Decarboxylation of Pyruvate to Acetaldehyde for Ethanol Production by Hyperthermophiles
Abstract
:1. Introduction
2. Microbial Production of Ethanol
3. Key Enzymes Involved in Ethanol Production
4. Pathways for the Production of Acetaldehyde from Pyruvate
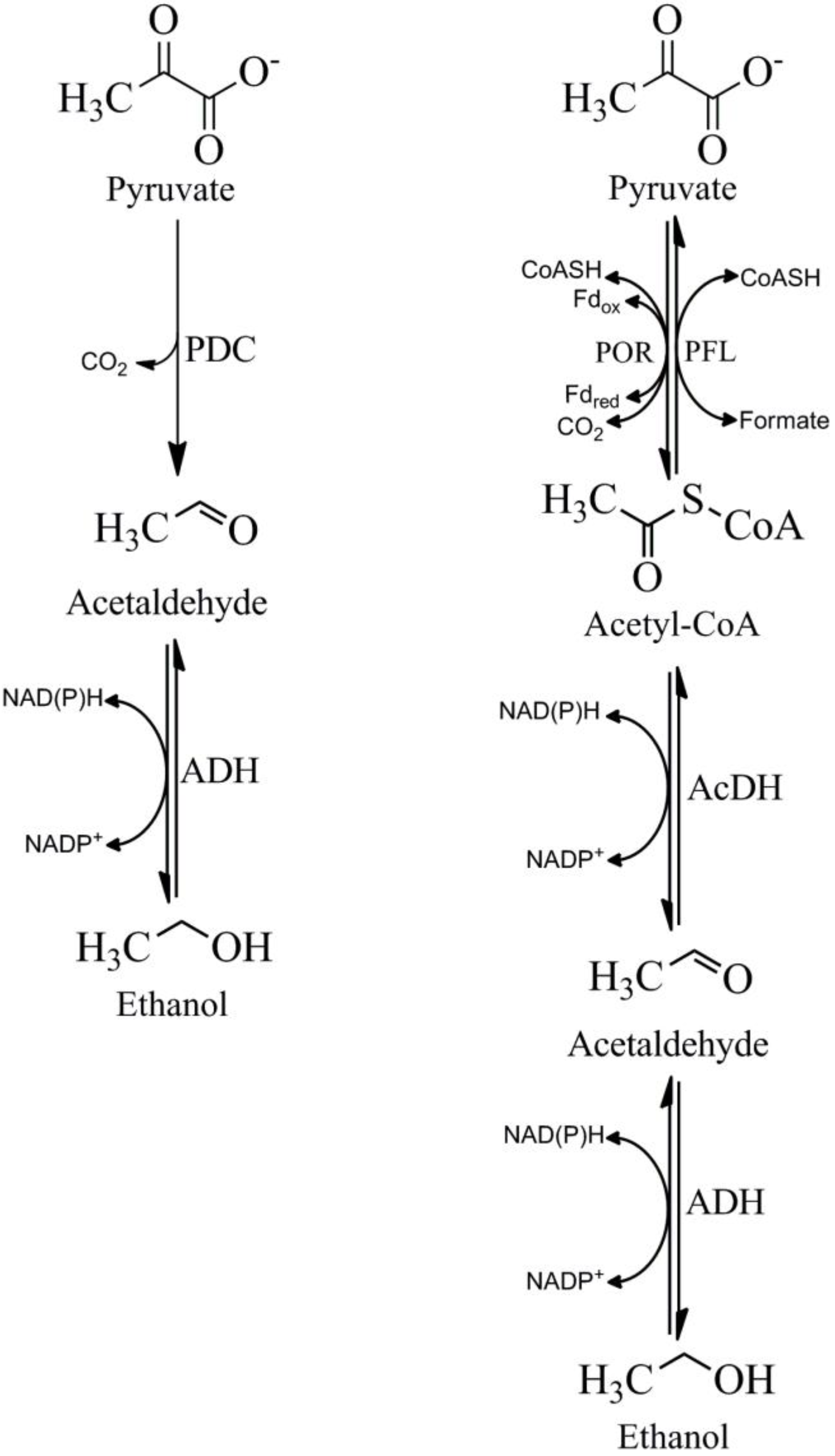
5. Pyruvate Decarboxylase (PDC)
6. Pyruvate Ferredoxin Oxidoreductase (POR)
7. POR/PDC Bi-Functional Enzyme
8. Acetaldehyde Dehydrogenase (CoA-Acetylating)
9. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wiegel, J. Temperature spans for growth: Hypothesis and discussion. FEMS Microbiol. Lett. 1990, 75, 155–169. [Google Scholar]
- Charlier, D.; Droogmans, L. Microbial Life at high temperature, the challenges, the strategies. Cell. Mol. Life Sci. 2005, 62, 2974–2984. [Google Scholar] [CrossRef]
- Stetter, K. History of discovery of the first hyperthermophiles. Extremophiles 2006, 10, 357–362. [Google Scholar] [CrossRef]
- Lebedinsky, A.; Chernyh, N.; Bonch-Osmolovskaya, E. Phylogenetic systematics of microorganisms inhabiting thermal environments. Biochemistry (Mosc.) 2007, 72, 1299–1312. [Google Scholar] [CrossRef]
- Wagner, I.D.; Wiegel, J. Diversity of Thermophilic Anaerobes. Ann. NY Acad. Sci. 2008, 1125, 1–43. [Google Scholar] [CrossRef]
- Morozkina, E.; Slutskaya, E.; Fedorova, T.; Tugay, T.; Golubeva, L.; Koroleva, O. Extremophilic microorganisms: Biochemical adaptation and biotechnological application. Appl. Biochem. Microbiol. 2010, 46, 1–14. [Google Scholar] [CrossRef]
- Sommer, P.; Georgieva, T.; Ahring, B.K. Potential for using thermophilic anaerobic bacteria for bioethanol production from hemicellulose. Biochem. Soc. Trans. 2004, 32, 283–289. [Google Scholar] [CrossRef]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Egorova, K.; Antranikian, G. Industrial relevance of thermophilic Archaea. Curr. Opin. Microbiol. 2005, 8, 649–655. [Google Scholar] [CrossRef]
- Van den Burg, B. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 2003, 6, 213–218. [Google Scholar] [CrossRef]
- Atomi, H.; Sato, T.; Kanai, T. Application of hyperthermophiles and their enzymes. Curr. Opin. Biotechnol. 2011, 22, 618–626. [Google Scholar] [CrossRef]
- Lynd, L.R.; Wyman, C.E.; Gerngross, T.U. Biocommodity engineering. Biotechnol. Prog. 1999, 15, 777–793. [Google Scholar] [CrossRef]
- Zaldivar, J.; Nielsen, J.; Olsson, L. Fuel ethanol production from lignocellulose: A challenge for metabolic engineering and process integration. Appl. Microbiol. Biotechnol. 2001, 56, 17–34. [Google Scholar] [CrossRef]
- Mabee, W.E.; Saddler, J.N. Bioethanol from lignocellulosics: Status and perspectives in Canada. Bioresour. Technol. 2010, 101, 4806–4813. [Google Scholar] [CrossRef]
- Dien, B.S.; Cotta, M.A.; Jeffries, T.W. Bacteria engineered for fuel ethanol production: Current status. Appl. Microbiol. Biotechnol. 2003, 63, 258–266. [Google Scholar] [CrossRef]
- Buschke, N.; Schäfer, R.; Becker, J.; Wittmann, C. Metabolic engineering of industrial platform microorganisms for biorefinery applications-optimization of substrate spectrum and process robustness by rational and evolutive strategies. Bioresour. Technol. 2012, 135, 544–554. [Google Scholar]
- Jang, Y.-S.; Park, J.M.; Choi, S.; Choi, Y.J.; Seung, D.Y.; Cho, J.H.; Lee, S.Y. Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol. Adv. 2012, 30, 989–1000. [Google Scholar] [CrossRef]
- Taylor, M.P.; Eley, K.L.; Martin, S.; Tuffin, M.I.; Burton, S.G.; Cowan, D.A. Thermophilic ethanologenesis: Future prospects for second-generation bioethanol production. Trends Biotechnol. 2009, 27, 398–405. [Google Scholar] [CrossRef]
- Bustard, M.T.; Burgess, J.G.; Meeyoo, V.; Wright, P.C. Novel opportunities for marine hyperthermophiles in emerging biotechnology and engineering industries. J. Chem. Technol. Biotechnol. 2000, 75, 1095–1109. [Google Scholar] [CrossRef]
- Huber, H.; Stetter, K.O. Hyperthermophiles and their possible potential in biotechnology. J. Biotechnol. 1998, 64, 39–52. [Google Scholar] [CrossRef]
- Schiraldi, C.; de Rosa, M. The production of biocatalysts and biomolecules from extremophiles. Trends Biotechnol. 2002, 20, 515–521. [Google Scholar] [CrossRef]
- Hough, D.W.; Danson, M.J. Extremozymes. Curr. Opin. Chem. Biol. 1999, 3, 39–46. [Google Scholar] [CrossRef]
- Lamed, R.; Zeikus, J.G. Ethanol production by thermophilic bacteria: Relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J. Bacteriol. 1980, 144, 569–578. [Google Scholar]
- Klapatch, T.R.; Hogsett, D.A.L.; Baskaran, S.; Pal, S.; Lynd, L.R. Organism development and characterization for ethanol production using thermophilic bacteria. Appl. Biochem. Biotechnol. 1994, 45–46, 209–223. [Google Scholar] [CrossRef]
- Chang, T.; Yao, S. Thermophilic, lignocellulolytic bacteria for ethanol production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 13–27. [Google Scholar] [CrossRef]
- Zeikus, J.G.; Ben-Bassat, A.; Ng, T.K.; Lamed, R.J. Thermophilic ethanol fermentations. In Trends in the Biology of Fermentations for Fuels and Chemicals; Hollaender, A., Rabson, R., Rogers, P., Pietro, A.S., Valentine, R., Wolfe, R., Eds.; Springer: New York, NY, USA, 1981; pp. 441–461. [Google Scholar]
- Demain, A.L.; Newcomb, M.; Wu, J.H.D. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 2005, 69, 124–154. [Google Scholar] [CrossRef]
- Barnard, D.; Casanueva, A.; Tuffin, M.; Cowan, D. Extremophiles in biofuel synthesis. Environ. Technol. 2010, 31, 871–888. [Google Scholar] [CrossRef]
- Tomás, A.F.; Karagöz, P.; Karakashev, D.; Angelidaki, I. Extreme thermophilic ethanol production from rapeseed straw: Using the newly isolated Thermoanaerobacter pentosaceus and combining it with Saccharomyces cerevisiae in a two-step process. Biotechnol. Bioeng. 2013, 110, 1574–1582. [Google Scholar] [CrossRef]
- Shaw, A.J.; Podkaminer, K.K.; Desai, S.G.; Bardsley, J.S.; Rogers, S.R.; Thorne, P.G.; Hogsett, D.A.; Lynd, L.R. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. USA 2008, 105, 13769–13774. [Google Scholar] [CrossRef]
- Yao, S.; Mikkelsen, M.J. Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in Thermoanaerobacter mathranii. J. Mol. Microbiol. Biotechnol. 2010, 19, 123–133. [Google Scholar] [CrossRef]
- Svetlitchnyi, V.; Kensch, O.; Falkenhan, D.; Korseska, S.; Lippert, N.; Prinz, M.; Sassi, J.; Schickor, A.; Curvers, S. Single-step ethanol production from lignocellulose using novel extremely thermophilic bacteria. Biotechnol. Biofuels 2013, 6, 1–15. [Google Scholar] [CrossRef]
- Yao, S.; Mikkelsen, M. Metabolic engineering to improve ethanol production in Thermoanaerobacter mathranii. Appl. Microbiol. Biotechnol. 2010, 88, 199–208. [Google Scholar] [CrossRef]
- Thompson, A.; Studholme, D.; Green, E.; Leak, D. Heterologous expression of pyruvate decarboxylase in Geobacillus thermoglucosidasius. Biotechnol. Lett. 2008, 30, 1359–1365. [Google Scholar] [CrossRef]
- Cripps, R.E.; Eley, K.; Leak, D.J.; Rudd, B.; Taylor, M.; Todd, M.; Boakes, S.; Martin, S.; Atkinson, T. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab. Eng. 2009, 11, 398–408. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Patel, B.K.; Mah, R.A.; Baresi, L. Caldicellulosiruptor owensensis sp. nov., an anaerobic, extremely thermophilic, xylanolytic bacterium. Int. J. Syst. Bacteriol. 1998, 48, 91–97. [Google Scholar] [CrossRef]
- Bredholt, S.; Sonne-Hansen, J.; Nielsen, P.; Mathrani, I.M.; Ahring, B.K. Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int. J. Syst. Bacteriol. 1999, 49, 991–996. [Google Scholar] [CrossRef]
- Van Niel, E.W.J.; Claassen, P.A.M.; Stams, A.J.M. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 2003, 81, 255–262. [Google Scholar] [CrossRef]
- Kengen, S.; de Bok, F.; van Loo, N.; Dijkema, C.; Stams, A.; de Vos, W. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J. Biol. Chem. 1994, 269, 17537–17541. [Google Scholar]
- Ma, K.; Loessner, H.; Heider, J.; Johnson, M.; Adams, M. Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: Characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. J. Bacteriol. 1995, 177, 4748–4756. [Google Scholar]
- Ying, X.; Ma, K. Characterization of a zinc-containing alcohol dehydrogenase with stereoselectivity from the hyperthermophilic archaeon Thermococcus guaymasensis. J. Bacteriol. 2011, 193, 3009–3019. [Google Scholar] [CrossRef]
- Moon, Y.-J.; Kwon, J.; Yun, S.-H.; Lim, H.L.; Kim, M.-S.; Kang, S.G.; Lee, J.-H.; Choi, J.-S.; Kim, S.L.; Chung, Y.-H. Proteome analyses of hydrogen-producing hyperthermophilic archaeon Thermococcus onnurineus NA1 in different one-carbon substrate culture conditions. Mol. Cell. Proteomics 2012. [Google Scholar] [CrossRef]
- Fardeau, M.L.; Ollivier, B.; Patel, B.K.C.; Magot, M.; Thomas, P.; Rimbault, A.; Rocchiccioli, F.; Garcia, J.L. Thermotoga hypogea sp. nov., a xylanolytic, thermophilic bacterium from an oil-producing well. Int. J. Syst. Bacteriol. 1997, 47, 1013–1019. [Google Scholar] [CrossRef]
- Balk, M.; Weijma, J.; Stams, A.J.M. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int. J. Syst. Evol. Microbiol. 2002, 52, 1361–1368. [Google Scholar] [CrossRef]
- De Vrije, T.; Bakker, R.; Budde, M.; Lai, M.; Mars, A.; Claassen, P. Efficient hydrogen production from the lignocellulosic energy crop Miscanthus by the extreme thermophilic bacteria Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Biotechnol. Biofuels 2009, 2, e12. [Google Scholar] [CrossRef]
- DiPippo, J.L.; Nesbo, C.L.; Dahle, H.; Doolittle, W.F.; Birkland, N.-K.; Noll, K.M. Kosmotoga olearia gen. nov., sp. nov., a thermophilic, anaerobic heterotroph isolated from an oil production fluid. Int. J. Syst. Evol. Microbiol. 2009, 59, 2991–3000. [Google Scholar] [CrossRef]
- Podosokorskaya, O.A.; Kublanov, I.V.; Reysenbach, A.L.; Kolganova, T.V.; Bonch-Osmolovskaya, E.A. Thermosipho affectus sp. nov., a thermophilic, anaerobic, cellulolytic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2011, 61, 1160–1164. [Google Scholar] [CrossRef]
- Reid, M.F.; Fewson, C.A. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 1994, 20, 13–56. [Google Scholar] [CrossRef]
- Littlechild, J.A.; Guy, J.E.; Isupov, M.N. Hyperthermophilic dehydrogenase enzymes. Biochem. Soc. Trans. 2004, 32, 255–258. [Google Scholar] [CrossRef]
- Radianingtyas, H.; Wright, P.C. Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol. Rev. 2003, 27, 593–616. [Google Scholar] [CrossRef]
- Korkhin, Y.; Kalb, A.J.; Peretz, M.; Bogin, O.; Burstein, Y.; Frolow, F. NADP-dependent bacterial alcohol dehydrogenases: Crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii. J. Mol. Biol. 1998, 278, 967–981. [Google Scholar] [CrossRef]
- Machielsen, R.; Uria, A.R.; Kengen, S.W.M.; van der Oost, J. Production and characterization of a thermostable alcohol dehydrogenase that belongs to the aldo-keto reductase superfamily. Appl. Environ. Microbiol. 2006, 72, 233–238. [Google Scholar] [CrossRef]
- Van der Oost, J.; Voorhorst, W.G.B.; Kengen, S.W.M.; Geerling, A.C.M.; Wittenhorst, V.; Gueguen, Y.; de Vos, W.M. Genetic and biochemical characterization of a short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur. J. Biochem. 2001, 268, 3062–3068. [Google Scholar] [CrossRef]
- Antoine, E.; Rolland, J.-L.; Raffin, J.-P.; Dietrich, J. Cloning and over-expression in Escherichia coli of the gene encoding NADPH group III alcohol dehydrogenase from Thermococcus hydrothermalis. Eur. J. Biochem. 1999, 264, 880–889. [Google Scholar] [CrossRef]
- Bashir, Q.; Rashid, N.; Jamil, F.; Imanaka, T.; Akhtar, M. Highly thermostable l-threonine dehydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Biochem. (Tokyo) 2009, 146, 95–102. [Google Scholar] [CrossRef]
- Bowyer, A.; Mikolajek, H.; Stuart, J.W.; Wood, S.P.; Jamil, F.; Rashid, N.; Akhtar, M.; Cooper, J.B. Structure and function of the l-threonine dehydrogenase (TkTDH) from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Struct. Biol. 2009, 168, 294–304. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, C.; Orita, I.; Imanaka, T.; Fukui, T.; Xing, X.-H. Thermostable alcohol dehydrogenase from Thermococcus kodakarensis KOD1 for enantioselective bioconversion of aromatic secondary alcohols. Appl. Environ. Microbiol. 2013, 79, 2209–2217. [Google Scholar] [CrossRef]
- Stekhanova, T.N.; Mardanov, A.V.; Bezsudnova, E.Y.; Gumerov, V.M.; Ravin, N.V.; Skryabin, K.G.; Popov, V.O. Characterization of a thermostable short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Thermococcus sibiricus. Appl. Environ. Microbiol. 2010, 76, 4096–4098. [Google Scholar] [CrossRef]
- Lyashenko, A.V.; Bezsudnova, E.Y.; Gumerov, V.M.; Lashkov, A.A.; Mardanov, A.V.; Mikhailov, A.M.; Polyakov, K.M.; Popov, V.O.; Ravin, N.V.; Skryabin, K.G.; et al. Expression, purification and crystallization of a thermostable short-chain alcohol dehydrogenase from the archaeon Thermococcus sibiricus. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2010, 66, 655–657. [Google Scholar] [CrossRef]
- Pennacchio, A.; Giordano, A.; Pucci, B.; Rossi, M.; Raia, C. Biochemical characterization of a recombinant short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius. Extremophiles 2010, 14, 193–204. [Google Scholar] [CrossRef]
- Ying, X.; Grunden, A.; Nie, L.; Adams, M.; Ma, K. Molecular characterization of the recombinant iron-containing alcohol dehydrogenase from the hyperthermophilic Archaeon, Thermococcus strain ES1. Extremophiles 2009, 13, 299–311. [Google Scholar] [CrossRef]
- Guy, J.E.; Isupov, M.N.; Littlechild, J.A. The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix. J. Mol. Biol. 2003, 331, 1041–1051. [Google Scholar] [CrossRef]
- Ying, X.; Wang, Y.; Badiei, H.; Karanassios, V.; Ma, K. Purification and characterization of an iron-containing alcohol dehydrogenase in extremely thermophilic bacterium Thermotoga hypogea. Arch. Microbiol. 2007, 187, 499–510. [Google Scholar] [CrossRef]
- Vitale, A.; Thorne, N.; Lovell, S.; Battaile, K.P.; Hu, X.; Shen, M.; D’Auria, S.; Auld, D.S. Physicochemical characterization of a thermostable alcohol dehydrogenase from Pyrobaculum aerophilum. PLoS One 2013, 8, e63828. [Google Scholar]
- Verhees, C.H.; Kengen, S.W.M.; Tuininga, J.E.; Schut, G.J.; Adams, M.W.W.; de Vos, W.M.; van der Oost, J. The unique features of glycolytic pathways in Archaea. Biochem. J. 2003, 375, 231–246. [Google Scholar] [CrossRef]
- Siebers, B.; Schönheit, P. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 2005, 8, 695–705. [Google Scholar] [CrossRef]
- Buchholz, S.E.; Dooley, M.M.; Eveleigh, D.E. Zymomonas—An alcoholic enigma. Trends Biotechnol. 1987, 5, 199–204. [Google Scholar] [CrossRef]
- Canale-Parola, E. Biology of the sugar-fermenting Sarcinae. Microbiol. Mol. Biol. Rev. 1970, 34, 82–97. [Google Scholar]
- Duggleby, R.G. Domain relationships in thiamine diphosphate-dependent enzymes. Acc. Chem. Res. 2006, 39, 550–557. [Google Scholar] [CrossRef]
- Costelloe, S.; Ward, J.; Dalby, P. Evolutionary analysis of the TPP-dependent enzyme family. J. Mol. Evol. 2008, 66, 36–49. [Google Scholar] [CrossRef]
- Kluger, R.; Tittmann, K. Thiamin diphosphate catalysis: Enzymic and nonenzymic covalent intermediates. Chem. Rev. 2008, 108, 1797–1833. [Google Scholar] [CrossRef]
- Schellenberger, A. Sixty years of thiamin diphosphate biochemistry. Biochim. Biophys. Acta 1998, 1385, 177–186. [Google Scholar] [CrossRef]
- Iding, H.; Siegert, P.; Mesch, K.; Pohl, M. Application of alpha-keto acid decarboxylases in biotransformations. Biochim. Biophys. Acta 1998, 1385, 307–322. [Google Scholar] [CrossRef]
- Bringer-Meyer, S.; Schimz, K.L.; Sahm, H. Pyruvate decarboxylase from Zymomonas mobilis. Isolation and partial characterization. Arch. Microbiol. 1986, 146, 105–110. [Google Scholar] [CrossRef]
- Raj, K.C.; Ingram, L.O.; Maupin-Furlow, J.A. Pyruvate decarboxylase: A key enzyme for the oxidative metabolism of lactic acid by Acetobacter pasteurianus. Arch. Microbiol. 2001, 176, 443–451. [Google Scholar] [CrossRef]
- Raj, K.C.; Talarico, L.A.; Ingram, L.O.; Maupin-Furlow, J.A. Cloning and characterization of the Zymobacter palmae pyruvate decarboxylase gene (pdc) and comparison to bacterial homologues. Appl. Environ. Microbiol. 2002, 68, 2869–2876. [Google Scholar] [CrossRef]
- Talarico, L.A.; Ingram, L.O.; Maupin-Furlow, J.A. Production of the Gram-positive Sarcina ventriculi pyruvate decarboxylase in Escherichia coli. Microbiology 2001, 147, 2425–2435. [Google Scholar]
- Wang, Q.; He, P.; Lu, D.; Shen, A.; Jiang, N. Purification, characterization, cloning and expression of pyruvate decarboxylase from Torulopsis glabrata IFO005. J. Biochem. (Tokyo) 2004, 136, 447–455. [Google Scholar] [CrossRef]
- Ma, K.; Hutchins, A.; Sung, S.-J.S.; Adams, M.W.W. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. USA 1997, 94, 9608–9613. [Google Scholar]
- Dobritzsch, D.; Konig, S.; Schneider, G.; Lu, G. High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases. J. Biol. Chem. 1998, 273, 20196–20204. [Google Scholar] [CrossRef]
- Jordan, F. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 2003, 20, 184–201. [Google Scholar] [CrossRef]
- Kluger, R. Thiamin diphosphate: A mechanistic update on enzymic and nonenzymic catalysis of decarboxylation. Chem. Rev. 1987, 87, 863–876. [Google Scholar] [CrossRef]
- Candy, J.M.; Duggleby, R.G. Structure and properties of pyruvate decarboxylase and site-directed mutagenesis of the Zymomonas mobilis enzyme. Biochim. Biophys. Acta 1998, 1385, 323–338. [Google Scholar] [CrossRef]
- Kutter, S.; Weiss, M.S.; Wille, G.; Golbik, R.; Spinka, M.; König, S. Covalently bound substrate at the regulatory site of yeast pyruvate decarboxylases triggers allosteric enzyme activation. J. Biol. Chem. 2009, 284, 12136–12144. [Google Scholar]
- Siegert, P.; McLeish, M.J.; Baumann, M.; Iding, H.; Kneen, M.M.; Kenyon, G.L.; Pohl, M. Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein Eng. Des. Sel. 2005, 18, 345–357. [Google Scholar] [CrossRef]
- rjunan, P.; Umland, T.; Dyda, F.; Swaminathan, S.; Furey, W.; Sax, M.; Farrenkopf, B.; Gao, Y.; Zhang, D.; Jordan, F. Crystal structure of the thiamin diphosphate-dependent enzyme pyruvate decarboxylase from the yeast Saccharomyces cerevisiae at 2.3 Å resolution. J. Mol. Biol. 1996, 256, 590–600. [Google Scholar] [CrossRef]
- Pohl, M. Protein design on pyruvate decarboxylase (PDC) by site-directed mutagenesis. Adv. Biochem. Eng. Biotechnol. 1997, 58, 15–43. [Google Scholar]
- Liu, M.; Sergienko, E.A.; Guo, F.; Wang, J.; Tittmann, K.; Hubner, G.; Furey, W.; Jordan, F. Catalytic acid-base groups in yeast pyruvate decarboxylase. 1. Site-directed mutagenesis and steady-state kinetic studies on the enzyme with the D28A, H114F, H115F, and E477Q substitutions. Biochemistry (Mosc.) 2001, 40, 355–7368. [Google Scholar]
- Schenk, G.; Layfield, R.; Candy, J.M.; Duggleby, R.G.; Nixon, P.F. Molecular evolutionary analysis of the thiamine-diphosphate-dependent enzyme, transketolase. J. Mol. Evol. 1997, 44, 552–572. [Google Scholar] [CrossRef]
- Raeburn, S.; Rabinowitz, J.C. Pyruvate: Ferredoxin oxidoreductase: II. Characteristics of the forward and reverse reactions and properties of the enzyme. Arch. Biochem. Biophys. 1971, 146, 21–33. [Google Scholar] [CrossRef]
- Uyeda, K.; Rabinowitz, J.C. Pyruvate-ferredoxin oxidoreductase. IV. Studies on the reaction mechanism. J. Biol. Chem. 1971, 246, 3120–3125. [Google Scholar]
- Uyeda, K.; Rabinowitz, J.C. Pyruvate-ferredoxin oxidoreductase. III. Purification and properties of the enzyme. J. Biol. Chem. 1971, 246, 3111–3119. [Google Scholar]
- Ragsdale, S.W. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev. 2003, 103, 2333–2346. [Google Scholar] [CrossRef]
- Ragsdale, S.W.; Pierce, E. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 2008, 1784, 1873–1898. [Google Scholar] [CrossRef]
- Tittmann, K. Reaction mechanisms of thiamin diphosphate enzymes: Redox reactions. FEBS J. 2009, 276, 2454–2468. [Google Scholar] [CrossRef]
- Kletzin, A.; Adams, M. Molecular and phylogenetic characterization of pyruvate and 2- ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 1996, 178, 248–257. [Google Scholar]
- Zhang, Q.; Iwasaki, T.; Wakagi, T.; Oshima, T. 2-Oxoacid: Ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J. Biochem. (Tokyo) 1996, 120, 587–599. [Google Scholar] [CrossRef]
- Townson, S.M.; Upcroft, J.A.; Upcroft, P. Characterisation and purification of pyruvate: Ferredoxin oxidoreductase from Giardia duodenalis. Mol. Biochem. Parasitol. 1996, 79, 183–193. [Google Scholar] [CrossRef]
- Horner, D.S.; Hirt, R.P.; Embley, T.M. A single eubacterial origin of eukaryotic pyruvate: Ferredoxin oxidoreductase genes: Implications for the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 1999, 16, 1280–1291. [Google Scholar] [CrossRef]
- Pineda, E.; Encalada, R.; Rodríguez-Zavala, J.S.; Olivos-García, A.; Moreno-Sánchez, R.; Saavedra, E. Pyruvate: Ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 2010, 277, 3382–3395. [Google Scholar] [CrossRef]
- Wahl, R.C.; Orme-Johnson, W.H. Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J. Biol. Chem. 1987, 262, 10489–10496. [Google Scholar]
- Meinecke, B.; Bertram, J.; Gottschalk, G. Purification and characterization of the pyruvate-ferredoxin oxidoreductase from Clostridium acetobutylicum. Arch. Microbiol. 1989, 152, 244–250. [Google Scholar] [CrossRef]
- Pieulle, L.; Magro, V.; Hatchikian, E. Isolation and analysis of the gene encoding the pyruvate-ferredoxin oxidoreductase of Desulfovibrio africanus, production of the recombinant enzyme in Escherichia coli, and effect of carboxy-terminal deletions on its stability. J. Bacteriol. 1997, 179, 5684–5692. [Google Scholar]
- Pieulle, L.; Guigliarelli, B.; Asso, M.; Dole, F.; Bernadac, A.; Hatchikian, E.C. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim. Biophys. Acta 1995, 1250, 49–59. [Google Scholar] [CrossRef]
- Pieulle, L.; Charon, M.-H.; Bianco, P.; Bonicel, J.; Petillot, Y.; Hatchikian, E.C. Structural and kinetic studies of the pyruvate-ferredoxin oxidoreductase/ferredoxin complex from Desulfovibrio africanus. Eur. J. Biochem. 1999, 264, 500–508. [Google Scholar] [CrossRef]
- Pieulle, L.; Chabriere, E.; Hatchikian, C.; Fontecilla-Camps, J.C.; Charon, M.-H. Crystallization and preliminary crystallographic analysis of the pyruvate-ferredoxin oxidoreductase from Desulfovibrio africanus. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 329–331. [Google Scholar] [CrossRef]
- Blamey, J.M.; Adams, M.W. Characterization of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium, Thermotoga maritima. Biochemistry (Mosc.) 1994, 33, 1000–1007. [Google Scholar]
- Blamey, J.M.; Adams, M.W.W. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1993, 1161, 19–27. [Google Scholar] [CrossRef]
- Kunow, J.; Linder, D.; Thauer, R.K. Pyruvate: Ferredoxin oxidoreductase from the sulfate-reducing Archaeoglobus fulgidus: Molecular composition, catalytic properties, and sequence alignments. Arch. Microbiol. 1995, 163, 21–28. [Google Scholar]
- Bock, A.-K.; Prieger-Kraft, A.; Schönheit, P. Pyruvate a novel substrate for growth and methane formation in Methanosarcina barkeri. Arch. Microbiol. 1994, 161, 33–46. [Google Scholar]
- Bock, A.K.; Kunow, J.; Glasemacher, J.; Schönheit, P. Catalytic properties, molecular composition and sequence alignments of pyruvate: Ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina Barkeri (Strain Fusaro). Eur. J. Biochem. 1996, 237, 35–44. [Google Scholar]
- Tersteegen, A.; Dietmar, L.R.K.; Thauer, R.H. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 1997, 244, 862–868. [Google Scholar]
- Chabrière, E.; Vernede, X.; Guigliarelli, B.; Charon, M.-H.; Hatchikian, E.C.; Fontecilla-Camps, J.C. Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase. Science 2001, 294, 2559–2563. [Google Scholar] [CrossRef]
- Chabrière, E.; Charon, M.-H.; Volbeda, A.; Pieulle, L.; Hatchikian, E.C.; Fontecilla-Camps, J.-C. Crystal structures of the key anaerobic enzyme pyruvate: Ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Mol. Biol. 1999, 6, 182–190. [Google Scholar] [CrossRef]
- Garczarek, F.; Dong, M.; Typke, D.; Witkowska, H.E.; Hazen, T.C.; Nogales, E.; Biggin, M.D.; Glaeser, R.M. Octomeric pyruvate-ferredoxin oxidoreductase from Desulfovibrio vulgaris. J. Struct. Biol. 2007, 159, 9–18. [Google Scholar] [CrossRef]
- Bock, A.-K.; Schönheit, P.; Teixeira, M. The iron-sulfur centers of the pyruvate:ferredoxin oxidoreductase from Methanosarcina barkeri (Fusaro). FEBS Lett. 1997, 414, 209–212. [Google Scholar] [CrossRef]
- Charon, M.-H.; Volbeda, A.; Chabriere, E.; Pieulle, L.; Fontecilla-Camps, J.C. Structure and electron transfer mechanism of pyruvate: Ferredoxin oxidoreductase. Curr. Opin. Struct. Biol. 1999, 9, 663–669. [Google Scholar] [CrossRef]
- Shiba, H.; Kawasumi, T.; Igarashi, Y.; Kodama, T.; Minoda, Y. The CO2 assimilation via. the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 1985, 141, 198–203. [Google Scholar]
- Ikeda, T.; Ochiai, T.; Morita, S.; Nishiyama, A.; Yamada, E.; Arai, H.; Ishii, M.; Igarashi, Y. Anabolic five subunit-type pyruvate: Ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Biochem. Biophys. Res. Commun. 2006, 340, 76–82. [Google Scholar] [CrossRef]
- Ikeda, T.; Yamamoto, M.; Arai, H.; Ohmori, D.; Ishii, M.; Igarashi, Y. Enzymatic and electron paramagnetic resonance studies of anabolic pyruvate synthesis by pyruvate: Ferredoxin oxidoreductase from Hydrogenobacter thermophilus. FEBS J. 2010, 277, 501–510. [Google Scholar] [CrossRef]
- Yamamoto, M.; Ikeda, T.; Arai, H.; Ishii, M.; Igarashi, Y. Carboxylation reaction catalyzed by 2-oxoglutarate: Ferredoxin oxidoreductases from Hydrogenobacter thermophilus. Extremophiles 2010, 14, 79–85. [Google Scholar] [CrossRef]
- Lin, W.C.; Yang, Y.-L.; Whitman, W.B. The anabolic pyruvate oxidoreductase from Methanococcus maripaludis. Arch. Microbiol. 2003, 179, 444–456. [Google Scholar]
- Lin, W.; Whitman, W. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch. Microbiol. 2004, 181, 68–73. [Google Scholar] [CrossRef]
- Imlay, A.J. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef]
- Linn, T.C.; Pettit, F.H.; Reed, L.J. α-Keto acid dehydrogenase complexes, X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc. Natl. Acad. Sci. USA 1969, 62, 234–241. [Google Scholar] [CrossRef]
- Patel, M.S.; Roche, T.E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990, 4, 3224–3233. [Google Scholar]
- Witzmann, S.; Bisswanger, H. The pyruvate dehydrogenase complex from thermophilic organisms: Thermal stability and re-association from the enzyme components. Biochim. Biophys. Acta 1998, 1385, 341–352. [Google Scholar] [CrossRef]
- Potter, S.; Fothergill-Gilmore, L.A. Purification and properties of pyruvate kinase from Thermoplasma acidophilum. FEMS Microbiol. Lett. 1992, 94, 235–239. [Google Scholar] [CrossRef]
- Heath, C.; Jeffries, A.C.; Hough, D.W.; Danson, M.J. Discovery of the catalytic function of a putative 2-oxoacid dehydrogenase multienzyme complex in the thermophilic archaeon Thermoplasma acidophilum. FEBS Lett. 2004, 577, 523–527. [Google Scholar] [CrossRef]
- Selig, M.; Schönheit, P. Oxidation of organic compounds to CO2 with sulfur or thiosulfate as electron acceptor in the anaerobic hyperthermophilic archaea Thermoproteus tenax and Pyrobaculum islandicum proceeds via. the citric acid cycle. Arch. Microbiol. 1994, 162, 286–294. [Google Scholar] [CrossRef]
- Ma, K.; Weiss, R.; Adams, M.W.W. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 2000, 182, 1864–1871. [Google Scholar] [CrossRef]
- Jenney, F.E.; Adams, M.W.W. Hydrogenases of the model hyperthermophiles. Ann. NY Acad. Sci. 2008, 1125, 252–266. [Google Scholar] [CrossRef]
- Rudolph, F.B.; Purich, D.L.; Fromm, H.J. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. J. Biol. Chem. 1968, 243, 5539–5545. [Google Scholar]
- Lurz, R.; Mayer, F.; Gottschalk, G. Electron microscopic study on the quaternary structure of the isolated particulate alcohol-acetaldehyde dehydrogenase complex and on its identity with the polygonal bodies of Clostridium kluyveri. Arch. Microbiol. 1979, 120, 255–262. [Google Scholar] [CrossRef]
- Nair, R.V.; Bennett, G.N.; Papoutsakis, E.T. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 1994, 176, 871–885. [Google Scholar]
- Tóth, J.; Ismaiel, A.A.; Chen, J.-S. The ald gene, encoding a coenzyme A-acylating aldehyde dehydrogenase, distinguishes Clostridium beijerinckii and two other solvent-producing clostridia from Clostridium acetobutylicum. Appl. Environ. Microbiol. 1999, 65, 4973–4980. [Google Scholar]
- Sánchez, L.B. Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch. Biochem. Biophys. 1998, 354, 57–64. [Google Scholar] [CrossRef]
- Bruchhaus, I.; Tannich, E. Purification and molecular characterization of the NAD(+)-dependent acetaldehyde/alcohol dehydrogenase from Entamoeba histolytica. Biochem. J. 1994, 303, 743–748. [Google Scholar]
- Burdette, D.; Zeikus, J.G. Purification of acetaldehyde dehydrogenase and alcohol dehydrogenases from Thermoanaerobacter ethanolicus 39E and characterization of the secondary-alcohol dehydrogenase (2 degrees Adh) as a bifunctional alcohol dehydrogenase--acetyl-CoA reductive thioesterase. Biochem. J. 1994, 302, 163–170. [Google Scholar]
- Brown, S.D.; Guss, A.M.; Karpinets, T.V.; Parks, J.M.; Smolin, N.; Yang, S.; Land, M.L.; Klingeman, D.M.; Bhandiwad, A.; Rodriguez, M. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 2011, 108, 13752–13757. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Eram, M.S.; Ma, K. Decarboxylation of Pyruvate to Acetaldehyde for Ethanol Production by Hyperthermophiles. Biomolecules 2013, 3, 578-596. https://doi.org/10.3390/biom3030578
Eram MS, Ma K. Decarboxylation of Pyruvate to Acetaldehyde for Ethanol Production by Hyperthermophiles. Biomolecules. 2013; 3(3):578-596. https://doi.org/10.3390/biom3030578
Chicago/Turabian StyleEram, Mohammad S., and Kesen Ma. 2013. "Decarboxylation of Pyruvate to Acetaldehyde for Ethanol Production by Hyperthermophiles" Biomolecules 3, no. 3: 578-596. https://doi.org/10.3390/biom3030578
APA StyleEram, M. S., & Ma, K. (2013). Decarboxylation of Pyruvate to Acetaldehyde for Ethanol Production by Hyperthermophiles. Biomolecules, 3(3), 578-596. https://doi.org/10.3390/biom3030578



