, 284, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Aha, P.; Gu, K.; Kuimelis, R.G.; Kurz, M.; Lam, T.; Lim, A.C.; Liu, H.; Lohse, P.A.; Sun, L.; et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 2002, 9, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Wojcik, J.; Gilbreth, R.N.; Hoey, R.J.; Koide, S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 2012, 415, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Hackel, B.J.; Kapila, A.; Wittrup, K.D. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J. Mol. Biol. 2008, 381, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Koide, S. Monobodies: Antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol. Biol. 2007, 352, 95–109. [Google Scholar] [PubMed]
- Koide, A.; Gilbreth, R.N.; Esaki, K.; Tereshko, V.; Koide, S. High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl. Acad. Sci. USA 2007, 104, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Fleishman, S.J.; Whitehead, T.A.; Ekiert, D.C.; Dreyfus, C.; Corn, J.E.; Strauch, E.M.; Wilson, I.A.; Baker, D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 2011, 332, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Strauch, E.M.; Fleishman, S.J.; Baker, D. Computational design of a pH-sensitive IgG binding protein. Proc. Natl. Acad. Sci. USA 2014, 111, 675–680. [Google Scholar] [CrossRef] [PubMed]
- McConnell, S.J.; Hoess, R.H. Tendamistat as a scaffold for conformationally constrained phage peptide libraries. J. Mol. Biol. 1995, 250, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Xue, Y.; Zhang, J.; Gao, B.; Feng, J.; Mao, C.; Zheng, L.; Liu, N.; Wang, F.; Wang, H. A conformation-constrained peptide library based on insect defensin A. Peptides 2004, 25, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Legendre, D.; Soumillion, P.; Fastrez, J. Engineering a regulatable enzyme for homogeneous immunoassays. Nat. Biotechnol. 1999, 17, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Folgori, A.; Wallace, A.; Nicotra, M.; Acali, S.; Phalipon, A.; Barbato, G.; Bazzo, R.; Cortese, R.; Felici, F.; et al. A conformationally homogeneous combinatorial peptide library. J. Mol. Biol. 1995, 247, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Murray, K.S.; van Cleave, V.; LaVallie, E.R.; Stahl, M.L.; McCoy, J.M. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: A system designed for exploring protein-protein interactions. Nat. Biotechnol. 1995, 13, 366–372. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mouratou, B.; Béhar, G.; Pecorari, F. Artificial Affinity Proteins as Ligands of Immunoglobulins. Biomolecules 2015, 5, 60-75. https://doi.org/10.3390/biom5010060
Mouratou B, Béhar G, Pecorari F. Artificial Affinity Proteins as Ligands of Immunoglobulins. Biomolecules. 2015; 5(1):60-75. https://doi.org/10.3390/biom5010060
Chicago/Turabian StyleMouratou, Barbara, Ghislaine Béhar, and Frédéric Pecorari. 2015. "Artificial Affinity Proteins as Ligands of Immunoglobulins" Biomolecules 5, no. 1: 60-75. https://doi.org/10.3390/biom5010060
APA StyleMouratou, B., Béhar, G., & Pecorari, F. (2015). Artificial Affinity Proteins as Ligands of Immunoglobulins. Biomolecules, 5(1), 60-75. https://doi.org/10.3390/biom5010060




