Bile Acids Are Potential Negative Allosteric Modulators of M1 Muscarinic Receptors
Abstract
1. Introduction
2. Materials and Methods
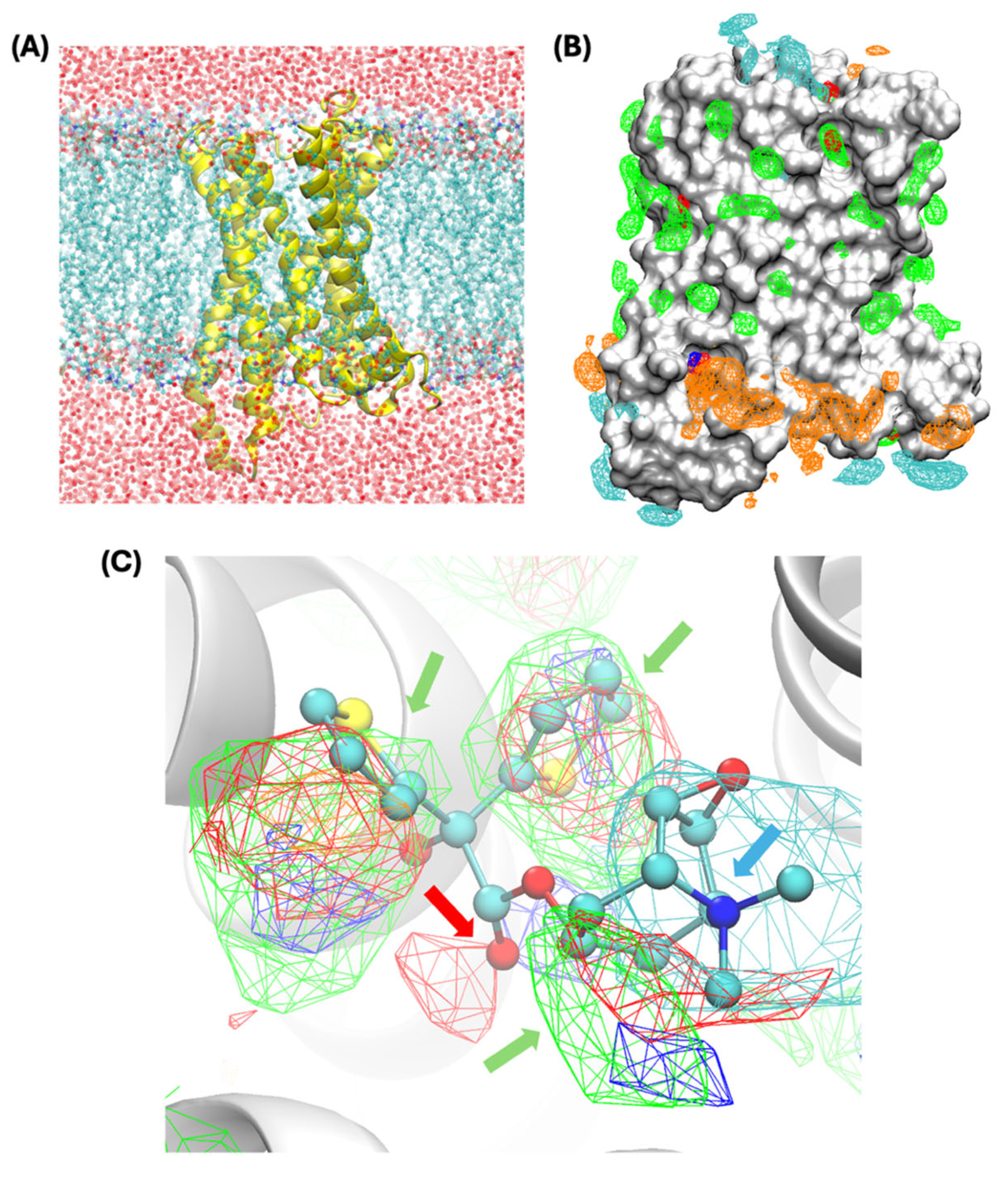
2.1. SILCS Simulations
2.2. SILCS-MC Docking
2.3. SILCS-Hotspots Analyses
2.4. MD Simulations of ACh Bound Complexes with and Without Bile Acids
2.5. Analyses of MD Trajectories
3. Results
3.1. Functional Group Binding Patterns Mapped for M1R
3.2. Prediction of Allosteric Binding Sites and Binding Poses of Bile Acids
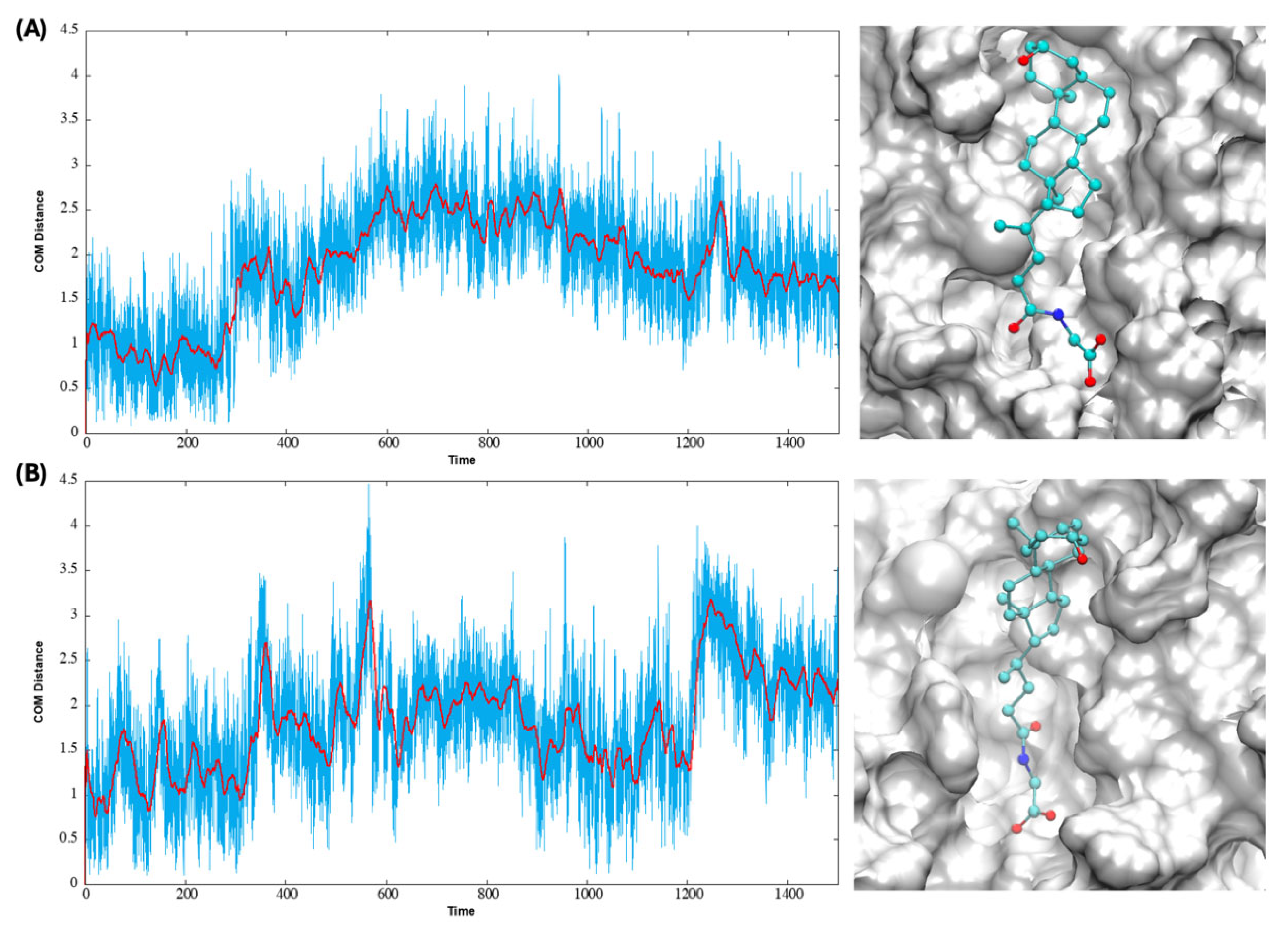
3.3. Lithocholic Bile Acids Stay in the Two Predicted Binding Sites Through Microsecond MD on the M1R-ACh-Bile Acid Complexes
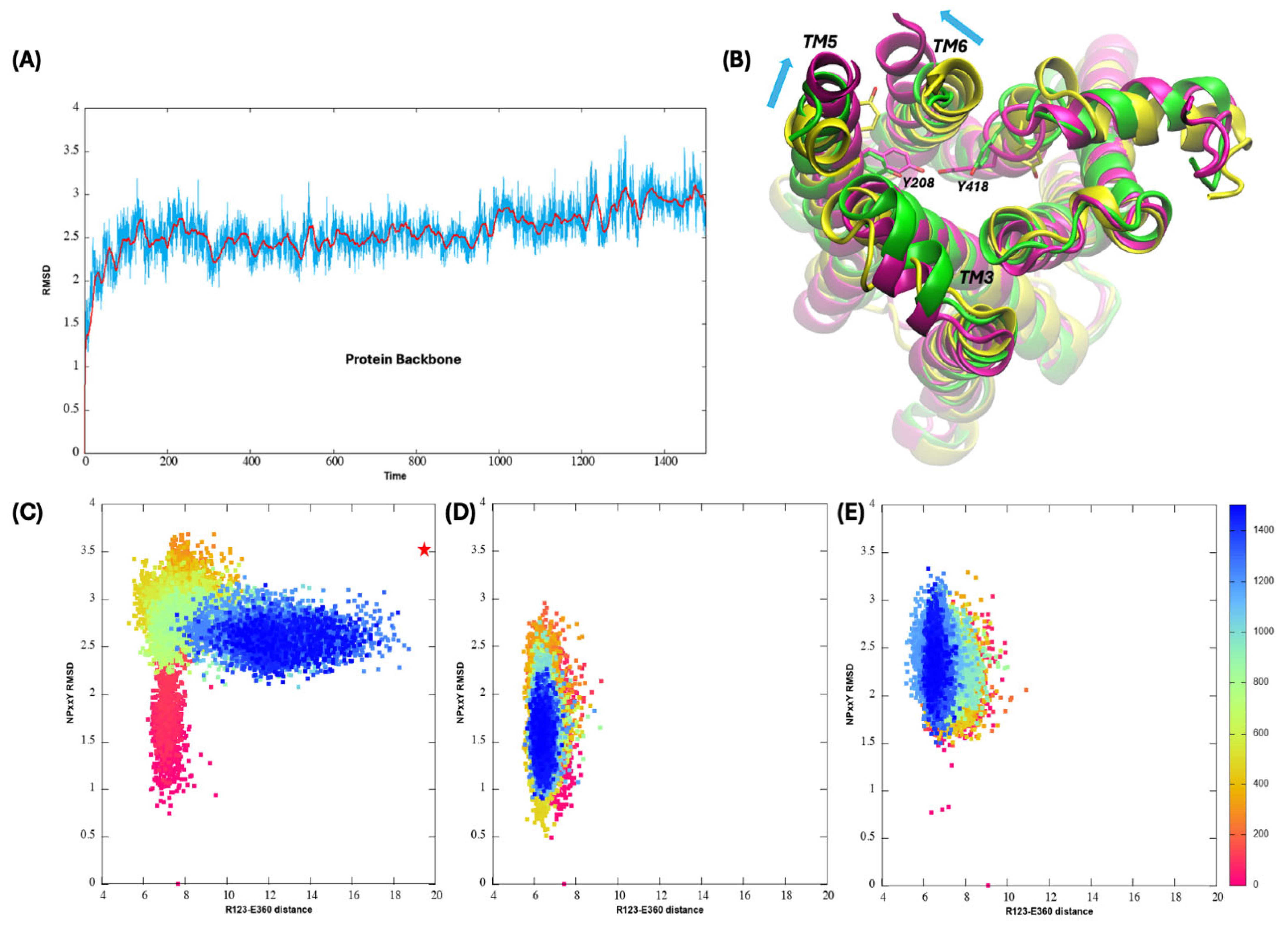
3.4. Transition Toward the Active State upon Agonist Binding with and Without the Presence of Bile Acids
3.5. Activation Mechanism of M1R upon ACh Binding
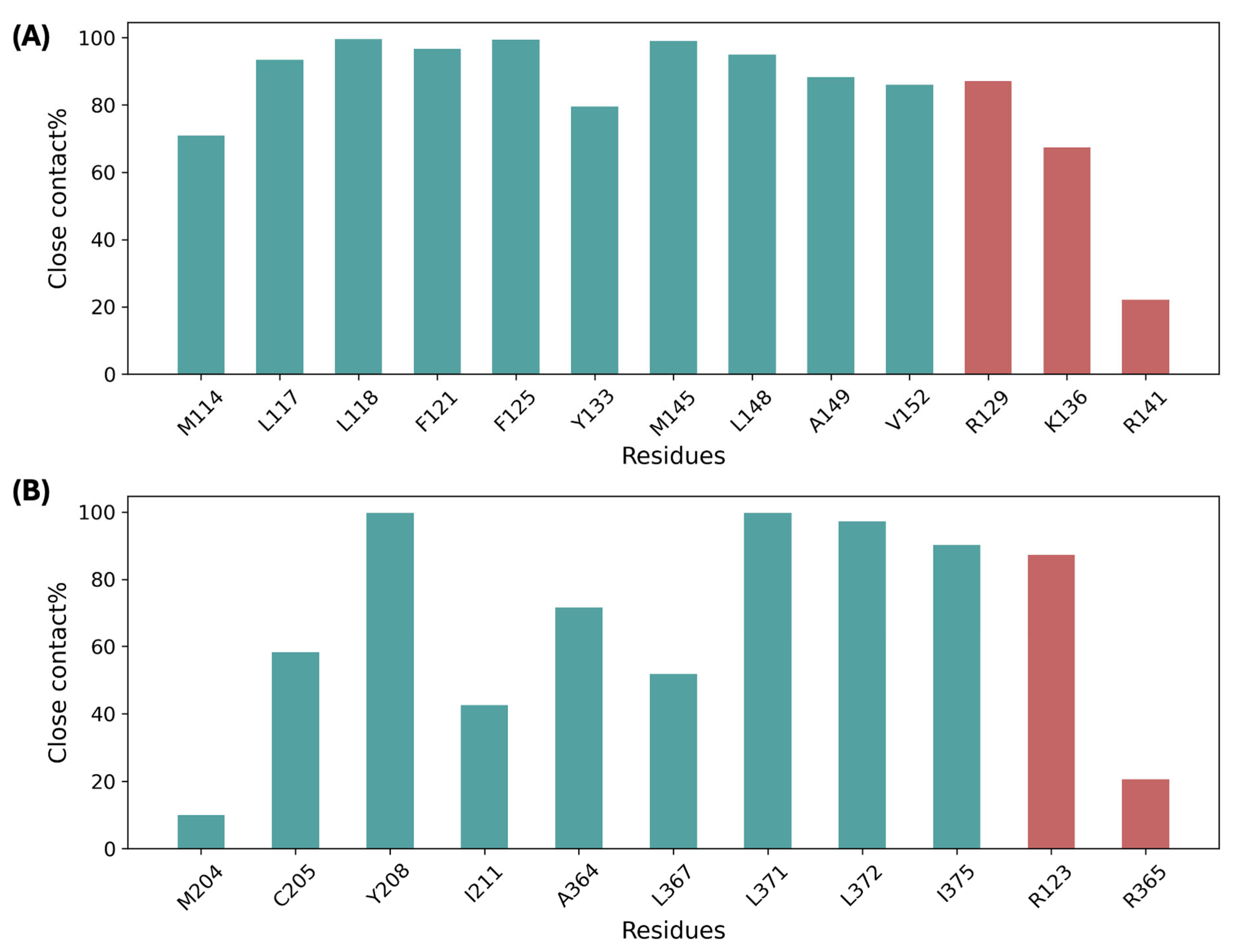
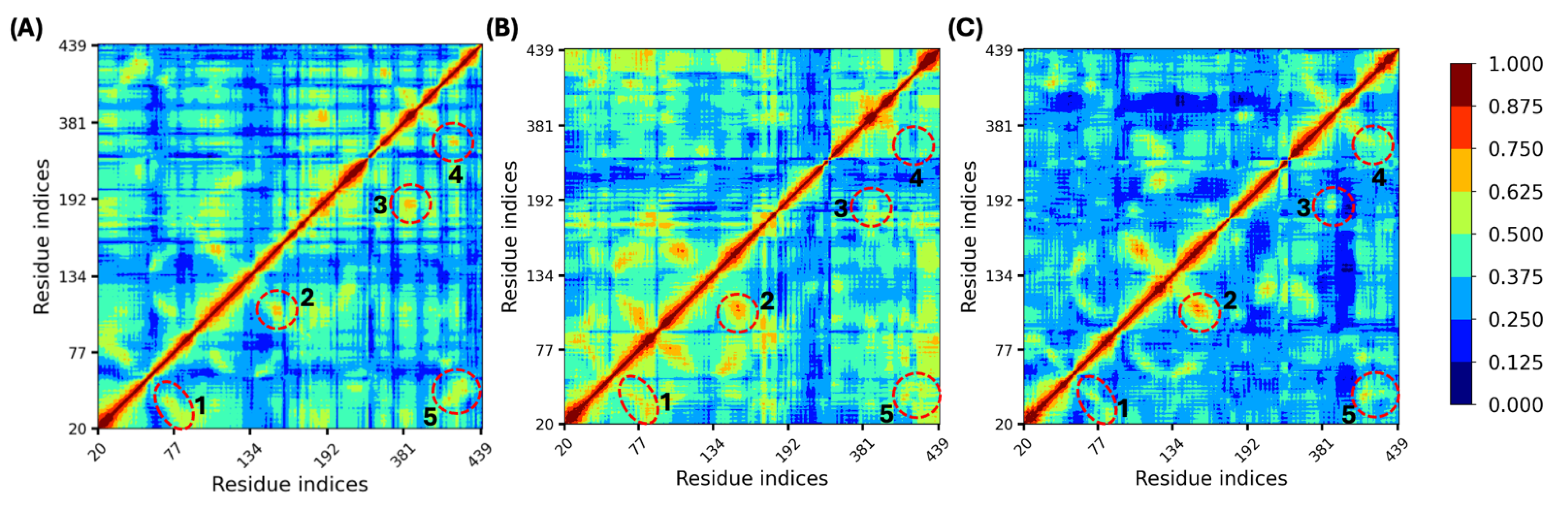
3.6. Bile Acid Binding Stabilized the Allosteric Sites and Other TM Regions
3.7. Allosteric Site Stabilization Weakened Cooperativity Between TM Domains
3.8. Allosteric Bile Acid Binding Distorts Agonist ACh Binding
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| M1R | M1 muscarinic receptor |
| ACh | Acetylcholine |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| SILCS | Site identification by ligand competitive saturation |
| MD | Molecular dynamics |
| FEP | Free energy perturbation |
| LGFE | Ligand grid free energy |
| MC | Monte Carlo |
| PDB | Protein data bank |
| LMI | Linear mutual information |
| RMSD | Root-mean-square deviation |
| RMSF | Root-mean-square fluctuation |
| COM | Center-of-mass |
References
- Perino, A.; Demagny, H.; Velazquez-Villegas, L.; Schoonjans, K. Molecular Physiology of Bile Acid Signaling in Health, Disease, and Aging. Physiol. Rev. 2021, 101, 683–731. [Google Scholar] [CrossRef]
- Fleishman, J.S.; Kumar, S. Bile Acid Metabolism and Signaling in Health and Disease: Molecular Mechanisms and Therapeutic Targets. Signal Transduct. Target. Ther. 2024, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Kiriyama, Y.; Nochi, H. The Biosynthesis, Signaling, and Neurological Functions of Bile Acids. Biomolecules 2019, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Raufman, J.-P.; Zimniak, P.; Bartoszko-Malik, A. Lithocholyltaurine Interacts with Cholinergic Receptors on Dispersed Chief Cells from Guinea Pig Stomach. Am. J. Physiol. Gastrointest. Liver Physiol. 1998, 274, G997–G1004. [Google Scholar] [CrossRef]
- Cheng, K.; Chen, Y.; Zimniak, P.; Raufman, J.-P.; Xiao, Y.; Frucht, H. Functional Interaction of Lithocholic Acid Conjugates with M3 Muscarinic Receptors on a Human Colon Cancer Cell Line. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2002, 1588, 48–55. [Google Scholar] [CrossRef]
- Khurana, S.; Raufman, J.-P.; Pallone, T.L. Bile Acids Regulate Cardiovascular Function. Clin. Transl. Sci. 2011, 4, 210–218. [Google Scholar] [CrossRef]
- Sheikh Abdul Kadir, S.H.; Miragoli, M.; Abu-Hayyeh, S.; Moshkov, A.V.; Xie, Q.; Keitel, V.; Nikolaev, V.O.; Williamson, C.; Gorelik, J. Bile Acid-Induced Arrhythmia Is Mediated by Muscarinic M2 Receptors in Neonatal Rat Cardiomyocytes. PLoS ONE 2010, 5, e9689. [Google Scholar] [CrossRef]
- Urso, A.; D’Ovidio, F.; Xu, D.; Emala, C.W.; Bunnett, N.W.; Perez-Zoghbi, J.F. Bile Acids Inhibit Cholinergic Constriction in Proximal and Peripheral Airways from Humans and Rodents. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L264–L275. [Google Scholar] [CrossRef]
- Kruse, A.C.; Hu, J.; Kobilka, B.K.; Wess, J. Muscarinic Acetylcholine Receptor X-Ray Structures: Potential Implications for Drug Development. Curr. Opin. Pharmacol. 2014, 16, 24–30. [Google Scholar] [CrossRef]
- Kruse, A.C.; Kobilka, B.K.; Gautam, D.; Sexton, P.M.; Christopoulos, A.; Wess, J. Muscarinic Acetylcholine Receptors: Novel Opportunities for Drug Development. Nat. Rev. Drug Discov. 2014, 13, 549–560. [Google Scholar] [CrossRef]
- Cheng, K.; Shang, A.C.; Drachenberg, C.B.; Zhan, M.; Raufman, J.-P. Differential Expression of M3 Muscarinic Receptors in Progressive Colon Neoplasia and Metastasis. Oncotarget 2017, 8, 21106–21114. [Google Scholar] [CrossRef]
- Cheng, K.; Xie, G.; Khurana, S.; Heath, J.; Drachenberg, C.B.; Timmons, J.; Shah, N.; Raufman, J.-P. Divergent Effects of Muscarinic Receptor Subtype Gene Ablation on Murine Colon Tumorigenesis Reveals Association of M3R and Zinc Finger Protein 277 Expression in Colon Neoplasia. Mol. Cancer 2014, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Sundel, M.H.; Sampaio Moura, N.; Cheng, K.; Chatain, O.; Hu, S.; Drachenberg, C.B.; Xie, G.; Raufman, J.-P. Selective Activation of M1 Muscarinic Receptors Attenuates Human Colon Cancer Cell Proliferation. Cancers 2023, 15, 4766. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, M.; Schledwitz, A.; Cheng, K.; Raufman, J.-P. Mechanistic Clues Provided by Concurrent Changes in the Expression of Genes Encoding the M1 Muscarinic Receptor, β-Catenin Signaling Proteins, and Downstream Targets in Adenocarcinomas of the Colon. Front. Physiol. 2022, 13, 857563. [Google Scholar] [CrossRef] [PubMed]
- Glinghammar, B.; Rafter, J. Carcinogenesis in the Colon: Interaction between Luminal Factors and Genetic Factors. Eur. J. Cancer Prev. 1999, 8, S87–S94. [Google Scholar]
- Bernstein, C.; Holubec, H.; Bhattacharyya, A.K.; Nguyen, H.; Payne, C.M.; Zaitlin, B.; Bernstein, H. Carcinogenicity of Deoxycholate, a Secondary Bile Acid. Arch. Toxicol. 2011, 85, 863–871. [Google Scholar] [CrossRef]
- Bernstein, H.; Bernstein, C. Bile Acids as Carcinogens in the Colon and at Other Sites in the Gastrointestinal System. Exp. Biol. Med. 2023, 248, 79–89. [Google Scholar] [CrossRef]
- Raufman, J.-P.; Chen, Y.; Cheng, K.; Compadre, C.; Compadre, L.; Zimniak, P. Selective Interaction of Bile Acids with Muscarinic Receptors: A Case of Molecular Mimicry. Eur. J. Pharmacol. 2002, 457, 77–84. [Google Scholar] [CrossRef]
- Jakubík, J.; El-Fakahany, E.E. Allosteric Modulation of Muscarinic Acetylcholine Receptors. Pharmaceuticals 2010, 3, 2838–2860. [Google Scholar] [CrossRef]
- Jakubík, J.; El-Fakahany, E.E. Allosteric Modulation of GPCRs of Class A by Cholesterol. Int. J. Mol. Sci. 2021, 22, 1953. [Google Scholar] [CrossRef]
- Russell, D.W. Fifty Years of Advances in Bile Acid Synthesis and Metabolism. J. Lipid Res. 2009, 50, S120–S125. [Google Scholar] [CrossRef]
- Schlick, T. Molecular Modeling and Simulation: An Interdisciplinary Guide; Springer: New York, NY, USA, 2010; Volume 2. [Google Scholar]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Goel, H.; Hazel, A.; Ustach, V.D.; Jo, S.; Yu, W.; MacKerell, A.D. Rapid and Accurate Estimation of Protein–Ligand Relative Binding Affinities Using Site-Identification by Ligand Competitive Saturation. Chem. Sci. 2021, 12, 8844–8858. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D.; Jo, S.; Lakkaraju, S.K.; Lind, C.; Yu, W. Identification and Characterization of Fragment Binding Sites for Allosteric Ligand Design Using the Site Identification by Ligand Competitive Saturation Hotspots Approach (SILCS-Hotspots). Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129519. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of Three-Dimensional Structural Information of Biological Macromolecules. Acta Crystallogr. Sect. D 1998, 54, 1078–1084. [Google Scholar] [CrossRef]
- Thal, D.M.; Sun, B.; Feng, D.; Nawaratne, V.; Leach, K.; Felder, C.C.; Bures, M.G.; Evans, D.A.; Weis, W.I.; Bachhawat, P.; et al. Crystal Structures of the M1 and M4 Muscarinic Acetylcholine Receptors. Nature 2016, 531, 335–340. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Jo, S.; Cheng, X.; Lee, J.; Kim, S.; Park, S.-J.; Patel, D.S.; Beaven, A.H.; Lee, K.I.; Rui, H.; Park, S.; et al. CHARMM-GUI 10 Years for Biomolecular Modeling and Simulation. J. Comput. Chem. 2017, 38, 1114–1124. [Google Scholar] [CrossRef]
- Mezei, M. A Cavity-Biased (T, V, μ) Monte Carlo Method for the Computer Simulation of Fluids. Mol. Phys. 1980, 40, 901–906. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; He, X.; Vanommeslaeghe, K.; MacKerell, A.D., Jr. Extension of the CHARMM General Force Field to Sulfonyl-Containing Compounds and Its Utility in Biomolecular Simulations. J. Comput. Chem. 2012, 33, 2451–2468. [Google Scholar] [CrossRef] [PubMed]
- Durell, S.R.; Brooks, B.R.; Ben-Naim, A. Solvent-Induced Forces between Two Hydrophilic Groups. J. Phys. Chem. 1994, 98, 2198–2202. [Google Scholar] [CrossRef]
- Metropolis, N.; Ulam, S. The Monte Carlo Method. J. Am. Stat. Assoc. 1949, 44, 335–341. [Google Scholar] [CrossRef]
- Kirkpatrick, S.; Gelatt, C.D.; Vecchi, M.P. Optimization by Simulated Annealing. Science 1983, 220, 671–680. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Miao, Y.; Nichols, S.E.; Gasper, P.M.; Metzger, V.T.; McCammon, J.A. Activation and Dynamic Network of the M2 Muscarinic Receptor. Proc. Natl. Acad. Sci. USA 2013, 110, 10982–10987. [Google Scholar] [CrossRef]
- Miao, Y.; Caliman, A.D.; McCammon, J.A. Allosteric Effects of Sodium Ion Binding on Activation of the M3 Muscarinic G-Protein-Coupled Receptor. Biophys. J. 2015, 108, 1796–1806. [Google Scholar] [CrossRef]
- Nosé, S. A Unified Formulation of the Constant Temperature Molecular Dynamics Methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A Flexible Algorithm for Calculating Pair Interactions on SIMD Architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef]
- Steinbach, P.J.; Brooks, B.R. New Spherical-Cutoff Methods for Long-Range Forces in Macromolecular Simulation. J. Comput. Chem. 1994, 15, 667–683. [Google Scholar] [CrossRef]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids; Oxford University Press: Oxford, UK, 2017; ISBN 0-19-252470-4. [Google Scholar]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Lange, O.F.; Grubmüller, H. Generalized Correlation for Biomolecular Dynamics. Proteins Struct. Funct. Bioinform. 2006, 62, 1053–1061. [Google Scholar] [CrossRef]
- Tekpinar, M.; Neron, B.; Delarue, M. Extracting Dynamical Correlations and Identifying Key Residues for Allosteric Communication in Proteins by Correlationplus. J. Chem. Inf. Model. 2021, 61, 4832–4838. [Google Scholar] [CrossRef]
- Yu, W.; Weber, D.J.; MacKerell, A.D. Computer-Aided Drug Design: An Update. In Antibiotics: Methods and Protocols; Sass, P., Ed.; Springer: New York, NY, USA, 2023; pp. 123–152. ISBN 978-1-0716-2855-3. [Google Scholar]
- Randáková, A.; Dolejší, E.; Rudajev, V.; Zimčík, P.; Doležal, V.; El-Fakahany, E.E.; Jakubík, J. Role of Membrane Cholesterol in Differential Sensitivity of Muscarinic Receptor Subtypes to Persistently Bound Xanomeline. Neuropharmacology 2018, 133, 129–144. [Google Scholar] [CrossRef]
- Miao, Y.; McCammon, J.A. Graded Activation and Free Energy Landscapes of a Muscarinic G-Protein–Coupled Receptor. Proc. Natl. Acad. Sci. USA 2016, 113, 12162–12167. [Google Scholar] [CrossRef]
- Gong, Z.; Zhang, X.; Liu, M.; Jin, C.; Hu, Y. Visualizing the M2 Muscarinic Acetylcholine Receptor Activation Regulated by Aromatic Ring Dynamics. bioRxiv 2024. bioRxiv:2024.02.18.580854. [Google Scholar] [CrossRef]
- Maeda, S.; Qu, Q.; Robertson, M.J.; Skiniotis, G.; Kobilka, B.K. Structures of the M1 and M2 Muscarinic Acetylcholine Receptor/G-Protein Complexes. Science 2019, 364, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.P.; Xie, G.; Raufman, J.-P.; Hogan, S.; Griffin, T.L.; Packard, C.A.; Chatfield, D.A.; Hagey, L.R.; Steinbach, J.H.; Hofmann, A.F. Human Cecal Bile Acids: Concentration and Spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G256–G263. [Google Scholar] [CrossRef]
- Zhu, L.; Rossi, M.; Cohen, A.; Pham, J.; Zheng, H.; Dattaroy, D.; Mukaibo, T.; Melvin, J.E.; Langel, J.L.; Hattar, S.; et al. Allosteric Modulation of β-Cell M3 Muscarinic Acetylcholine Receptors Greatly Improves Glucose Homeostasis in Lean and Obese Mice. Proc. Natl. Acad. Sci. USA 2019, 116, 18684–18690. [Google Scholar] [CrossRef]
- Tobin, A.B. A Golden Age of Muscarinic Acetylcholine Receptor Modulation in Neurological Diseases. Nat. Rev. Drug Discov. 2024, 23, 743–758. [Google Scholar] [CrossRef]
- Guzior, D.V.; Okros, M.; Shivel, M.; Armwald, B.; Bridges, C.; Fu, Y.; Martin, C.; Schilmiller, A.L.; Miller, W.M.; Ziegler, K.M.; et al. Bile Salt Hydrolase Acyltransferase Activity Expands Bile Acid Diversity. Nature 2024, 626, 852–858. [Google Scholar] [CrossRef]



| Acid | Glycine | Taurine | ||||
|---|---|---|---|---|---|---|
| Name | AS34 | AS56 | AS34 | AS56 | AS34 | AS56 |
| Cholic | −10.30 | −7.41 | −11.25 | −8.61 | −11.33 | −9.75 |
| Chenodeoxycholic | −10.64 | −7.70 | −11.94 | −9.94 | −11.53 | −10.30 |
| Deoxycholic | −9.40 | −8.19 | −10.99 | −9.56 | −11.29 | −9.95 |
| Lithocholic | −9.91 | −7.48 | −12.30 | −10.29 | −12.12 | −10.72 |
| Off-Diagonal | Domain 1 | Domain 2 | ||
|---|---|---|---|---|
| Region | TM | Residues | TM | Residues |
| 1 | TM1 | 27/29/31 | TM2 | 83 |
| 37/40 | 75/76 | |||
| 39/40/41/43 | 71/72 | |||
| 2 | TM3 | 105–114 | TM4 | 151–157 |
| 99 | 161 | |||
| 3 | TM5 | 183–191 | TM6 | 383–390 |
| 4 | TM6 | 364–370 | TM7 | 416–421 |
| 5 | TM1 | 39 | TM7 | 412 |
| 41–43 | 414–416 | |||
| 48/49 | 425/427/428 | |||
| 55/56 | 424/425 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; MacKerell, A.D., Jr.; Weber, D.J.; Raufman, J.-P. Bile Acids Are Potential Negative Allosteric Modulators of M1 Muscarinic Receptors. Biomolecules 2025, 15, 1326. https://doi.org/10.3390/biom15091326
Yu W, MacKerell AD Jr., Weber DJ, Raufman J-P. Bile Acids Are Potential Negative Allosteric Modulators of M1 Muscarinic Receptors. Biomolecules. 2025; 15(9):1326. https://doi.org/10.3390/biom15091326
Chicago/Turabian StyleYu, Wenbo, Alexander D. MacKerell, Jr., David J. Weber, and Jean-Pierre Raufman. 2025. "Bile Acids Are Potential Negative Allosteric Modulators of M1 Muscarinic Receptors" Biomolecules 15, no. 9: 1326. https://doi.org/10.3390/biom15091326
APA StyleYu, W., MacKerell, A. D., Jr., Weber, D. J., & Raufman, J.-P. (2025). Bile Acids Are Potential Negative Allosteric Modulators of M1 Muscarinic Receptors. Biomolecules, 15(9), 1326. https://doi.org/10.3390/biom15091326








