MYC and Metabolomics: Can We Use What We Know for DLBCL Subtyping and Diagnosis?
Abstract
1. Introduction
2. DLBCL Molecular Subtyping
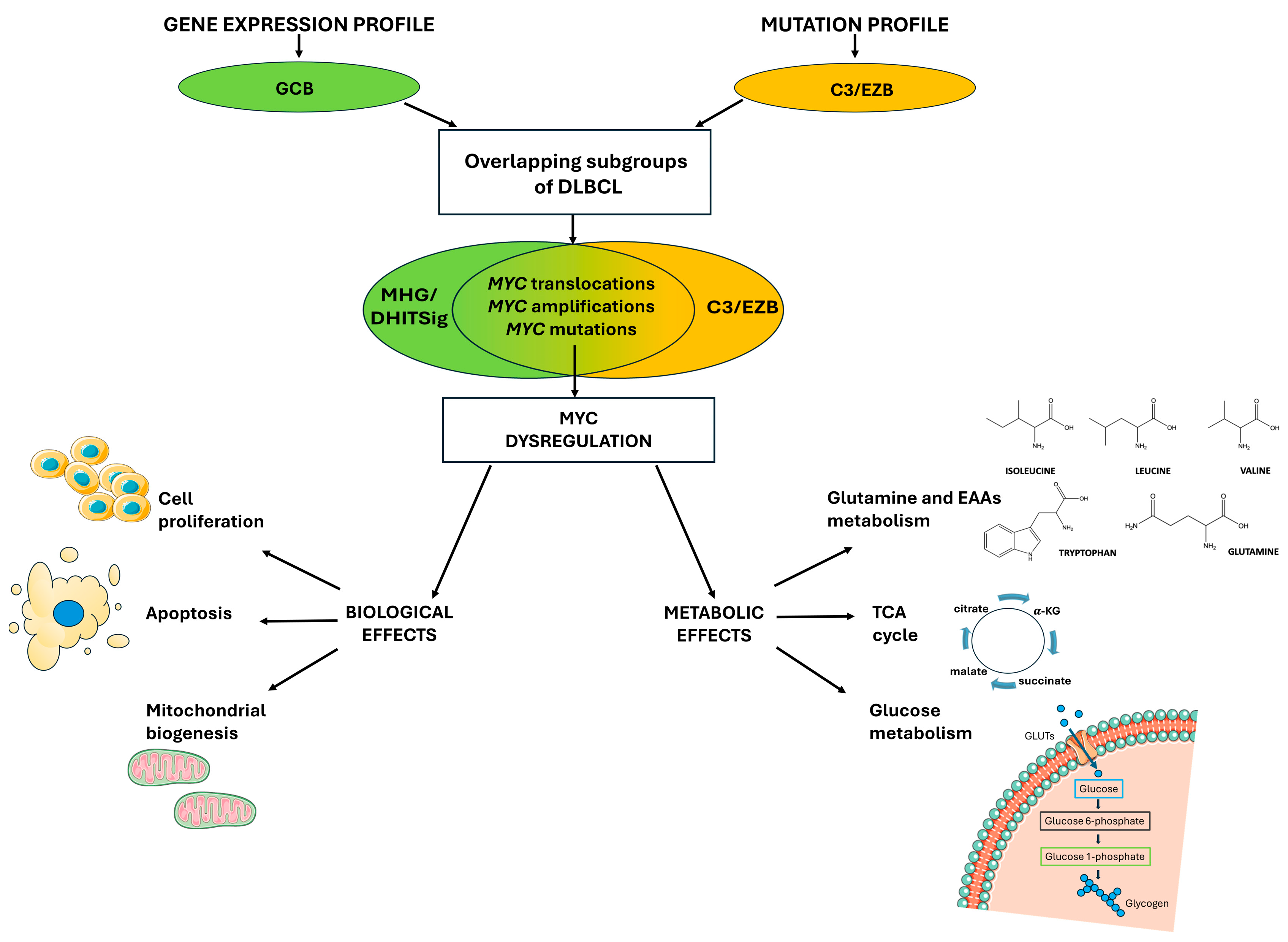
3. MYC-Driven Lymphomas
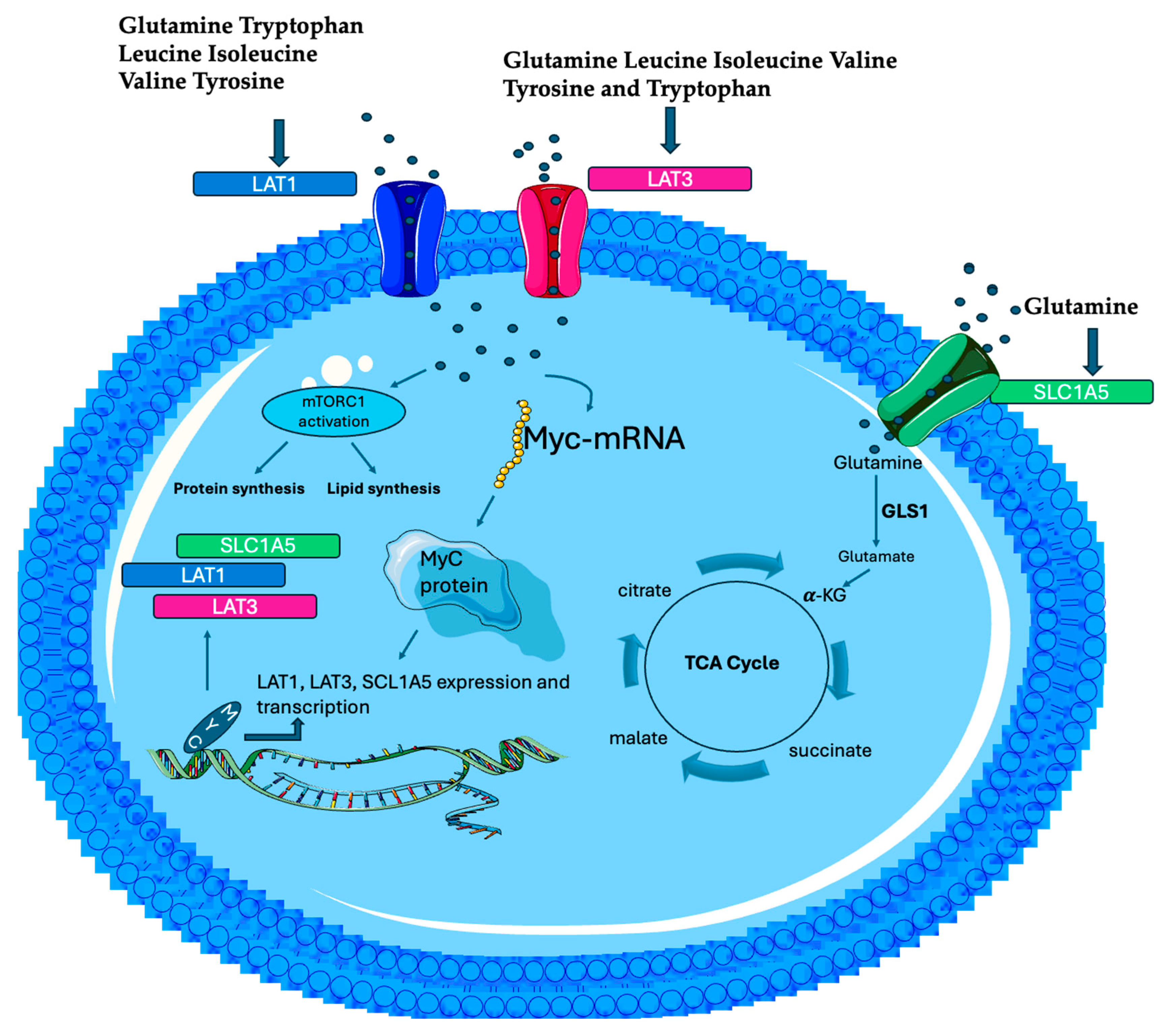
4. MYC Dysregulation: From Gene Expression to Metabolic Reprogramming
5. The Use of Metabolomic Techniques in B-Cell Lymphoma for Diagnosis and Prognosis
6. MYC-Driven Amino Acid Metabolism in Cancer Cells
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DLBCL | Diffuse large B-cell lymphoma |
| AAs | Amino acids |
| BCL2 | B-cell lymphoma 2 |
| BCL6 | B-cell lymphoma 6 |
| FISH | Fluorescence in situ hybridization |
| WHO | World Health Organization |
| BL | Burkitt lymphoma |
| IHC | Immunohistochemistry |
| HIF-1α | Hypoxia-inducible factor-1 alpha |
| BCAAs | Branched amino acids |
| SLC | Solute carrier |
| SLC1A5 | Solute carrier family 1 member 5 |
| BCAT1 | Branched-chain amino acid transaminase 1 |
| TCA | Tricarboxylic acid cycle |
| GEP | Gene expression profile |
| GCB | Germinal center B-cell |
| ABC | Activated B-cell |
| COO | Cell-of-origin |
| MHG | Molecular high-grade |
| GC | Germinal center |
| DHITSig | Double-hit gene expression signature |
| HGBL | High grade B-cell lymphoma |
| WES | Whole exome sequencing |
| CSR | Class-switch recombination |
| LBCLs | Large B-cell lymphomas |
| aSHM | Aberrant somatic hypermutation machinery |
| TSS | Transcription start site |
| Ig | Immunoglobulin |
| IgH | Immunoglobulin heavy chain |
| MCL | Mantle cell lymphoma |
| MBI | MYC Box I |
| FBXW7 | F-Box and WD repeat domain containing 7 |
| NMR | Nuclear magnetic resonance |
| bHLH-LZ | basic helix-loop-helix leucine zipper |
| CDKN2B | Cyclin dependent kinase inhibitor 2B |
| ARF | Alternate reading frame |
| MDM2 | Mouse double minute 2 homolog |
| TP53 | Tumor protein p53 |
| TFAM | Transcription factor A |
| PPARGC1A | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| HK2 | Hexokinase 2 |
| PFKM | Phosphofructokinase |
| ENO1 | Enolase 1 |
| LDHA | Lactate dehydrogenase A |
| PKM2 | Pyruvate kinase M2 |
| LC-MS | Liquid chromatography–mass spectrometry |
| GC-MS | Gas chromatography–mass spectrometry |
| ND | Recently diagnosed |
| CR | Complete remission |
| α-KG | Alpha-ketoglutarate |
| REF | Refractory disease |
| REL | Early relapse |
| EAAs | Essential amino acids |
| NEAAs | Non-essential amino acids |
| LAT | L-type amino acid transporter |
| OAA | Oxaloacetate |
| GLS1 | Glutaminase 1 |
| GLS2 | Glutaminase 2 |
| TFEB | Transcription factor EB |
| Gln | Glutamine |
References
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A Report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Rosenwald, A.; Bens, S.; Advani, R.; Barrans, S.; Copie-Bergman, C.; Elsensohn, M.-H.; Natkunam, Y.; Calaminici, M.; Sander, B.; Baia, M.; et al. Prognostic Significance of MYC Rearrangement and Translocation Partner in Diffuse Large B-Cell Lymphoma: A Study by the Lunenburg Lymphoma Biomarker Consortium. J. Clin. Oncol. 2019, 37, 3359–3368. [Google Scholar] [CrossRef] [PubMed]
- Cucco, F.; Barrans, S.; Sha, C.; Clipson, A.; Crouch, S.; Dobson, R.; Chen, Z.; Thompson, J.S.; Care, M.A.; Cummin, T.; et al. Distinct Genetic Changes Reveal Evolutionary History and Heterogeneous Molecular Grade of DLBCL with MYC/BCL2 Double-Hit. Leukemia 2020, 34, 1329–1341. [Google Scholar] [CrossRef] [PubMed]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-Wide Discovery of Somatic Coding and Noncoding Mutations in Pediatric Endemic and Sporadic Burkitt Lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef]
- Salghetti, S.E.; Young Kim, S.; Tansey, W.P. Destruction of Myc by Ubiquitin-mediated Proteolysis: Cancer-associated and Transforming Mutations Stabilize Myc. EMBO J. 1999, 18, 717–726. [Google Scholar] [CrossRef]
- Ott, G.; Rosenwald, A.; Campo, E. Understanding MYC-Driven Aggressive B-Cell Lymphomas: Pathogenesis and Classification. Blood 2013, 122, 3884–3891. [Google Scholar] [CrossRef]
- Wu, J.; Meng, F.; Ran, D.; Song, Y.; Dang, Y.; Lai, F.; Yang, L.; Deng, M.; Song, Y.; Zhu, J. The Metabolism and Immune Environment in Diffuse Large B-Cell Lymphoma. Metabolites 2023, 13, 734. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC Oncogene—The Grand Orchestrator of Cancer Growth and Immune Evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Yang, W.; Guo, Q.; Quan, S.; Chalmers, Z.R.; Parker, J.B.; Truica, M.; Dufficy, M.F.; Kerber, M.M.; Vasan, K.; Gupta, D.G.; et al. Impaired Mitochondrial Metabolism Is a Critical Cancer Vulnerability for MYC Inhibitors. Sci. Adv. 2025, 11, eadw5228. [Google Scholar] [CrossRef]
- Venkatraman, S.; Balasubramanian, B.; Thuwajit, C.; Meller, J.; Tohtong, R.; Chutipongtanate, S. Targeting MYC at the Intersection between Cancer Metabolism and Oncoimmunology. Front. Immunol. 2024, 15, 1324045. [Google Scholar] [CrossRef]
- Vettore, L.; Westbrook, R.L.; Tennant, D.A. New Aspects of Amino Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Feng, X.; Ren, C.; Jiang, X.; Liu, W.; Huang, W.; Liu, Z.; Li, Z.; Zeng, L.; Wang, L.; et al. Over-Expression of BCAT1, a c-Myc Target Gene, Induces Cell Proliferation, Migration and Invasion in Nasopharyngeal Carcinoma. Mol. Cancer 2013, 12, 53. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.-C.; Lee, Y.-S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. C-Myc Suppression of miR-23a/b Enhances Mitochondrial Glutaminase Expression and Glutamine Metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct Types of Diffuse Large B-Cell Lymphoma Identified by Gene Expression Profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Shipp, M.A.; Ross, K.N.; Tamayo, P.; Weng, A.P.; Kutok, J.L.; Aguiar, R.C.T.; Gaasenbeek, M.; Angelo, M.; Reich, M.; Pinkus, G.S.; et al. Diffuse Large B-Cell Lymphoma Outcome Prediction by Gene-Expression Profiling and Supervised Machine Learning. Nat. Med. 2002, 8, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Sha, C.; Barrans, S.; Cucco, F.; Bentley, M.A.; Care, M.A.; Cummin, T.; Kennedy, H.; Thompson, J.S.; Uddin, R.; Worrillow, L.; et al. Molecular High-Grade B-Cell Lymphoma: Defining a Poor-Risk Group That Requires Different Approaches to Therapy. J. Clin. Oncol. 2019, 37, 202–212. [Google Scholar] [CrossRef]
- Sha, C.; Barrans, S.; Care, M.A.; Cunningham, D.; Tooze, R.M.; Jack, A.; Westhead, D.R. Transferring Genomics to the Clinic: Distinguishing Burkitt and Diffuse Large B Cell Lymphomas. Genome Med. 2015, 7, 64. [Google Scholar] [CrossRef]
- Ennishi, D.; Jiang, A.; Boyle, M.; Collinge, B.; Grande, B.M.; Ben-Neriah, S.; Rushton, C.; Tang, J.; Thomas, N.; Slack, G.W.; et al. Double-Hit Gene Expression Signature Defines a Distinct Subgroup of Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2019, 37, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.; Zhang, J.; Davis, N.S.; Moffitt, A.B.; Love, C.L.; Waldrop, A.; Leppa, S.; Pasanen, A.; Meriranta, L.; Karjalainen-Lindsberg, M.-L.; et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017, 171, 481–494.e15. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular Subtypes of Diffuse Large B Cell Lymphoma Are Associated with Distinct Pathogenic Mechanisms and Outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Wright, G.W.; Huang, D.W.; Phelan, J.D.; Coulibaly, Z.A.; Roulland, S.; Young, R.M.; Wang, J.Q.; Schmitz, R.; Morin, R.D.; Tang, J.; et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020, 37, 551–568.e14. [Google Scholar] [CrossRef]
- Sewastianik, T.; Prochorec-Sobieszek, M.; Chapuy, B.; Juszczyński, P. MYC Deregulation in Lymphoid Tumors: Molecular Mechanisms, Clinical Consequences and Therapeutic Implications. Biochim. Biophys. Acta 2014, 1846, 457–467. [Google Scholar] [CrossRef]
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the C-Myc Gene into the Immunoglobulin Heavy Chain Locus in Human Burkitt Lymphoma and Murine Plasmacytoma Cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef]
- Molyneux, E.M.; Rochford, R.; Griffin, B.; Newton, R.; Jackson, G.; Menon, G.; Harrison, C.J.; Israels, T.; Bailey, S. Burkitt’s Lymphoma. Lancet 2012, 379, 1234–1244. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Pagani, C.; Rusconi, C.; Dalla Pria, A.; Ravano, E.; Schommers, P.; Bastos-Oreiro, M.; Verga, L.; Gini, G.; Spina, M.; Arcaini, L.; et al. MYC Rearrangements in HIV-Associated Large B-Cell Lymphomas: EUROMYC, a European Retrospective Study. Blood Adv. 2024, 8, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Zak, T.; Santana-Santos, L.; Gao, J.; Behdad, A.; Aqil, B.; Wolniak, K.; Lu, X.; Ji, P.; Chen, Q.; Chen, Y.-H.; et al. Prognostic Significance of Copy Number Gains of MYC Detected by Fluorescence in Situ Hybridization in Large B-Cell Lymphoma. Leuk. Lymphoma 2024, 65, 26–36. [Google Scholar] [CrossRef]
- Rabbitts, T.H.; Hamlyn, P.H.; Baer, R. Altered Nucleotide Sequences of a Translocated C-Myc Gene in Burkitt Lymphoma. Nature 1983, 306, 760–765. [Google Scholar] [CrossRef]
- Schaub, F.X.; Dhankani, V.; Berger, A.C.; Trivedi, M.; Richardson, A.B.; Shaw, R.; Zhao, W.; Zhang, X.; Ventura, A.; Liu, Y.; et al. Pan-Cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018, 6, 282–300.e2. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- De Luca, D.; Munafò, C.; Lorenzi, L.; Cucco, F. MYC Point Mutations in Cancer: A Reboot and a Sequel. Clin. Cancer Res. 2025. accepted. [Google Scholar] [CrossRef] [PubMed]
- Khodabakhshi, A.H.; Morin, R.D.; Fejes, A.P.; Mungall, A.J.; Mungall, K.L.; Bolger-Munro, M.; Johnson, N.A.; Connors, J.M.; Gascoyne, R.D.; Marra, M.A.; et al. Recurrent Targets of Aberrant Somatic Hypermutation in Lymphoma. Oncotarget 2012, 3, 1308–1319. [Google Scholar] [CrossRef]
- Hemann, M.T.; Bric, A.; Teruya-Feldstein, J.; Herbst, A.; Nilsson, J.A.; Cordon-Cardo, C.; Cleveland, J.L.; Tansey, W.P.; Lowe, S.W. Evasion of the P53 Tumour Surveillance Network by Tumour-Derived MYC Mutants. Nature 2005, 436, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Arthur, N.B.J.; Christensen, K.A.; Mannino, K.; Ruzinova, M.B.; Kumar, A.; Gruszczynska, A.; Day, R.B.; Erdmann-Gilmore, P.; Mi, Y.; Sprung, R.; et al. Missense Mutations in Myc Box I Influence Nucleocytoplasmic Transport to Promote Leukemogenesis. Clin. Cancer Res. 2024, 30, 3622–3639. [Google Scholar] [CrossRef]
- Freie, B.; Carroll, P.A.; Varnum-Finney, B.J.; Ramsey, E.L.; Ramani, V.; Bernstein, I.; Eisenman, R.N. A Germline Point Mutation in the MYC-FBW7 Phosphodegron Initiates Hematopoietic Malignancies. Genes. Dev. 2024, 38, 253–272. [Google Scholar] [CrossRef]
- Papadimitropoulou, A.; Makri, M.; Zoidis, G. MYC the Oncogene from Hell: Novel Opportunities for Cancer Therapy. Eur. J. Med. Chem. 2024, 267, 116194. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Guo, C.; Das, S.K.; Chow, C.C.; Batchelor, E.; Simons, S.S.; Levens, D. Dissecting Transcriptional Amplification by MYC. Elife 2020, 9, e52483. [Google Scholar] [CrossRef]
- Levens, D. You Don’t Muck with MYC. Genes. Cancer 2010, 1, 547–554. [Google Scholar] [CrossRef]
- Mainwaring, O.J.; Weishaupt, H.; Zhao, M.; Rosén, G.; Borgenvik, A.; Breinschmid, L.; Verbaan, A.D.; Richardson, S.; Thompson, D.; Clifford, S.C.; et al. ARF Suppression by MYC but Not MYCN Confers Increased Malignancy of Aggressive Pediatric Brain Tumors. Nat. Commun. 2023, 14, 1221. [Google Scholar] [CrossRef]
- Clipson, A.; Barrans, S.; Zeng, N.; Crouch, S.; Grigoropoulos, N.F.; Liu, H.; Kocialkowski, S.; Wang, M.; Huang, Y.; Worrillow, L.; et al. The Prognosis of MYC Translocation Positive Diffuse Large B-Cell Lymphoma Depends on the Second Hit. J. Pathol. Clin. Res. 2015, 1, 125–133. [Google Scholar] [CrossRef]
- Purhonen, J.; Klefström, J.; Kallijärvi, J. MYC-an Emerging Player in Mitochondrial Diseases. Front. Cell Dev. Biol. 2023, 11, 1257651. [Google Scholar] [CrossRef]
- Zhao, J.; Jin, D.; Huang, M.; Ji, J.; Xu, X.; Wang, F.; Zhou, L.; Bao, B.; Jiang, F.; Xu, W.; et al. Glycolysis in the Tumor Microenvironment: A Driver of Cancer Progression and a Promising Therapeutic Target. Front. Cell Dev. Biol. 2024, 12, 1416472. [Google Scholar] [CrossRef]
- Jin, J.; Byun, J.-K.; Choi, Y.-K.; Park, K.-G. Targeting Glutamine Metabolism as a Therapeutic Strategy for Cancer. Exp. Mol. Med. 2023, 55, 706–715. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s Contributions to Current Concepts of Cancer Metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.R.; Lane, A.N.; Robertson, B.; Kemp, S.; Liu, Y.; Hill, B.G.; Dean, D.C.; Clem, B.F. Control of Glutamine Metabolism by the Tumor Suppressor Rb. Oncogene 2014, 33, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An Emerging but Powerful Tool for Precision Medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef]
- Alfaifi, A.; Refai, M.Y.; Alsaadi, M.; Bahashwan, S.; Malhan, H.; Al-Kahiry, W.; Dammag, E.; Ageel, A.; Mahzary, A.; Albiheyri, R.; et al. Metabolomics: A New Era in the Diagnosis or Prognosis of B-Cell Non-Hodgkin’s Lymphoma. Diagnostics 2023, 13, 861. [Google Scholar] [CrossRef]
- Li, H.; Tennessen, J.M. Methods for Studying the Metabolic Basis of Drosophila Development. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e280. [Google Scholar] [CrossRef]
- Want, E.J.; Cravatt, B.F.; Siuzdak, G. The Expanding Role of Mass Spectrometry in Metabolite Profiling and Characterization. Chembiochem 2005, 6, 1941–1951. [Google Scholar] [CrossRef]
- Barberini, L.; Noto, A.; Fattuoni, C.; Satta, G.; Zucca, M.; Cabras, M.G.; Mura, E.; Cocco, P. The Metabolomic Profile of Lymphoma Subtypes: A Pilot Study. Molecules 2019, 24, 2367. [Google Scholar] [CrossRef] [PubMed]
- Bueno Duarte, G.H.; de Piloto Fernandes, A.M.A.; Silva, A.A.R.; Zamora-Obando, H.R.; Amaral, A.G.; de Sousa Mesquita, A.; Schmidt-Filho, J.; Cordeiro de Lima, V.C.; D’Almeida Costa, F.; Andrade, V.P.; et al. Gas Chromatography-Mass Spectrometry Untargeted Profiling of Non-Hodgkin’s Lymphoma Urinary Metabolite Markers. Anal. Bioanal. Chem. 2020, 412, 7469–7480. [Google Scholar] [CrossRef]
- Fei, F.; Zheng, M.; Xu, Z.; Sun, R.; Chen, X.; Cao, B.; Li, J. Plasma Metabolites Forecast Occurrence and Prognosis for Patients With Diffuse Large B-Cell Lymphoma. Front. Oncol. 2022, 12, 894891. [Google Scholar] [CrossRef] [PubMed]
- Mi, M.; Liu, Z.; Zheng, X.; Wen, Q.; Zhu, F.; Li, J.; Mungur, I.D.; Zhang, L. Serum Metabolomic Profiling Based on GC/MS Helped to Discriminate Diffuse Large B-Cell Lymphoma Patients with Different Prognosis. Leuk. Res. 2021, 111, 106693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, L.; Wang, C.; Kong, R.; Lu, T.; Ding, M.; Jiang, Y.; Wang, X.; Zhou, X. Comprehensive Metabolome Analysis Identified Novel Biomarkers for the Diagnosis and Prognosis of Diffuse Large B-Cell Lymphoma. Blood 2022, 140, 11934–11935. [Google Scholar] [CrossRef]
- Stenson, M.; Pedersen, A.; Hasselblom, S.; Nilsson-Ehle, H.; Karlsson, B.G.; Pinto, R.; Andersson, P.-O. Serum Nuclear Magnetic Resonance-Based Metabolomics and Outcome in Diffuse Large B-Cell Lymphoma Patients—A Pilot Study. Leuk Lymphoma 2016, 57, 1814–1822. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, G.; Wang, M.C. Host and Microbiota Metabolic Signals in Aging and Longevity. Nat Chem Biol 2021, 17, 1027–1036. [Google Scholar] [CrossRef]
- Shigeta, K.; Hasegawa, M.; Hishiki, T.; Naito, Y.; Baba, Y.; Mikami, S.; Matsumoto, K.; Mizuno, R.; Miyajima, A.; Kikuchi, E.; et al. IDH2 Stabilizes HIF-1α-Induced Metabolic Reprogramming and Promotes Chemoresistance in Urothelial Cancer. EMBO J 2023, 42, e110620. [Google Scholar] [CrossRef]
- Chen, D.; Xia, S.; Zhang, R.; Li, Y.; Famulare, C.A.; Fan, H.; Wu, R.; Wang, M.; Zhu, A.C.; Elf, S.E.; et al. Lysine Acetylation Restricts Mutant IDH2 Activity to Optimize Transformation in AML Cells. Mol Cell 2021, 81, 3833–3847.e11. [Google Scholar] [CrossRef] [PubMed]
- Showalter, M.R.; Cajka, T.; Fiehn, O. Epimetabolites: Discovering Metabolism beyond Building and Burning. Curr Opin Chem Biol 2017, 36, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Yin, Y.; Wu, G. Dietary Essentiality of “Nutritionally Non-Essential Amino Acids” for Animals and Humans. Exp. Biol. Med. 2015, 240, 997–1007. [Google Scholar] [CrossRef]
- Chen, J.; Cui, L.; Lu, S.; Xu, S. Amino Acid Metabolism in Tumor Biology and Therapy. Cell Death Dis. 2024, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Weng, H.; Zhou, H.; Qu, L. Attacking C-Myc: Targeted and Combined Therapies for Cancer. Curr. Pharm. Des. 2014, 20, 6543–6554. [Google Scholar] [CrossRef]
- Wang, Q.; Holst, J. L-Type Amino Acid Transport and Cancer: Targeting the mTORC1 Pathway to Inhibit Neoplasia. Am. J. Cancer Res. 2015, 5, 1281–1294. [Google Scholar]
- Babu, E.; Kanai, Y.; Chairoungdua, A.; Kim, D.K.; Iribe, Y.; Tangtrongsup, S.; Jutabha, P.; Li, Y.; Ahmed, N.; Sakamoto, S.; et al. Identification of a Novel System L Amino Acid Transporter Structurally Distinct from Heterodimeric Amino Acid Transporters. J. Biol. Chem. 2003, 278, 43838–43845. [Google Scholar] [CrossRef]
- Semba, R.D.; Trehan, I.; Gonzalez-Freire, M.; Kraemer, K.; Moaddel, R.; Ordiz, M.I.; Ferrucci, L.; Manary, M.J. Perspective: The Potential Role of Essential Amino Acids and the Mechanistic Target of Rapamycin Complex 1 (mTORC1) Pathway in the Pathogenesis of Child Stunting. Adv. Nutr. 2016, 7, 853–865. [Google Scholar] [CrossRef]
- Yue, M.; Jiang, J.; Gao, P.; Liu, H.; Qing, G. Oncogenic MYC Activates a Feedforward Regulatory Loop Promoting Essential Amino Acid Metabolism and Tumorigenesis. Cell Rep. 2017, 21, 3819–3832. [Google Scholar] [CrossRef]
- Fernandez, M.R.; Schaub, F.X.; Yang, C.; Li, W.; Yun, S.; Schaub, S.K.; Dorsey, F.C.; Liu, M.; Steeves, M.A.; Ballabio, A.; et al. Disrupting the MYC-TFEB Circuit Impairs Amino Acid Homeostasis and Provokes Metabolic Anergy. Cancer Res. 2022, 82, 1234–1250. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Pi, M.; Kuang, H.; Yue, C.; Yang, Q.; Wu, A.; Li, Y.; Assaraf, Y.G.; Yang, D.-H.; Wu, S. Targeting Metabolism to Overcome Cancer Drug Resistance: A Promising Therapeutic Strategy for Diffuse Large B Cell Lymphoma. Drug Resist. Updates 2022, 61, 100822. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.-Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc Regulates a Transcriptional Program That Stimulates Mitochondrial Glutaminolysis and Leads to Glutamine Addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Peña, E.; Arnold, J.; Shivakumar, V.; Joseph, R.; Vidhya Vijay, G.; den Hollander, P.; Bhangre, N.; Allegakoen, P.; Prasad, R.; Conley, Z.; et al. The Epithelial to Mesenchymal Transition Promotes Glutamine Independence by Suppressing GLS2 Expression. Cancers 2019, 11, 1610. [Google Scholar] [CrossRef]
- Guo, J.Y.; Chen, H.-Y.; Mathew, R.; Fan, J.; Strohecker, A.M.; Karsli-Uzunbas, G.; Kamphorst, J.J.; Chen, G.; Lemons, J.M.S.; Karantza, V.; et al. Activated Ras Requires Autophagy to Maintain Oxidative Metabolism and Tumorigenesis. Genes. Dev. 2011, 25, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.; Vincelette, N.D.; Yu, X.; Watson, G.W.; Fernandez, M.R.; Yang, C.; Hitosugi, T.; Cheng, C.-H.; Freischel, A.R.; Zhang, L.; et al. TFEB Links MYC Signaling to Epigenetic Control of Myeloid Differentiation and Acute Myeloid Leukemia. Blood Cancer Discov. 2021, 2, 162–185. [Google Scholar] [CrossRef]
- Annunziata, I.; van de Vlekkert, D.; Wolf, E.; Finkelstein, D.; Neale, G.; Machado, E.; Mosca, R.; Campos, Y.; Tillman, H.; Roussel, M.F.; et al. MYC Competes with MiT/TFE in Regulating Lysosomal Biogenesis and Autophagy through an Epigenetic Rheostat. Nat. Commun. 2019, 10, 3623. [Google Scholar] [CrossRef]
- Choi, B.-H.; Coloff, J.L. The Diverse Functions of Non-Essential Amino Acids in Cancer. Cancers 2019, 11, 675. [Google Scholar] [CrossRef]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer Cell Metabolism: The Essential Role of the Nonessential Amino Acid, Glutamine. EMBO J. 2017, 36, 1302–1315. [Google Scholar] [CrossRef]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Hosoda, N.; Endo, H.; Saito, K.; Tsujihara, K.; Yamamura, M.; Sakata, T.; Anzai, N.; Wempe, M.F.; Kanai, Y.; et al. L-Type Amino Acid Transporter 1 Inhibitors Inhibit Tumor Cell Growth. Cancer Sci. 2010, 101, 173–179. [Google Scholar] [CrossRef] [PubMed]
| Technology | Advantages | Disadvantages | Most Metabolites Detected | Metabolites and References |
|---|---|---|---|---|
| GC-MS | High sensibility, resolution, and reproducibility Variety of commercial libraries Separation efficiency and quantitative accuracy | Requires derivatization Only suitable for small and volatile compounds | Low-molecular-weight molecules, volatile and polar such as alcohols, aldehydes ketones, lactate pyruvate amino acids, sugars, and free fatty acids | Glicyne, serine threonine glucose, aspartate, methionine, and cysteine valine pyroglutammic acid [53,54,55,56] |
| LC-MS | High sensitivity and specificity detects trace amounts of metabolites Does not require derivatization Suitable for polar and non-polar compounds Wide applicability | Ion suppression may affect accuracy Less standardization and reproducibility than GC-MS | High-molecular-weight molecules, non-volatile and less polar such as complex lipids (phospholipids sphingolipids triglycerides), peptides, proteins, and energy metabolites (NAD+/NAD) | Glutamine, glutamate [57] |
| NMR Spectroscopy | Non-destructive Simple sample preparation Highly reproducible Detailed structural information | Lower sensitivity than LC-MS and GC-MS Longer and more complex analysis times | Global profiling of lipids, proteins, nucleotides, and derivates (ADP, ATP, and AMP), urine metabolites, and small bioactive molecules (neurotransmitters) | Lysine, arginine [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suman, A.F.; De Luca, D.; Gaggini, M.; Cucco, F. MYC and Metabolomics: Can We Use What We Know for DLBCL Subtyping and Diagnosis? Biomolecules 2025, 15, 1346. https://doi.org/10.3390/biom15091346
Suman AF, De Luca D, Gaggini M, Cucco F. MYC and Metabolomics: Can We Use What We Know for DLBCL Subtyping and Diagnosis? Biomolecules. 2025; 15(9):1346. https://doi.org/10.3390/biom15091346
Chicago/Turabian StyleSuman, Adrian Florentin, Davide De Luca, Melania Gaggini, and Francesco Cucco. 2025. "MYC and Metabolomics: Can We Use What We Know for DLBCL Subtyping and Diagnosis?" Biomolecules 15, no. 9: 1346. https://doi.org/10.3390/biom15091346
APA StyleSuman, A. F., De Luca, D., Gaggini, M., & Cucco, F. (2025). MYC and Metabolomics: Can We Use What We Know for DLBCL Subtyping and Diagnosis? Biomolecules, 15(9), 1346. https://doi.org/10.3390/biom15091346






