Interaction Between Heparan Sulfate Oligosaccharide and the Receptor-Binding Domain of the Wild-Type and Omicron Variant of the SARS-CoV-2 Spike Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Dynamics Simulations
2.1.1. System Preparation and MD Simulation Protocol
2.1.2. Filtering and Analysis of the MD Trajectories: Bound State Condition of S1-RBD and Hexa
2.1.3. Model Validation by Reduced Matrix (RedMat)
2.1.4. Cluster Analysis
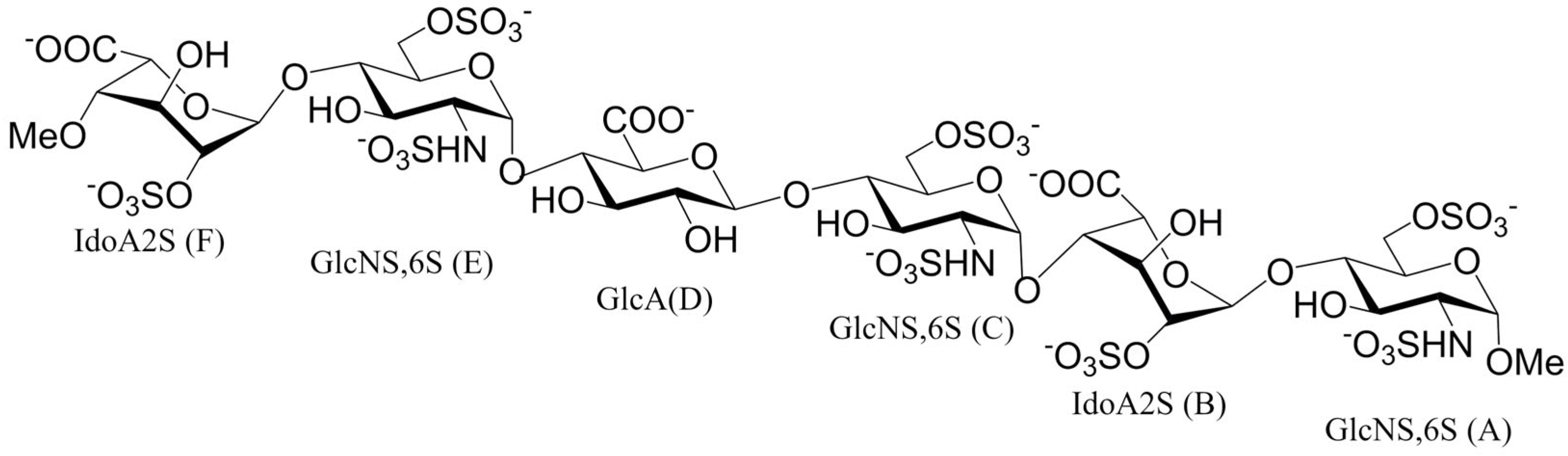
2.2. NMR Spectroscopy: Sample Preparation and Experimental Procedure
3. Results and Discussion
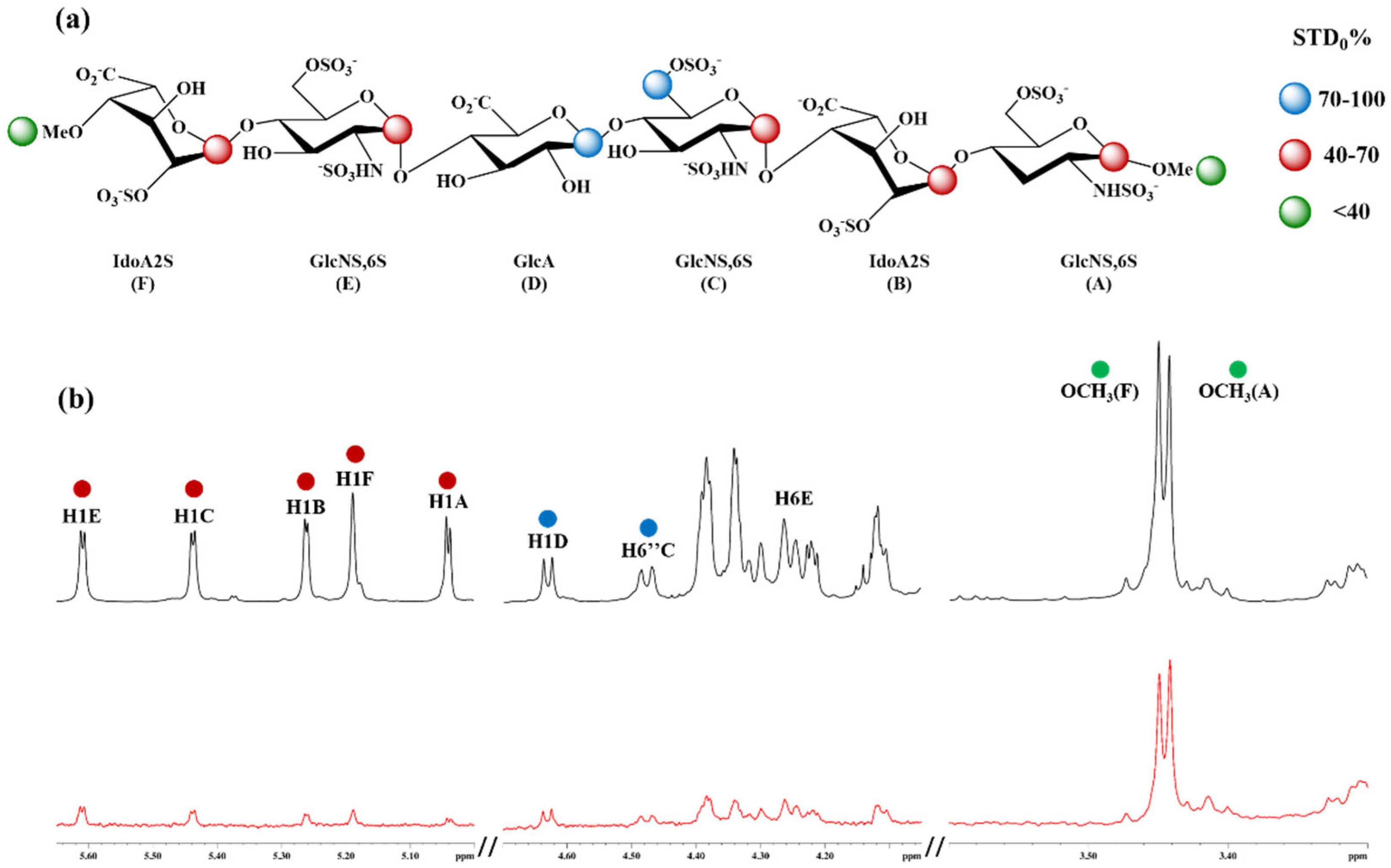
3.1. 1H-STD NMR Interaction Experiment Between Hexa and Omi S1-RBD
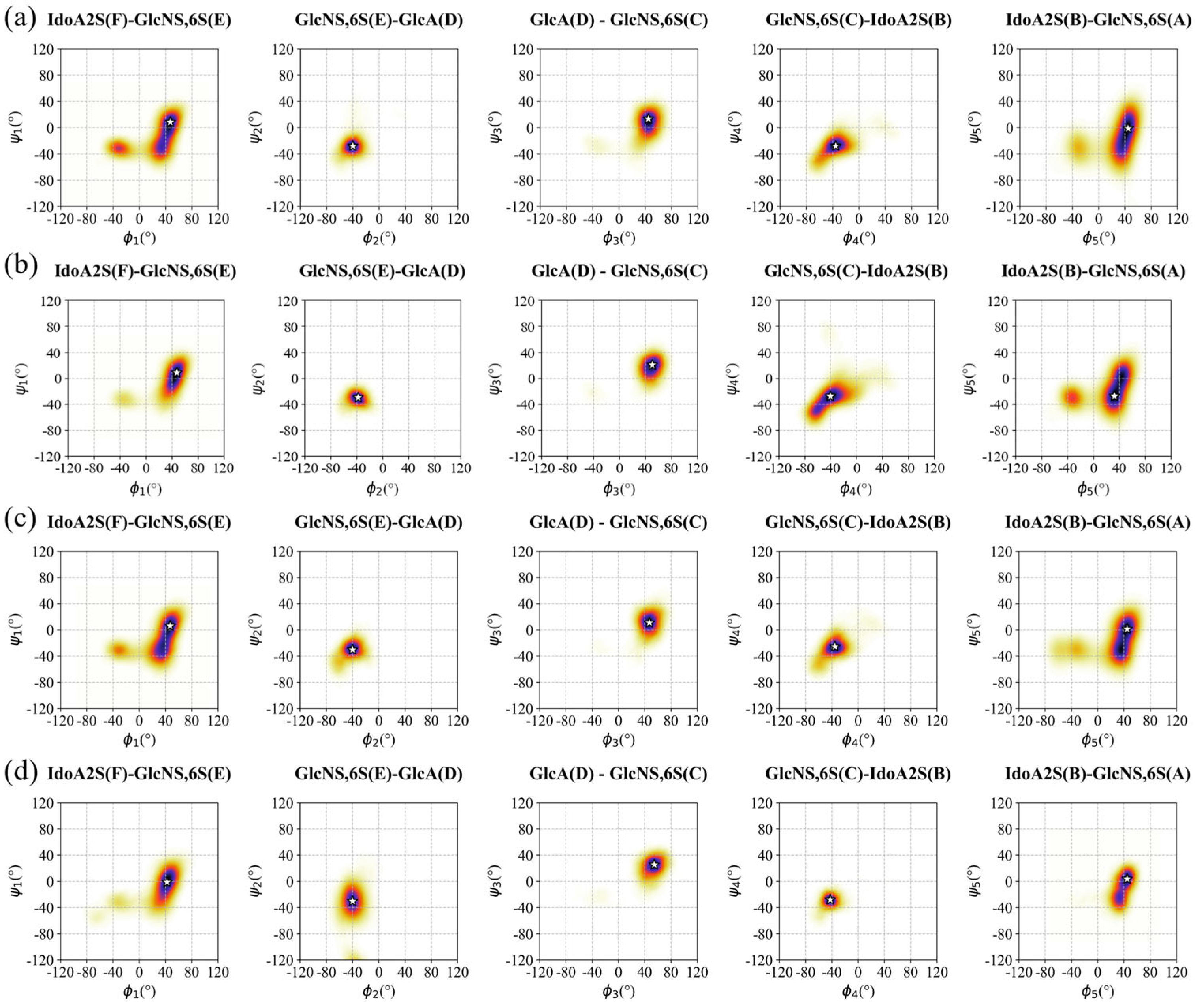
3.2. Conformation of Hexa in the Bound State with Omicron and WT S1-RBD
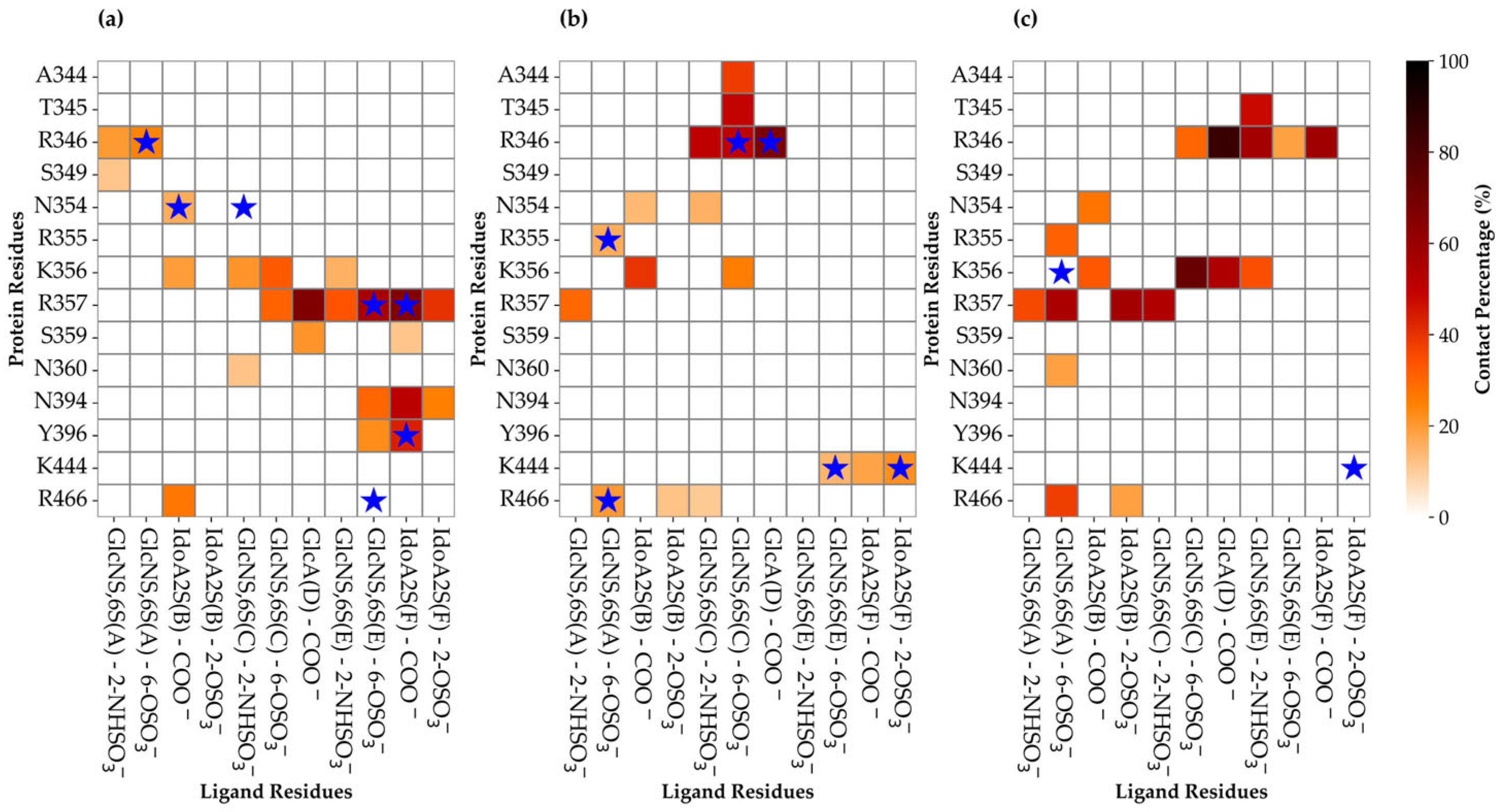
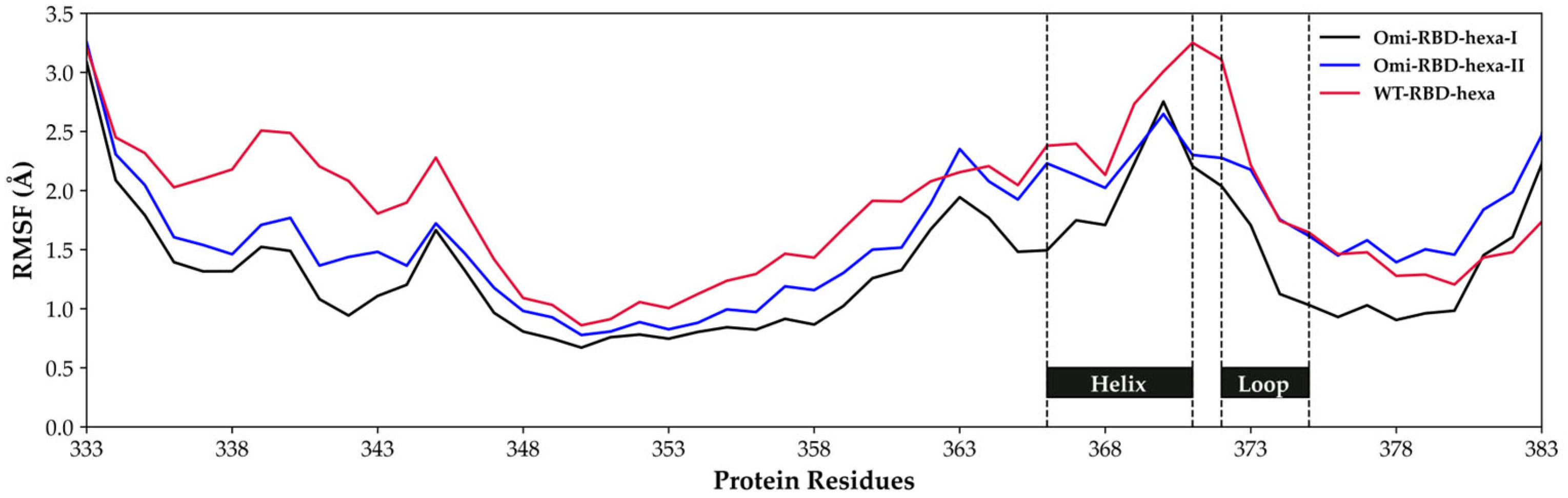
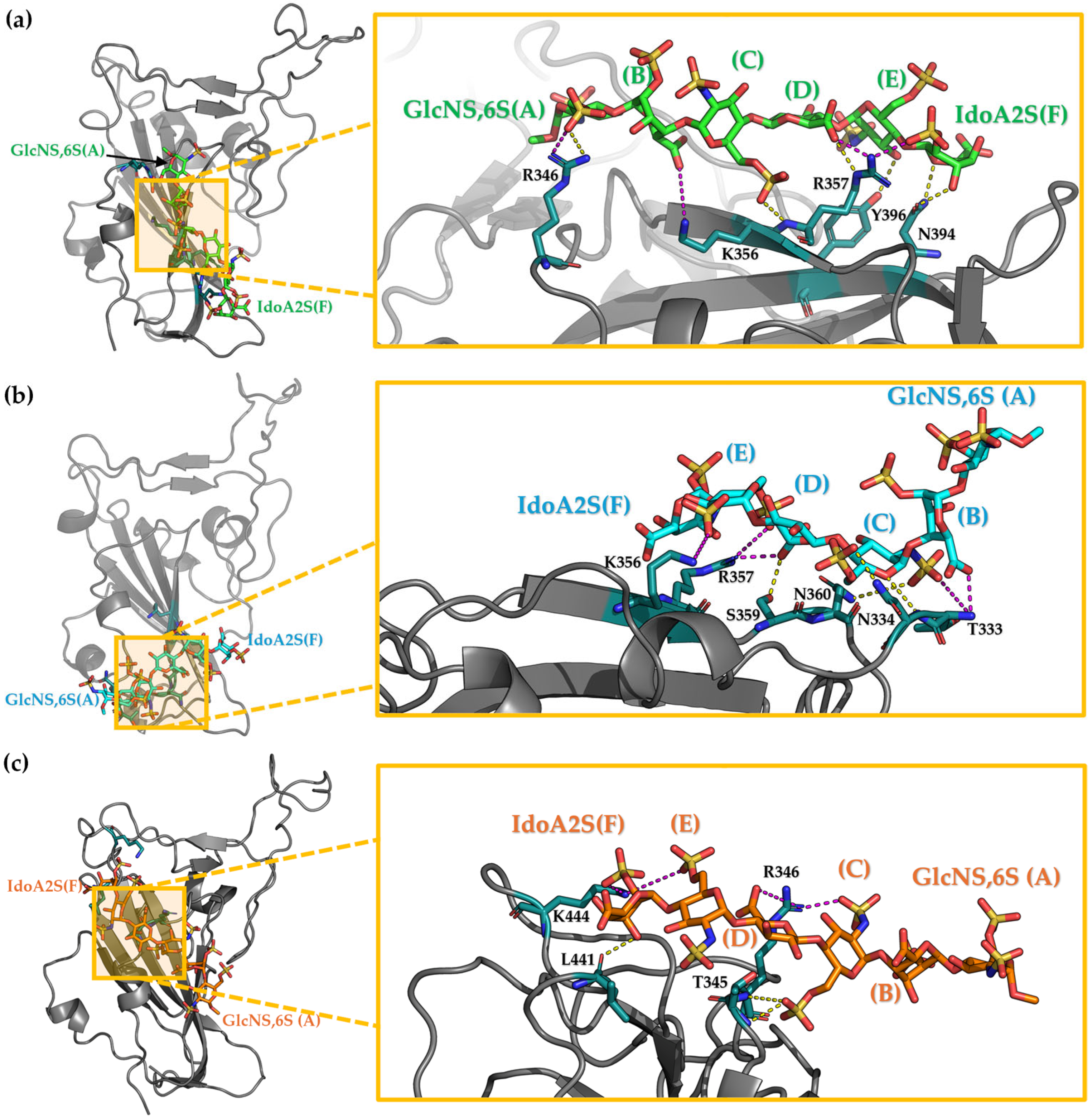
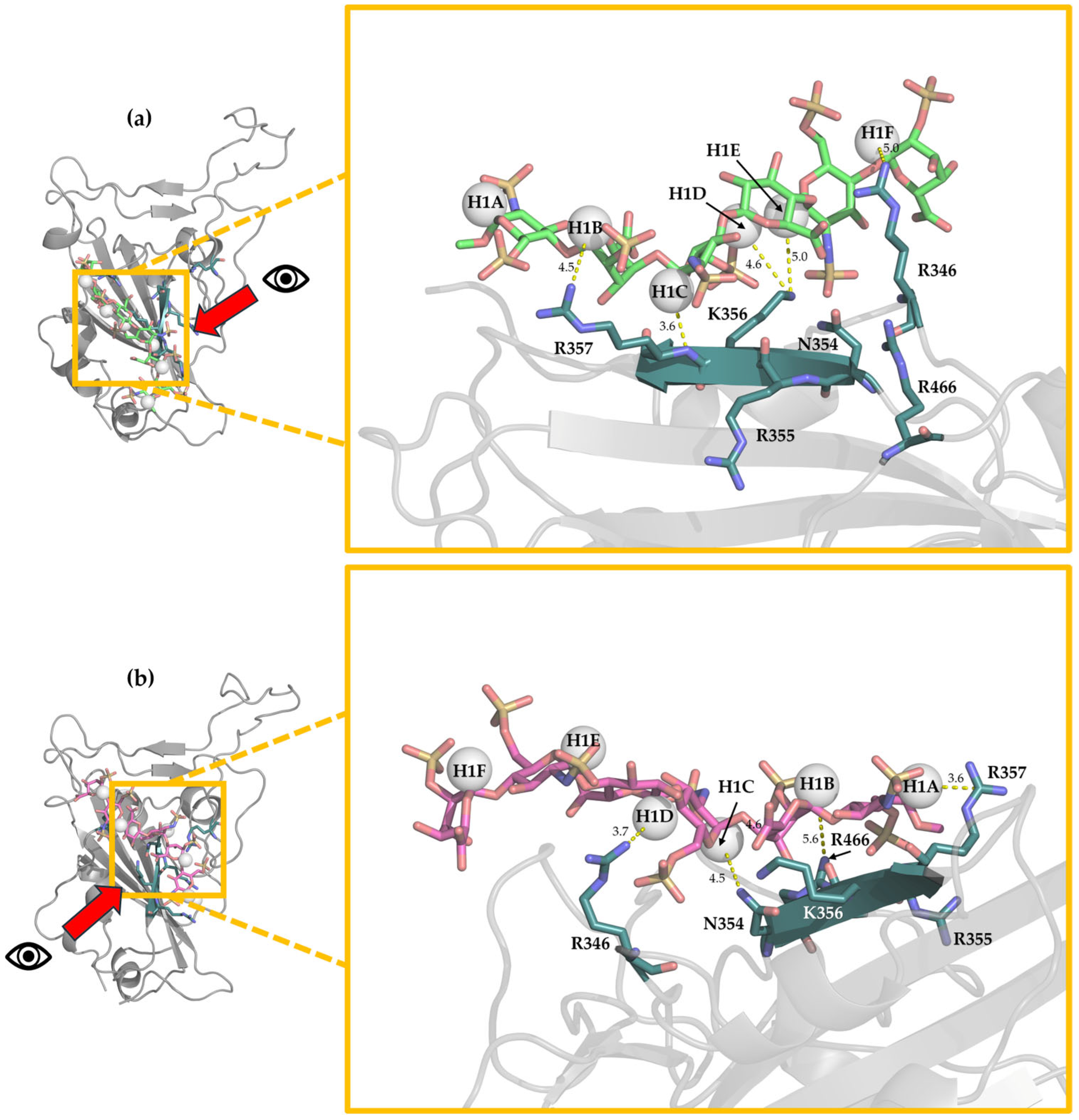
3.3. Characterization of the Interaction Between Hexa and S1-RBD of Wild-Type and Omicron Variant
Analysis of the MD Simulation Meta-Trajectories
3.4. The Mutation G339D in Omi-S1-RBD Reduces the Shielding Effect That the N343 Glycosyl Moiety Exerts on Site I
3.5. Selection of the Complexes Omi-S1-RBD-Hexa Using the Experimental STD0
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-Converting Enzyme 2 |
| GlcA | Glucuronic Acid |
| GlcNS,6S | Glucosamine, N-, 6-O-Disulfated |
| HS | Heparan Sulfate |
| IdoA2S | Iduronic Acid 2-O-Sulfated |
| MD | Molecular Dynamics |
| RBD | Receptor-Binding Domain |
| RMSD | Root Mean Square Deviation |
| RMSF | Root Mean Square Fluctuation |
| SARS | Severe Acute Respiratory Syndrome |
| STD-NMR | Saturation Transfer Difference Nuclear Magnetic Resonance |
References
- Kreuger, J.; Kjellén, L. Heparan Sulfate Biosynthesis: Regulation and Variability. J. Histochem. Cytochem. 2012, 60, 898. [Google Scholar] [CrossRef]
- Gallagher, J.T.; Lyon, M.; Steward, W.P. Structure and function of heparan sulphate proteoglycans. Biochem. J. 1986, 236, 313–325. [Google Scholar] [CrossRef]
- Lindahl, U.; Höök, M. Glycosaminoglycans and their binding to biological macromolecules. Annu. Rev. Biochem. 1978, 47, 385–417. [Google Scholar] [CrossRef]
- Casu, B.; Lindahl, U. Structure and biological interactions of heparin and heparan sulfate. Adv. Carbohydr. Chem. Biochem. 2001, 57, 159–206. [Google Scholar] [CrossRef] [PubMed]
- Lortat-Jacob, H.; Grosdidier, A.; Imberty, A. Structural diversity of heparan sulfate binding domains in chemokines. Proc. Natl. Acad. Sci. USA 2002, 99, 1229–1234. [Google Scholar] [CrossRef]
- Burgess, W.H.; Maciag, T. The Heparin-Binding (Fibroblast) Growth Factor Family of Proteins. Annu. Rev. Biochem. 1989, 58, 575–602. [Google Scholar] [CrossRef]
- Aquino, R.S.; Hayashida, K.; Hayashida, A.; Park, P.W. Role of HSPGs in Systemic Bacterial Infections. Methods Mol. Biol. 2022, 2303, 605. [Google Scholar] [CrossRef]
- Cagno, V.; Tseligka, E.D.; Jones, S.T.; Tapparel, C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses 2019, 11, 596. [Google Scholar] [CrossRef] [PubMed]
- Rudd, T.R.; Preston, M.D.; Yates, E.A. The nature of the conserved basic amino acid sequences found among 437 heparin binding proteins determined by network analysis. Mol. Biosyst. 2017, 13, 852–865. [Google Scholar] [CrossRef]
- Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, N.W.; Bai, X.; Esko, J.D.; Cohen, G.H.; Eisenberg, R.J.; Rosenberg, R.D.; Spear, P.G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 1999, 99, 13–22. [Google Scholar] [CrossRef]
- Artpradit, C.; Robinson, L.N.; Gavrilov, B.K.; Rurak, T.T.; Ruchirawat, M.; Sasisekharan, R. Recognition of heparan sulfate by clinical strains of dengue virus serotype 1 using recombinant subviral particles. Virus Res. 2013, 176, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Donalisio, M.; Rusnati, M.; Cagno, V.; Civra, A.; Bugatti, A.; Giuliani, A.; Pirri, G.; Volante, M.; Papotti, M.; Landolfo, S.; et al. Inhibition of human respiratory syncytial virus infectivity by a dendrimeric heparan sulfate-binding peptide. Antimicrob. Agents Chemother. 2012, 56, 5278–5288. [Google Scholar] [CrossRef]
- Cagno, V.; Donalisio, M.; Civra, A.; Volante, M.; Veccelli, E.; Oreste, P.; Rusnati, M.; Lembo, D. Highly sulfated K5 Escherichia coli polysaccharide derivatives inhibit respiratory syncytial virus infectivity in cell lines and human tracheal-bronchial histocultures. Antimicrob. Agents Chemother. 2014, 58, 4782–4794. [Google Scholar] [CrossRef]
- Johnson, S.M.; McNally, B.A.; Ioannidis, I.; Flano, E.; Teng, M.N.; Oomens, A.G.; Walsh, E.E.; Peeples, M.E.; Heise, M.T. Respiratory Syncytial Virus Uses CX3CR1 as a Receptor on Primary Human Airway Epithelial Cultures. PLoS Pathog. 2015, 11, e1005318. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Yue, J.; Jin, W.; Yang, H.; Faulkner, J.; Song, X.; Qiu, H.; Teng, M.; Azadi, P.; Zhang, F.; Linhardt, R.J.; et al. Heparan Sulfate Facilitates Spike Protein-Mediated SARS-CoV-2 Host Cell Invasion and Contributes to Increased Infection of SARS-CoV-2 G614 Mutant and in Lung Cancer. Front. Mol. Biosci. 2021, 8, 649575. [Google Scholar] [CrossRef]
- Kearns, F.L.; Sandoval, D.R.; Casalino, L.; Clausen, T.M.; Rosenfeld, M.A.; Spliid, C.B.; Amaro, R.E.; Esko, J.D. Spike-heparan sulfate interactions in SARS-CoV-2 infection. Curr. Opin. Struct. Biol. 2022, 76, 102439. [Google Scholar] [CrossRef]
- Parafioriti, M.; Ni, M.; Petitou, M.; Mycroft-West, C.J.; Rudd, T.R.; Gandhi, N.S.; Ferro, V.; Turnbull, J.E.; Lima, M.A.; Skidmore, M.A.; et al. Evidence for multiple binding modes in the initial contact between SARS-CoV-2 spike S1 protein and cell surface glycans. Chem. A Eur. J. 2022, 29, e202202599. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Sahoo, A.K.; Netz, R.R.; Herrmann, A.; Ballauff, M.; Haag, R. Charge Matters: Mutations in Omicron Variant Favor Binding to Cells. ChemBioChem 2022, 23, e202100681. [Google Scholar] [CrossRef]
- Gelbach, A.L.; Zhang, F.; Kwon, S.-J.; Bates, J.T.; Farmer, A.P.; Dordick, J.S.; Wang, C.; Linhardt, R.J. Interactions between heparin and SARS-CoV-2 spike glycoprotein RBD from omicron and other variants. Front. Mol. Biosci. 2022, 9, 912887. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, X.; Wang, X. Glycosylation: Mechanisms, biological functions and clinical implications. Signal Transduct. Target. Ther. 2024, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Berndsen, Z.T.; Raghwani, J.; Seabright, G.E.; Allen, J.D.; Pybus, O.G.; McLellan, J.S.; Wilson, I.A.; Bowden, T.A.; Ward, A.B.; et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688. [Google Scholar] [CrossRef]
- Ives, C.M.; Nguyen, L.; A Fogarty, C.; Harbison, A.M.; Durocher, Y.; Klassen, J.S.; Fadda, E. Role of N343 glycosylation on the SARS-CoV-2 S RBD structure and co-receptor binding across variants of concern. eLife 2024, 13, RP95708. [Google Scholar] [CrossRef]
- Nepravishta, R.; Ramírez-Cárdenas, J.; Rocha, G.; Walpole, S.; Hicks, T.; Monaco, S.; Muñoz-García, J.C.; Angulo, J. Fast Quantitative Validation of 3D Models of Low-Affinity Protein-Ligand Complexes by STD NMR Spectroscopy. J. Med. Chem. 2024, 67, 10025–10034. [Google Scholar] [CrossRef]
- Froese, J.; Mandalari, M.; Civera, M.; Elli, S.; Pagani, I.; Vicenzi, E.; Garcia-Monge, I.; Di Iorio, D.; Frank, S.; Bisio, A.; et al. Evolution of SARS-CoV-2 spike trimers towards optimized heparan sulfate cross-linking and inter-chain mobility. Sci. Rep. 2024, 14, 32174. [Google Scholar] [CrossRef]
- Amber18. Available online: https://ambermd.org/CiteAmber.php (accessed on 14 September 2025).
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Loncharich, R.J.; Brooks, B.R.; Pastor, R.W. Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 1992, 32, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Mayer, M.; James, T.L. NMR-Based Characterization of Phenothiazines as a RNA Binding Scaffoldt. J. Am. Chem. Soc. 2004, 126, 4453–4460. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640.e10. [Google Scholar] [CrossRef]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Gandhi, N.S.; Guimond, S.E.; Miller, G.J.; Meneghetti, M.C.Z.; Nader, H.B.; et al. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Thromb. Haemost. 2020, 120, 1700–1715. [Google Scholar] [CrossRef]
- Paiardi, G.; Ferraz, M.; Rusnati, M.; Wade, R.C. The accomplices: Heparan sulfates and N-glycans foster SARS-CoV-2 spike:ACE2 receptor binding and virus priming. Proc. Natl. Acad. Sci. USA 2024, 121, e2404892121. [Google Scholar] [CrossRef]
- Sanda, M.; Morrison, L.; Goldman, R. N-and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 2021, 93, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Sztain, T.; Ahn, S.-H.; Bogetti, A.T.; Casalino, L.; Goldsmith, J.A.; Seitz, E.; McCool, R.S.; Kearns, F.L.; Acosta-Reyes, F.; Maji, S.; et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021, 13, 963–968. [Google Scholar] [CrossRef] [PubMed]
- CHARMM-GUI. Available online: https://charmm-gui.org/?doc=archive&lib=covid19 (accessed on 10 July 2025).
- Woo, H.; Park, S.-J.; Choi, Y.K.; Park, T.; Tanveer, M.; Cao, Y.; Kern, N.R.; Lee, J.; Yeom, M.S.; Croll, T.I.; et al. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B 2020, 124, 7128–7137. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Xiao, Y.; Tong, B.; Mao, Y.; Ge, R.; Tian, F.; Dong, X.; Zheng, P. S373P Mutation Stabilizes the Receptor-Binding Domain of the Spike Protein in Omicron and Promotes Binding. JACS Au 2023, 3, 1902–1910. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandalari, M.; Parafioriti, M.; Ni, M.; Benevelli, F.; Civera, M.; Elli, S.; Guerrini, M. Interaction Between Heparan Sulfate Oligosaccharide and the Receptor-Binding Domain of the Wild-Type and Omicron Variant of the SARS-CoV-2 Spike Protein. Biomolecules 2025, 15, 1343. https://doi.org/10.3390/biom15091343
Mandalari M, Parafioriti M, Ni M, Benevelli F, Civera M, Elli S, Guerrini M. Interaction Between Heparan Sulfate Oligosaccharide and the Receptor-Binding Domain of the Wild-Type and Omicron Variant of the SARS-CoV-2 Spike Protein. Biomolecules. 2025; 15(9):1343. https://doi.org/10.3390/biom15091343
Chicago/Turabian StyleMandalari, Marco, Michela Parafioriti, Minghong Ni, Francesca Benevelli, Monica Civera, Stefano Elli, and Marco Guerrini. 2025. "Interaction Between Heparan Sulfate Oligosaccharide and the Receptor-Binding Domain of the Wild-Type and Omicron Variant of the SARS-CoV-2 Spike Protein" Biomolecules 15, no. 9: 1343. https://doi.org/10.3390/biom15091343
APA StyleMandalari, M., Parafioriti, M., Ni, M., Benevelli, F., Civera, M., Elli, S., & Guerrini, M. (2025). Interaction Between Heparan Sulfate Oligosaccharide and the Receptor-Binding Domain of the Wild-Type and Omicron Variant of the SARS-CoV-2 Spike Protein. Biomolecules, 15(9), 1343. https://doi.org/10.3390/biom15091343





